1. Introduction
Chronic Myeloid Leukemia (CML), is a chronic myeloproliferative malignancy of stem cells that is manifested in the blood (Senapati et al. 2023). It is caused by t(9;22), leading to formation of a chromosomal abnormality called as Philadelphia (Ph) chromosome (Narlı et al. 2023). This chromosomal abnormality results in BCR-ABL fusion oncogene responsible for onset of cancer in myeloid lineage of hematopoietic stem cells (Eden et al. 2023). Estimates of the annual prevalence range from 0.6 to 2.0 instances per 100,000 people monitored, or around 10-15% of newly diagnosed adult cases of leukemia (Sasaki et al, 2023, Siegel et al, 2017). The median age of diagnosis for CML is between 57 and 60 years old, and 1.2-1.7% more males than women get the disease (Apperley, 2015; Jabbour, 2020, Cortes, 2006, Lurlo et al, 2023).
The modern BCR-ABL inhibitor drugs called tyrosine kinase inhibitors (TKIs) CML have revolutionized CML treatment. However, there are certain challenges in managing patients with these drugs (Senapati et al. 2023). CML has three disease phases called as chronic phase (CP-), accelerated phase (AP-) and blast crisis (BC-) CML (Boucher et al., 2023). TKIs are very effective in CP-CML, resulting in the overall survival rate improving from 20% to 80–90% (Venkitaraman, 2004; Senapati et al. 2023). This led to overall survival of CP-CML patients equal to general public, at least in technologically advanced countries like USA, Canada, Europe and Japan (Busch et al, 2023). Nevertheless, patients in early progression phase (AP-CML) and terminal progression phase (BC-CML) show resistance to TKIs (Takahashi, 2023). Specifically in BC-CML, despite all advancements in treatment modalities, the average overall survival is less than a year, which provides a little time frame for oncologists to clinically intervene CML progression (Yoshimaru & Minami,, 2023). Early detection of CML patients at risk of disease progression can considerably help oncologists to delay or even avoid CML progression by timely treatment decision making specifically with the introduction of third and fourth generation TKIs (Shin et al., 2023). Nevertheless, no specific and universal molecular biomarkers exist for timely detection of CML patient groups at risk of disease progression (Li et al. 2023).
Cancer has several mechanisms involved in the initiation and progression of its different types, including DNA repair defects leading to genomic instability (Telliam et al., 2023). Fanconi anemia (FA) is a rare autosomal recessive hereditary illness characterized by gene mutations that are predominantly involved in DNA damage response or repair (Leung et al., 2023). The FANC genes play a crucial role in the FA pathway, regulating DNA damage responses through complicated processes like ubiquitination, phosphorylation, and degradation signals, all of which are required for genome stability and genomic integrity (Dong, 2015). Due to increased genomic instability, it is well known that the FANC gene dysfunction increases the chance of developing various hematological and solid malignancies (D’Andrea, 2003). FANCD2 mutations have recently been found associated with BC-CML as a biomarker of terminal CML progression (Absar et al, 2020). Accordingly, this study was designed to find out the potential of mutated FANCD2 as a biomarker of early CML progression in AP-CML. For the first time in literature, we hereby report mutations in major FA-pathway genes associated exclusively with early CML progression in AP-CML patients.
2. Materials and Methods
Patient selection and recruitment: Clinical Follow-up The study was conducted on CML patients enrolled in Hayatabad Medical Complex (HMC) Peshawar, Khyber Pakhtunkhawa (KP), Pakistan, from January 2020 to July 2023. The number of CP-CML patients included in this study was 123. The experimental group compromised 60 Accelerated Phase (AP-CML) patients, while 123 age/gender-matched Chronic Phase (CP-CML) patients were controls. All patients were initially treated with imatinib mesylate (IM), and patients with IM resistance were given nilotinib (NI). The European Leukemia Net guidelines 2020 were utilized to determine the criteria of treatment responses (Baccarani, 2013; Baccarani, 2015, Yilmaz et. 2023). Standard terminologies, version 4.03 was used to classify the hematological adverse events and other adverse events (Cortes, 2012).
