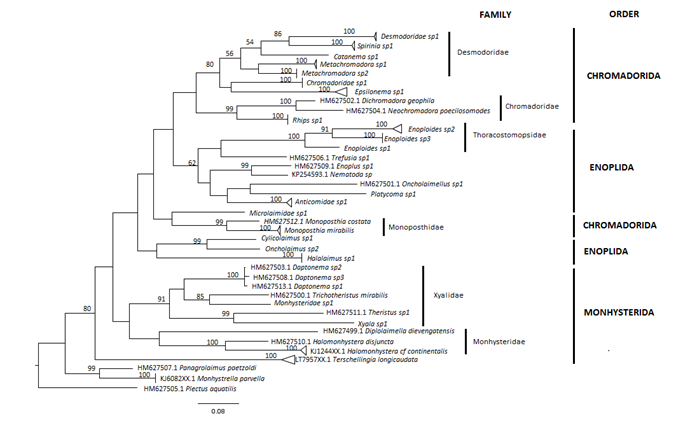
Nematode biodiversity is mostly unknown; while about 20,000 nematode species have been described, estimates for species diversity range from 0.1 to 100 million. The study of nematode diversity, like that of meiofaunal organisms in general, has been mostly based on morphology-based taxonomy, a time-consuming and costly task that requires well-trained specialists. This work represents the first on the taxonomy of Mexican nematodes to integrate morphological and molecular data. We add seven new morphological records for the Mexican Caribbean: Anticomid sp1, Cylicolaimus sp1, Oncholaimus sp2, Platycoma sp1, Catanema sp1, Enopliodes spp., and Metachromadora spp. We recover 55 COI sequences that represent 20 species. Of the studied sites, Cozumel had 12 species and Cancún had two species (Rhips sp1 and Monoposthia mirabilis) represented by several individuals. All sequences are new for the genetic international databases GenBank and BOLD. Phylogenetic analyses and species delineation methods support the occurrence of the 20 entities and confirm the high resolution of COI sequences in delimiting species. ABGD and mPTP methods disentangled 20 entities, whereas Barcode Index Numbers (BINs) recovered 22 genetic species. DNA taxonomy was demonstrated to be an efficient, fast, and low-cost method to address a taxonomical shortfall of meiofaunal organisms.