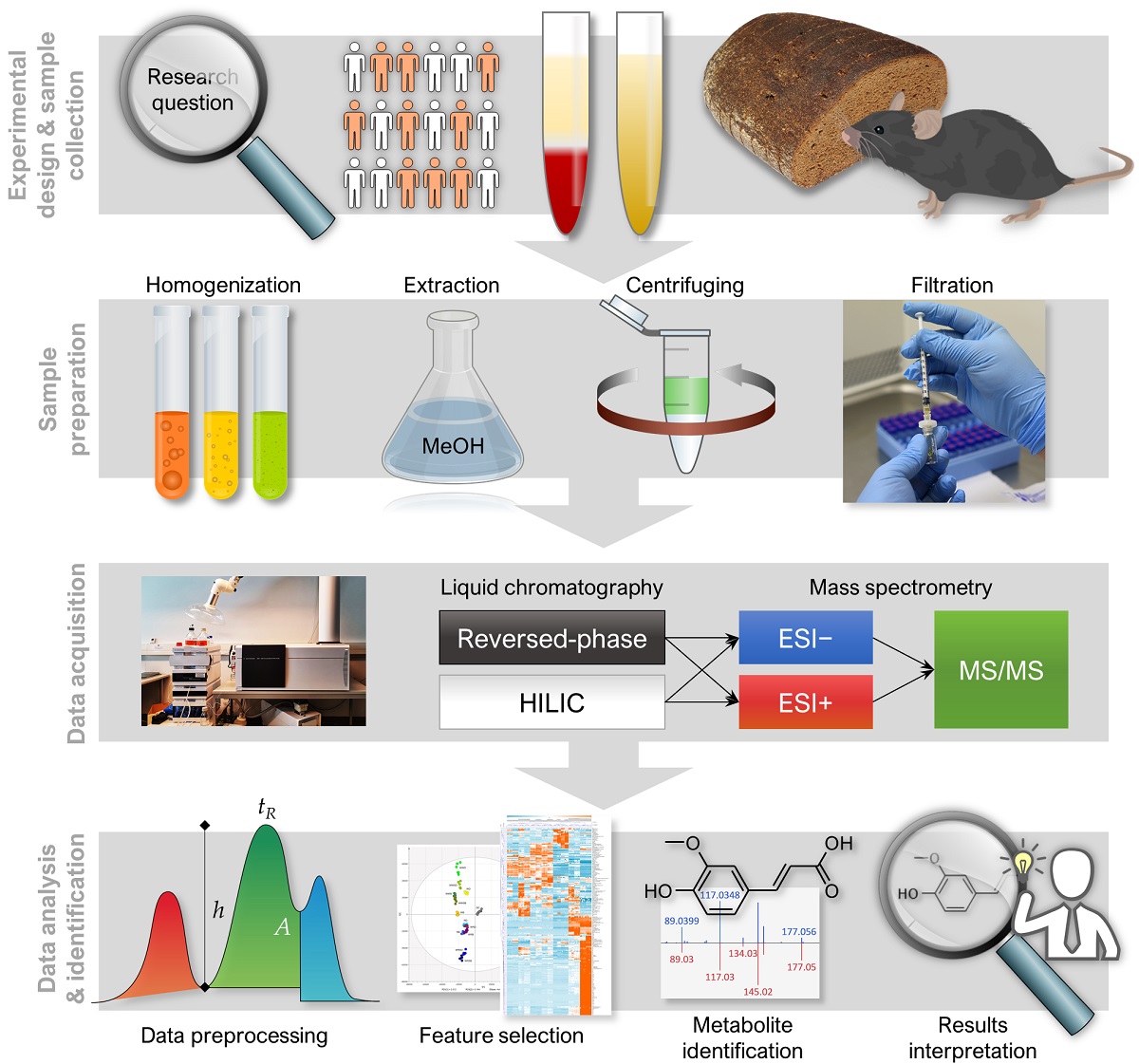
Metabolomics analysis generates vast arrays of data, necessitating comprehensive workflows involving expertise in analytics, biochemistry and bioinformatics, in order to provide coherent and high-quality data that enables discovery of robust and biologically significant metabolic findings. In this protocol article, we introduce NoTaMe, an analytical workflow for non-targeted metabolic profiling approaches utilizing liquid chromatography–mass spectrometry analysis. We provide an overview of lab protocols and statistical methods that we commonly practice for the analysis of nutritional metabolomics data. The paper is divided into three main sections: the first and second sections introducing the background and the study designs available for metabolomics research, and the third section describing in detail the steps of the main methods and protocols used to produce, preprocess and statistically analyze metabolomics data, and finally to identify and interpret the compounds that have emerged as interesting.