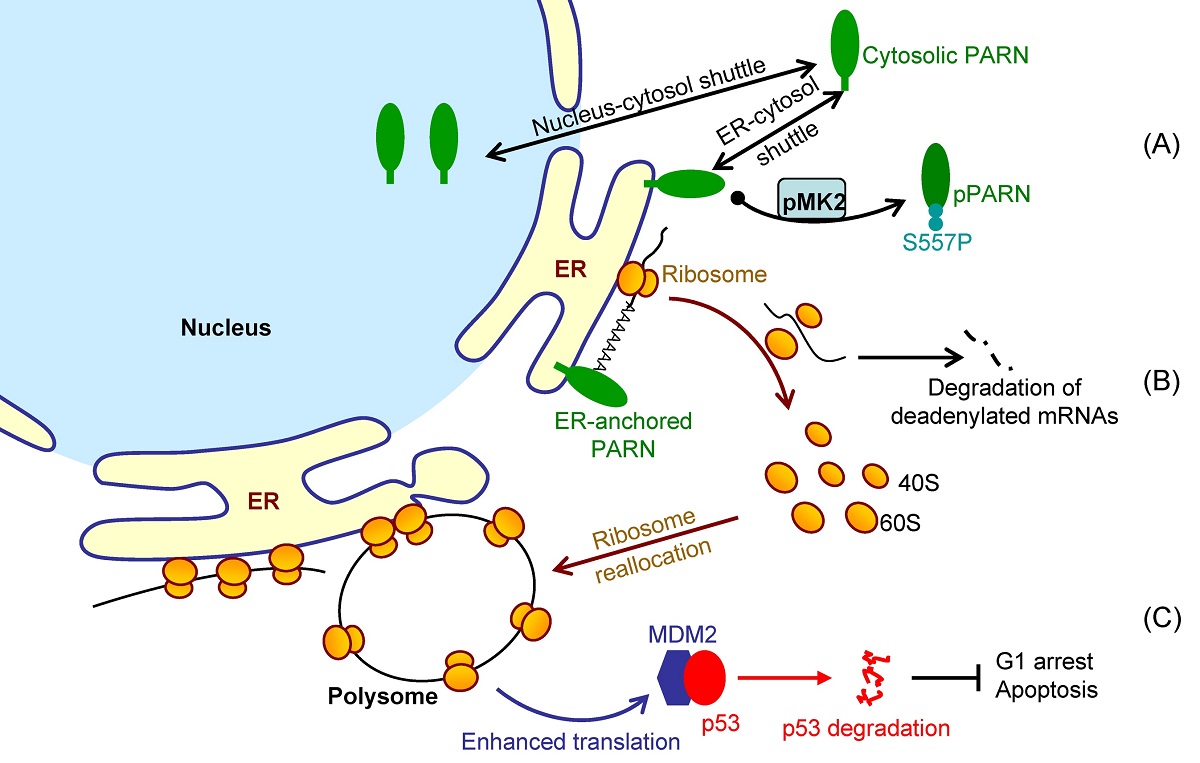
Translation is spatiotemporally regulated and ER-associated mRNAs are generally in efficient translation. It is unclear whether the ER-associated mRNAs are deadenylated or degraded on the ER surface in situ or in the cytosol. Here, we showed that ER possessed active deadenylases, particularly the poly(A)-specific ribonuclease (PARN), in common cell lines and mouse tissues. Consistently, purified recombinant PARN exhibited a strong ability to insert into the Langmuir monolayer and liposome. ER-anchored PARN was found to be able to reshape the poly(A) length profile of the ER-associated RNAs by suppressing long poly(A) tails without significantly influencing the cytosolic RNAs. The shortening of long poly(A) tails did not affect global translation efficiency, suggesting that the non-specific action of PARN towards long poly(A) tails was beyond the scope of translation regulation on the ER surface. Transcriptome sequencing analysis indicated that the ER-anchored PARN trigged the degradation of a small subset of ER-enriched transcripts. The ER-anchored PARN modulated the translation of its targets by redistributing ribosomes to heavy polysomes, suggesting that PARN may play a role in dynamic ribosome reallocation. During DNA damage response, MK2 phosphorylated PARN-Ser557 to modulate PARN translocation from the ER to cytosol. By promoting the decay of ER-associated MDM2 transcripts with low ribosome occupancy, the ER-anchored PARN modulated DNA damage response and thereby cell viability. These findings revealed that a highly regulated communication between mRNA degradation rate and translation efficiency is present on the ER surface in situ and that PARN may contribute to this communication by modulating the dynamic ribosome reallocation between transcripts with low and high ribosome occupancies.