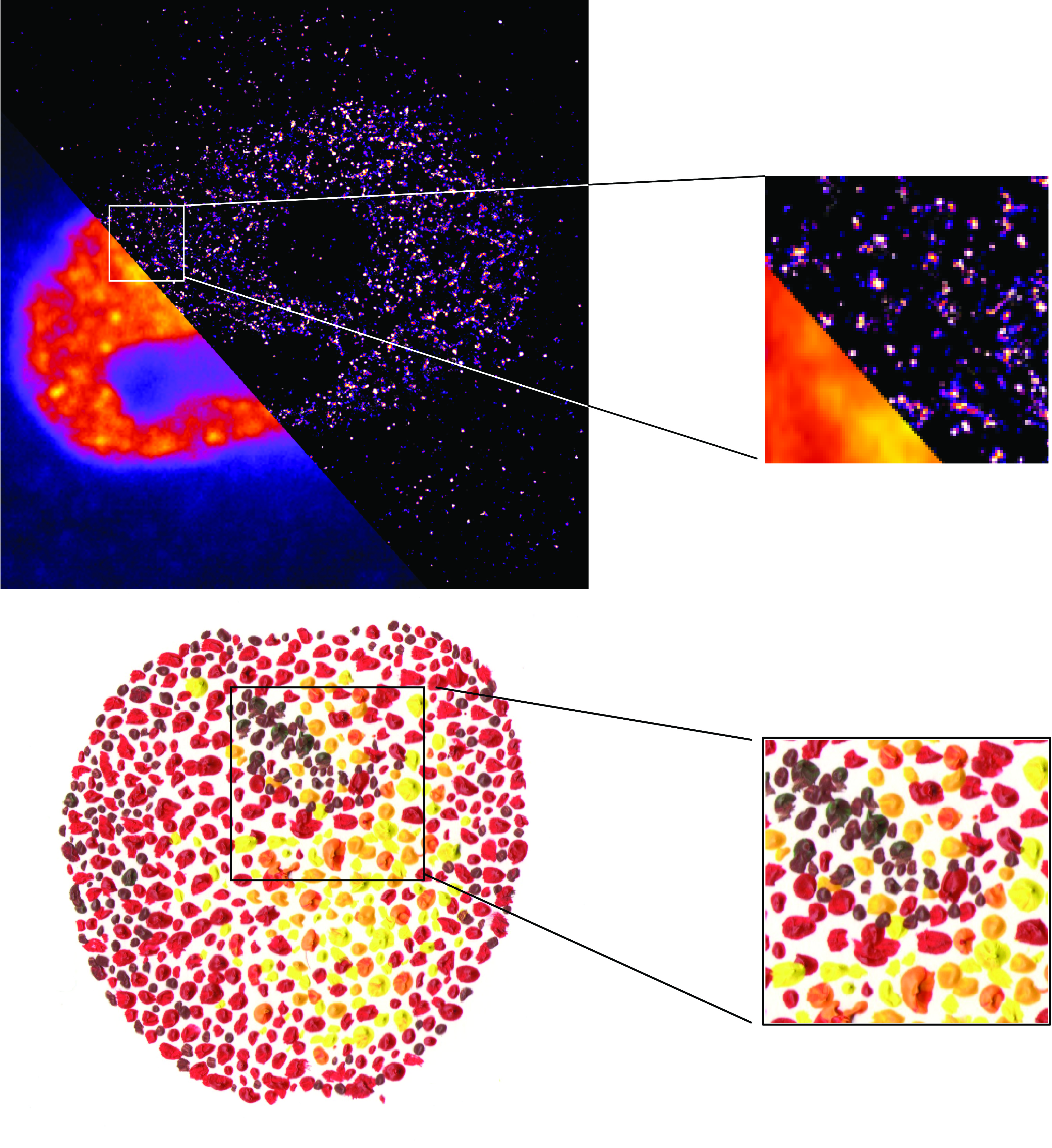
Classical models of gene expression were built using genetics and biochemistry. Although these approaches are powerful, they have very limited consideration of the spatial and temporal organization of gene expression. Although the spatial organization and dynamics of RNA polymerase II (RNAPII) transcription machinery has fundamental functional consequences for gene expression, its detailed studies have been for long time abrogated by the limits of classical light microscopy. The advent of super-resolution microscopy (SRM) techniques allowed for the visualization of the RNAPII transcription machinery with nanometer resolution and millisecond precision. In this review, we summarize the recent methodological advances in SRM, focus on its application for studies of the nanoscale organization in space and time of RNAPII transcription, and discuss its consequences for the mechanistic understanding of gene expression.