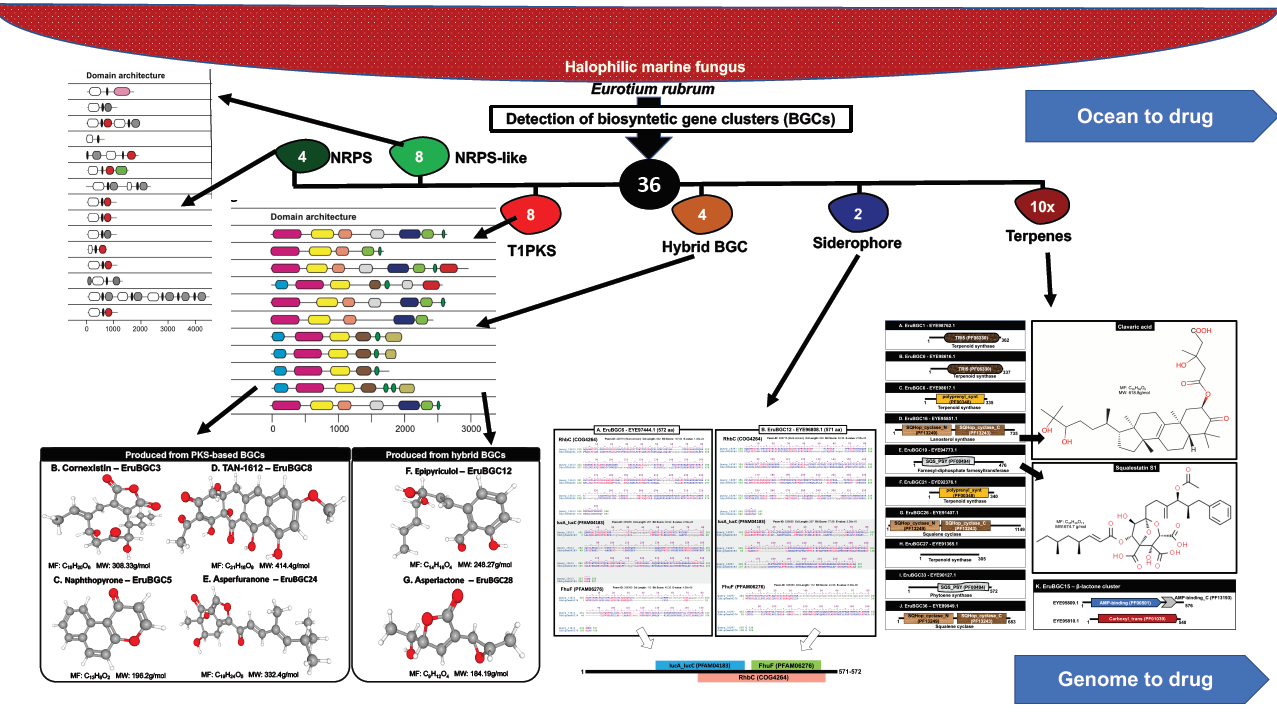
Eurotium rubrum is a halophilic marine ascomycete, which can bear the hypersalinities of the Red Sea and proliferate, while most living entities cannot bear this condition. Recently, a 26.2 Mb assembled genome of this fungus had become available. Marine fungi are fascinating organisms capable of harboring several biosynthetic gene clusters (BGCs), which enables them to produce several natural compounds with antibiotic and anticancerous properties. Understanding the BGCs are critically important for the development of biotechnological applications and the discovery of future drugs. There is no knowledge available on the BGCs of this halophilic marine ascomycete. Herein, we set out to explore and characterize BGCs and the corresponding genes from E. rubrum using bioinformatic methods. We deciphered 36 BGCs in the genome of E. rubrum. These 36 BGCs can be grouped into four non-ribosomal peptide synthetase (NRPS) clusters, eight NRPS-like (NRPSL) BGCs, eight type I polyketide synthase (T1PKS), 11 terpene BGCs including one β-lactone cluster, four hybrid BGCs, and two siderophore BGCs. This study is an example of marine genomics application into potential future drug-like compound discovery.