The regulations of the Declaration of Helsinki were followed throughout the study. All patients included in the study provided written informed consent (World Medical Association, 2007; Goodyear, 2007). The approval of study protocols was obtained from Scientific Committees and Ethical Review Boards (ERBs) of King Abdullah International Medical Research Center (KAIMRC); King Saud bin Abdulaziz University for Health Sciences (KSAU-HS), Hayatabad Medical Complex (HMC), Peshawar, Pakistan; and University of the Punjab, Lahore, Pakistan.
Sample Collection and DNA Extraction
Peripheral blood samples were collected in 3-5ml EDTA tubes (BD Vacutainer Systems, Franklin Lakes, N.J.) from all age groups and clinical phases of CML patients and stored at -70°C for further examination. Venous blood samples were obtained from registered CML patients biweekly, for follow-up and medication refills, during their visits to the outpatient department (OPD) of the medical oncology unit, HMC, Peshawar Khyber Pakhtunkhawa (KPK) Pakistan. All blood samples, DNA extraction kits, and reagents were set to room temperature (15-25°C) prior to DNA extraction by using a 56°C water bath. The extraction of genomic DNA from blood samples was performed using the QIAamp DNA Mini Kit (#51306) (Qiagen).
Targeted Resequencing of FANCD2 using Next Generation Sequencing (NGS):
To represent each clinical phase of the disease (Chronic Phase and Accelerated Phase), well-characterized CML samples were selected and processed for NGS (Gnirke, 2009). Illumina® DNA Prep with Enrichment, (S) Augmentation kit (Cat. # 20025523) was utilized for target enrichment (Gnirke, 2009; AlAsiri, 2015). The first step to NGS was DNA fragmentation, followed by tagmentation. Afterward, tagmented DNA fragments were amplified and then purified using magnetic beads. Next, Oligos were utilized to capture target regions. Enriched libraries were amplified by PCR and quantified using a Qubit fluorometer, while Agilent Bioanalyzer was equipped to measure the library size distribution. Finally, cluster generation and exon sequencing were performed using the Illumina NextSeq500 instrument by loading the quantified DNA libraries on the flow cell (AlAsiri, 2015, Absar et al., 2020).
Next Generation Sequencing (NGS) Data Analysis
The conversion of output files, BCL files to FASTQ files was done by BCL2FASTQ software. Alignment of FASTQ files to the human genome was performed by BWA Aligner, applying the BWA-MEM algorithm. Variants were called by the Genome analysis tool kit (GATK). Illumina Variant Studio was used for the annotation and filtration of genomic variants (AlAsiri, 2015).
Primary Analysis
FANCD2 gene was analyzed in all AP-CML patients to detect shared biomarkers of CML progression. Filtration strategies that relied on calling rare variants and excluding intron and synonymous variants were applied to modify the excel file presenting NGS. Furthermore, all variants with known prediction were removed, either benign (B) or tolerant (T). Some variants were considered as B when it has 70% or more of B, while other variants were classified as T when T’s frequency was 70% or more (Carson, 2014). Variants with more than 0.005 population frequency in the dbSNP and Exome Sequencing Project (ESP) database were also eliminated. Thus, variant calling was only limited to variants with intermediate and high protein effects along with splice variants, resulting in about 124 rare variants. Moreover, data was further analyzed to investigate novel gene mutations that are present in AP-CML patients but not in CP-CML patients and healthy controls, suggesting its role in disease progression (Branford, 2018; Xu, 2020). Access to data made by next-generation sequencing can be obtained from NCBI, to which it was submitted, at
https://www.ncbi.nlm.nih.gov/sra/PRJNA734 750 (SRA accession number PRJNA734750; accessed on 28 September 2022).
