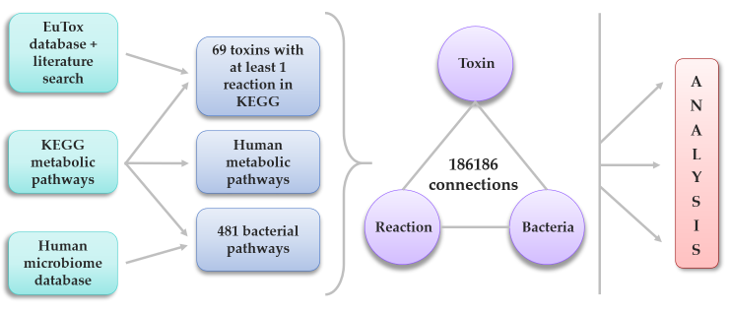
Uremic toxins are the compounds that emerge in the blood when kidney excretory function is impaired. The cumulative detrimental effect of uremic toxins results in numerous health problems and eventually death during acute or chronic uremia, especially in end-stage renal disease. More than 100 different solutes rise during uremia; however, the exact origin for most of them is still discussable. There are 3 main sources for such compounds: exogenous ones are consumed with food, whereas endogenous are produced by host metabolism or by symbiotic microbiota metabolism. In this article, we identified uremic toxins presumably of gut microbiota origin. We analyzed various databases to get information on enzymatic reactions in bacteria and human organism potentially yielded uremic toxins and to determine what toxins could be synthetized in bacteria residing in human gut. We selected biochemical pathways resulting in uremic toxins synthesis related to specific bacterial strains, and revealed links between toxin concentration in uremia and the proportion of different bacteria species, which can synthesize it. Moreover, we defined the relative abundance of human toxin-generating enzymes as well as the possibility of a particular toxin synthesis by the human metabolism. Finally, we analyzed which bacteria are potentially producing the biggest number of uremic toxins as well as which bacteria are decomposing them. Our study presents a bioinformatics-based approach for both elucidation of the origin of uremic toxins and search of the most likely human microbiome producers of toxins that can be targeted and used for the therapy of adverse consequences of uremia.