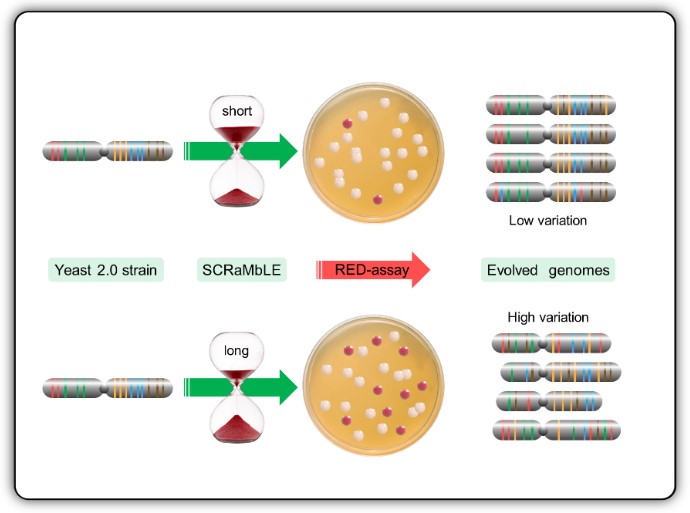
Genome-scale engineering and custom synthetic genomes are reshaping the next generation of industrial yeast strains. The Cre-recombinase mediated chromosomal rearrangement mechanism of designer synthetic Saccharomyces cerevisiae chromosomes, known as SCRaMbLE, is a powerful tool which allows rapid genome evolution upon command. This system is able to generate millions of novel genomes with potential valuable phenotypes, but the excessive loss of essential genes often results in poor growth or even the death of cells with useful phenotypes. In this study we expanded the versatility of SCRaMbLE to industrial strains, and evaluated different control measures to optimise genomic rearrangement, whilst limiting cell death. To achieve this, we have developed RED (Rapid Evolution Detection), a simple colorimetric plate-assay procedure to rapidly quantify the degree of genomic rearrangements within a post-SCRaMbLE yeast population. RED-enabled semi-synthetic strains were mated with haploid progeny of industrial yeast strains to produce stress tolerant heterozygous diploid strains. Analysis of these heterozygous strains with the RED-assay, genome sequencing and custom bioinformatics scripts demonstrated a correlation between RED-assay frequencies and physical genomic rearrangements. Here we show that RED is a fast and effective method to evaluate optimal SCRaMbLE induction times of different Cre-recombinse expression systems for the development of industrial strains.