Validation of Mutation by Sanger Sequencing
Samples were prepared using ABI Prism 3730 Genetic Analyzer (Applied Biosystems, California, USA) and ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kits, and Amplification of samples was done via PCR. Variants identified through NGS were validated by Sanger sequencing (Tsiatis, 2010). Forward and reverse sequencing of PCR amplified FANCD2 fragments were performed by the Sanger sequencer and mutational analysis carried out, as described earlier (Absar et al. 2020).
Statistical Analysis of Patient Clinical Data
Categorical variables were represented with percentages and absolute numbers, while continuous variables were measured with mean and median according to the normality test. Chi-Square and Fisher’s exact test were utilized to compare categorical data of two groups, depending on Applicability, while the comparison of two groups of continuous data was done by Two sample independent test or Mann Whitney U test, depending on the normality hypothesis. ANOVA or Kruskal-Wallis test were performed to analyze data from more than 3 groups. Assessment of survival outcome was made using Kaplan-Meier survival analysis curves, and the log-rank test was used to compare groups. [SAS/STAT] software version 9.4 (SAS Institute Inc., Cary, NC, USA.) and R foundation were used for data analysis and statistical computing (Vienna, Austria), accordingly (R Core Team., 2012). Calculations of the Sokal risk score, Eutos risk score, and Euro risk scores were also done (Sokal, 1984; Hasford, 1998; Hasford, 2011).
3. Results
This study comprised 183 CML patients. The overall mean age of all patients was 34.6. However, mean age for CP-CML and AP-CML patients was 33.5 and 35.6, respectively. Regarding gender, CML was more common in males as they constituted 60.5%, while females were only 39.5%, giving a significant male-to-female ratio of 1.6:1(p = 0.0200). Moreover, the male-to-female ratios for CP-CML and AP-CML patients were 1.5:1 and 2:1, accordingly. The