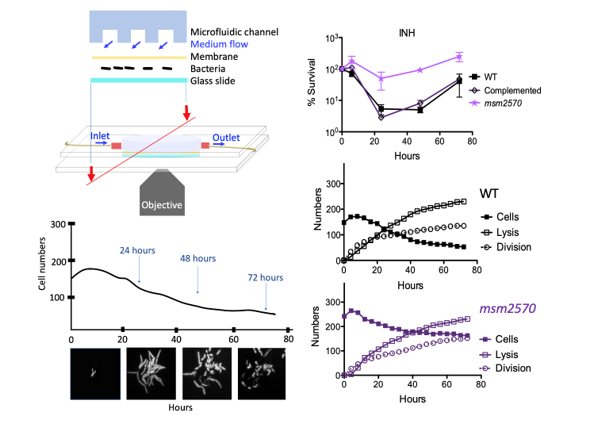
To reveal rare phenotypes in bacterial populations conventional microbiology tools should be advanced to generate rapid, quantitative, accurate and high-throughput data. The main drawbacks of widely used traditional methods for antibiotic studies include low sampling rate and averaging data for population measurements. To overcome these limitations microfluidic-microscopy systems have great promise to produce quantitative single-cell data with high sampling rates. Using Mycobacterium smegmatis cells we applied both conventional assays and a microfluidic-microscopy method to reveal antibiotic-tolerance mechanisms of wild type and the msm2570::Tnmutant cells. Our results revealed that the enhanced antibiotic tolerance mechanism of the msm2570::Tn mutant was due to the low number of lysed cells during the antibiotic exposure compared with wild-type cells. This is the first study that characterized the antibiotic-tolerance phenotype of the msm2570::Tn mutant that has a transposon insertion in the msm2570 gene encoding a putative xanthine/uracil permease, which enrolls in uptake of nitrogen compound during nitrogen limitation. The experimental results indicate that the msm2570::Tn mutant can be further interrogated to reveal antibiotic killing mechanisms, in particularly, antibiotics those targets cell wall integrity.