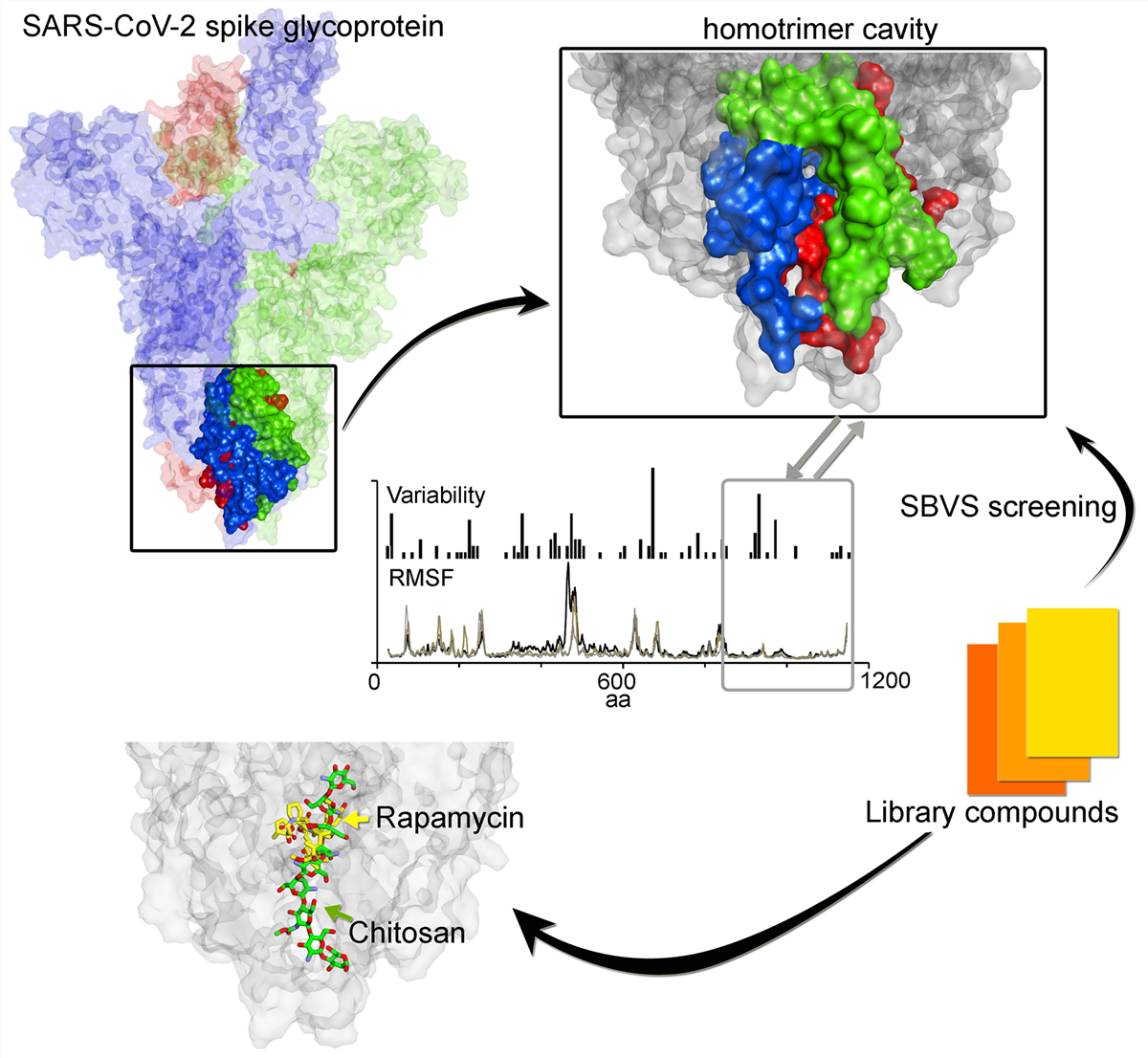
An important stage in SARS-CoV-2 life cycle is the fusion of spike(S) protein with the ACE2 host-cell receptor. Therefore, to explore conserved features in S protein dynamics and to identify potentially novel regions for drugging, we measured variability derived from 791 viral genomes and studied its properties by MD simulation. The findings indicated that S2 subunit (HR1, CH, and CD domains) showed low variability, low fluctuations in MD, and displayed a trimer cavity. By contrast, the RBD domain, which is typically targeted in drug discovery programmes, exhibits more sequence variability and flexibility. Interpretations from MD suggest that the monomer is in constant motion showing transitions up-to-down state, and the trimer cavity may function as a 'bouncing spring' that may facilitates S protein interactions with ACE2. Feasibility of trimer cavity for potential drug target was examined by SBVS screening. Several hits that have already been validated or suggested to inhibit the SARS-CoV-2 virus in cell systems were identified; in particular, the data suggest an action mechanism for such molecules including Chitosan and macrolide types. These findings identify a novel binding-site formed by the S protein, that might assist in future drug discovery programmes aimed at targeting the CoV family of viruses.