means of clinical characteristics calculated were 10.1 for hemoglobin, 317.9 for white blood cell count, and 400.2 for platelet count. Furthermore, anemia and leukocytosis of more than 50 × 109/L were observed in more than two third of the patients. In addition, various types of treatment were applied to patients, including Imatinib and chemotherapy. Overall, characteristics including male-to-female ratio, hemoglobin level, WBC count, platelet count, treatment type, hepatomegaly, splenomegaly, and survival status were significantly altered in AP-CML patients, compared to CP-CML patients. The comparison between CML phases in regard to patients’ demographic and laboratory characteristics are displayed in
Table 1.
Next Generation Sequencing (NGS)
The results of NGS indicated a novel splice site mutation at genomic position 10,106,408, corresponding to leading to cytosine to thymine substitution (c. 2022-5C>T), in the FANCD2 gene. This gene is an important member of FA-pathway genes located in chromosome 3. This mutation was shared by all AP-CML patients but not CP-CML patients, suggesting its association with disease progression.
Validation by Sanger Sequencing
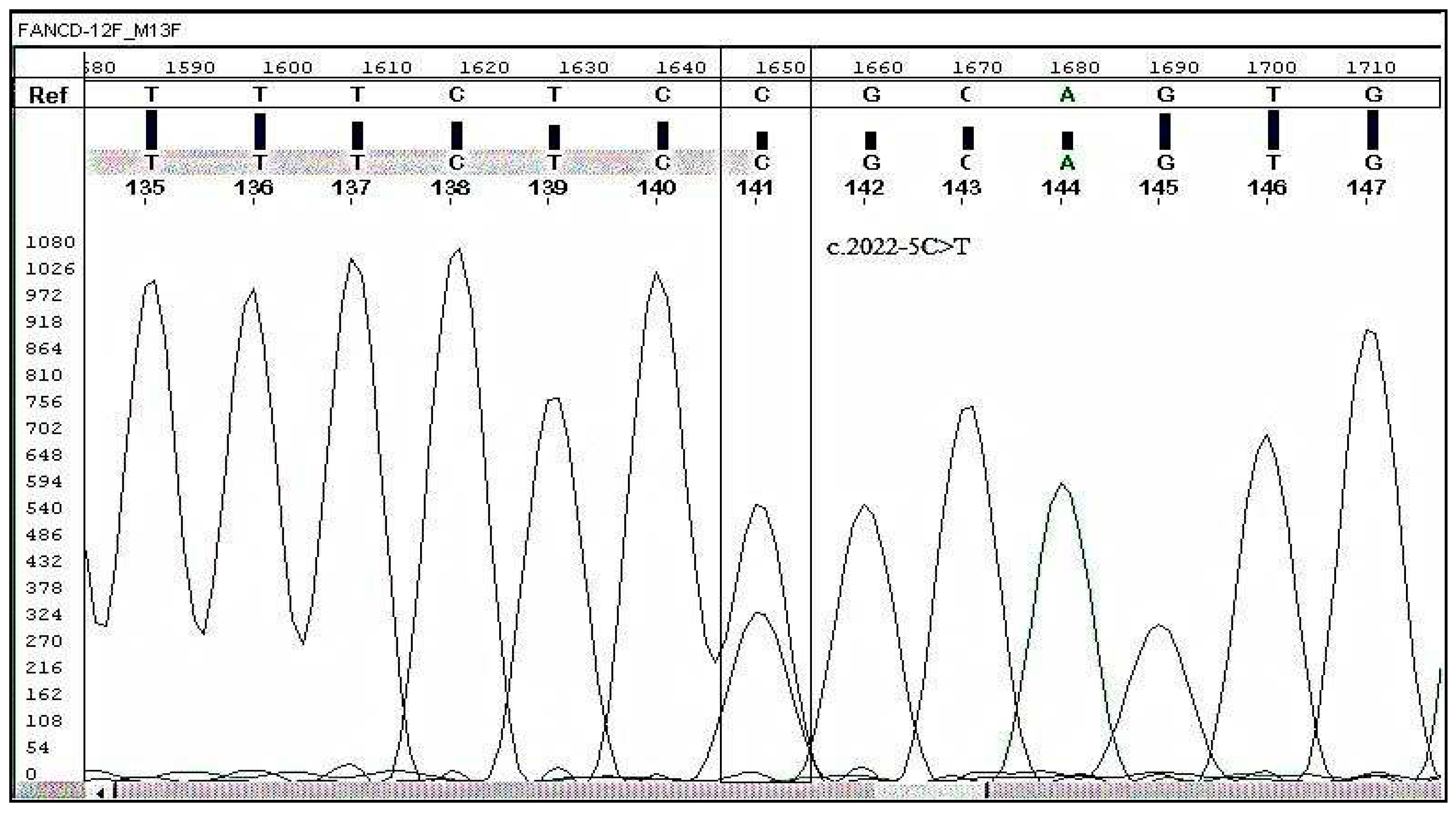
A heterozygous variant (C2022T) was found and validated by sanger sequencing. The FANCD2, c. 2022-5C>T (genomic position 10,106,408) detected by NGS was confirmed by Sanger sequencing as indicated in Figure 3. This demonstrates that the above-mentioned mutation is an indicator of early CML progression in CML patients.
As a result of our clinical validation investigations, it is concluded that the FANCD2 gene is exclusively mutated in all AP-CML patients but not in any of the control subjects. Our research indicates that the FANCD2 (c. 2022-5C>T) mutation can serve as a highly specific molecular biomarker for the early progression of CML.
Table 1.
A comparison between CP- and AP-CML patients in this study in regard to their demographic and laboratory characteristics.
Table 1.
