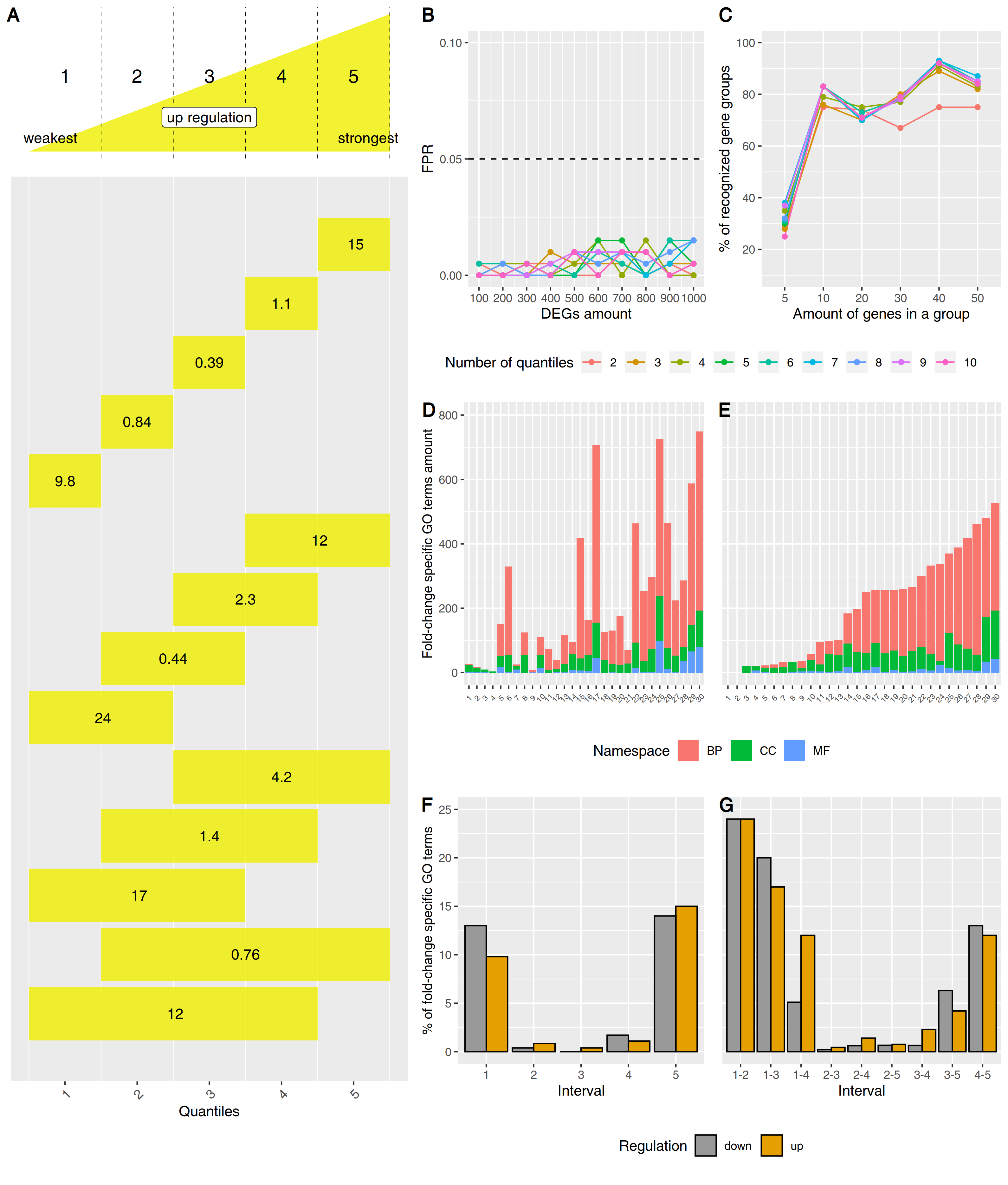
Gene expression profiling data contains more information than is routinely extracted with standard approaches. Here we present Fold-change-Specific Enrichment Analysis (FSEA), a new method for functional annotation of differentially expressed genes from transcriptome data with respect to their fold changes. FSEA identifies GO terms, which are shared by the group of genes with a similar magnitude of response, and assesses these changes. GO terms found by FSEA are fold-change-specifically (e.g. weakly, moderately or strongly) affected by a stimulus under investigation. We demonstrate that many responses to abiotic factors, mutations, treatments and diseases occur in a fold-change-specific manner. FSEA analyses suggest that there are two prevailing responses of functionally-related gene groups, either weak or strong. Notably, some of the fold-change-specific GO terms are invisible by classical algorithms for functional gene enrichment, SEA and GSEA. These are GO terms not enriched compared to the genome background but strictly regulated by a factor within specific fold-change intervals. FSEA analysis of a cancer-related transcriptome suggested that the gene groups with a tightly coordinated response can be the valuable source to search for possible regulators, markers and therapeutic targets in oncogenic processes. Availability and Implementation: FSEA is implemented as the FoldGO Bioconductor R package and a web-server https://webfsgor.sysbio.cytogen.ru/ .