A comparison between CP- and AP-CML patients in this study in regard to their demographic and laboratory characteristics.
| Characteristics |
Patient Groups (%) n |
|
| CP-CML |
AP-CML |
P value |
| |
(95.35) 123 |
(4.65) 60 |
|
| Mean age (Range) |
33.5 (range 7-69) |
35.6 (range=27-43) |
|
| Gender |
|
|
| Male |
(60.2) 74 |
(66.67) 40 |
0.60 |
| Female |
(39.8) 49 |
(33.33) 20 |
0.59 |
| Male: Female Ratio |
1.5:1 |
2:1 |
0.02 |
| Mean Hemoglobin (g/dL) |
10.1 |
|
| Mean WBC count (×109/L) |
313.7 |
315 |
|
| 50> |
(16.3) 20 |
(20) 10 |
0.82 |
| 50=/< |
(83.7) 103 |
(80) 50 |
0.02 |
| P value |
0.005 |
0.02752 |
|
| |
|
|
|
| Platelets (× 109/L) Mean |
400.2 |
| <450 |
75 (61) |
40 (66.7) |
|
| >/=450 |
33 (26.8) |
20 (33.3) |
|
| No data found |
15 (12.2) |
0 |
|
| P value |
0.0011 |
0.47 |
|
| Imatinib |
(66.7) 82 |
(66.7) 40 |
0.72 |
| Interferon |
(33.3) 41 |
(0) 0 |
0.0038 |
| Chemotherapy |
(8.1) 10 |
(66.7) 40 |
<0.0001 |
| Splenomegaly |
|
|
| cm 5> |
(3.3) 4 |
(0) 0 |
0.43 |
| cm 8–5 |
(7.3) 9 |
(16.7) 10 |
0.061 |
| cm 8< |
(56.9) 70 |
(83.3) 50 |
0.07 |
| No splenomegaly |
(32.5) 40 |
(0) 0 |
0.004 |
| Hepatomegaly |
|
|
| Yes |
(28.5) 35 |
(66.7) 40 |
0.001 |
| Survival Status |
|
|
| Confirmed Deaths |
0 |
1(1.7) |
0.0003 |
| Overall survival at last follow-up |
(100) 123 |
59 (98.3) |
0.0003 |
Figure 1.
Electropherogram showing mutation C > T in FANCD2 at splice site of intron 22 and its comparison with reference / wild-type sequence.
Figure 1.
Electropherogram showing mutation C > T in FANCD2 at splice site of intron 22 and its comparison with reference / wild-type sequence.
4. Discussion
In our study, 4.65% of 129 CML patients progressed to AP-CML. We found that the FANCD2 gene was mutated in all patients in the accelerated phase, compared to healthy individuals and Chronic phase CML patients. To consolidate our findings, we found another study suggesting that the crucial FA pathway protein, FANCD2, has been linked to the advancement of the CML illness (Valeri, 2012). In 2007, a study concluded that the frequency of FANCD2 was greater than that of any other FA complementation group (Kalb, 2007). Although FANCD2’s exact function is yet unclear, the upkeep of genomic stability is assumed to be mostly regulated by FANCD2(31). According to Valeri et al., BCR-ABL1 induces genetic instability in CML cells by inhibiting FANCD2 nuclear foci, leading to centrosomal amplification and DNA repair deficiencies. FANCD2, located at 3p25.3, plays a role in repairing impaired DNA (Valeri, 2012; Meetei, 2003). However, a mutated FANCD2 gene will result in its inability to continue the DNA-repairing cascade (Absar, 2023). The FANCD2 variant detected in this study was located in chromosome 3, and it was positioned at 10106408 along with a displacement of Cytosine to Thymine. By the end of 2015, A similar FANCD2 variant, NM_001018115.3(FANCD2): c.2022-5C>T), had its first publication. Conflicting theories about the pathogenicity of both comparable variants were present. They have been associated with FA as a pathogenic and with Hereditary Breast Ovarian Cancer Syndrome as benign (National Center for Biotechnology Information).
In our study, the splice site mutation between intron 22 and exon 23 resulted in the intron 22 variant. The function of the intron 22 variant on FANCD2 mono-ubiquitination remains uncertain, but it could be predicted since it is close to intron 19, which is the FANCD2 mono-ubiquitination site (Lewis, 2005). A study in China identified a missense mutation c.3713T>A; p.M1238K in the FANCD2 gene that leads to non-expression of the FANCD2 protein. Furthermore, function studies were applied to find that other splice site mutations in the FA gene cause exon skipping (Li, 2018). Another study in the US detected 25 intronic variants and 6 silent coding variants that lead to familial breast cancer. One of which was in exon 23 c.2148 C>G, resulting in T716T protein change (Lewis, 2005).
The FANCI-FANCD2 heterodimer is where the FA pathway comes together (Wang, 2004). It serves as a substrate for the FA core complex as well as a potential collecting site for proteins involved in downstream DNA repairs, such as FAN1 nuclease and other FANC proteins (MacKay, 2012). Despite only possessing a 14% conservation in their solenoidal structures, FANCI and FANCD2 were known to have striking similarities in the 2011 discovery of the crystal lattice of mouse FANCI-FANCD2 (Joo, 2011). More than 97% of FA patients have a deficiency caused by mutations in the genes encoding FANCD2 and FANCI (Wang, 2015). At the region of DNA damage, the FA proteins function as a ubiquitin E3 ligase to monoubiquitinate the FANCI-FANCD2 pair (van Twest, 2017). This results in enlisting downstream nucleases with ubiquitin-binding domains to restore the interstrand DNA bridge (Smogorzewska, 2010). Although monoubiquitination and FA pathway activation necessitate DNA binding of FANCI-FANCD2 (Liang, 2016). It is not certain how it triggers the repair of DNA (Li, 2020).
High FA gene expression is typically associated with chemo-resistance; the high expression level of FANCD2 is associated with reduced chemotherapy sensitivity and a higher tumor mutation rate. It was observed in breast, lung, and ovarian cancers, Thus, resulting in reduced survival time (Dan, 2021; Miao, 2022). FA pathway inhibition by targeted therapies is a promising approach for improving the efficacy of chemotherapy due to its role in chemoresistance across a wide range of cancers (Liu et al., 2020). In early studies, Curcumin, Wortmannin, H-9, and Alsterpaullone were found to inhibit FANCD2 by apoptosis through the NFκB pathway (Chirnomas, 2006). A study further assessed monoketone analogues of Curcumin and found that EF24 was more specific and active against monoubiquitination of FANCD2 (Shen et al. 2015). Lastly, a recent study has identified CU2 as a compound that shows potential biochemical ubiquitylation selectivity and activity against the FA pathway (Cornwell, 2019). Currently, three PARP inhibitors that target the FA pathway are FDA-approved for treating relapsed breast and ovarian cancers, olaparib, rucaparib, and niraparib (Niraj, 2019).
In addition, there have been reports that linked cancer incidence with FA pathway mutations (Shen, 2015). The previously mentioned mutations were reported to cause FA bone marrow failure. Also, the most well-known genes that predispose to breast cancer are BRCA1 and BRCA2, both of which are considered part of the FANC gene family. This system is frequently referred to as the FA-BRCA pathway given the growing relationship between FA and the genes for breast cancer (Williams, 2011). In addition, two studies were investigating the relationship between FANCD2 and breast cancer. In China, a poor prognosis was observed in sporadic breast cancer patients with high levels of FANCD2 (Feng, 2019). A study in Finland supports these findings by showing a significant association of variant (c.2715 + 1G > A) in the FANCD2 gene with breast cancer (Mantere, 2017). Also, studies conducted in the United Kingdom and Poland on ovarian carcinoma samples verified that the risk of recurrence and death are highly associated with the expression level of FANCD2 (Moes-Sosnowska, 2019; Mani, 2021). On the other hand, in the United States, a study conducted on 181 ovarian cancer patients provided evidence of an increased survival rate in patients with FANCD2 mutation (Joshi, 2020). On the basis of its upregulation, FANCD2 has an unsatisfactory prognosis for other types of cancer, which may contribute to tumorigenesis. Such cancers are esophageal squamous cell carcinoma, head and neck carcinoma, nasopharyngeal carcinoma, and lung adenocarcinoma (Lei, 2020; Chandrasekharappa, 2017; Xu, 2019).
In addition, the FANCD2 gene mutation has also been observed in non-cancer diseases. Two studies have proven the association of FANCD2 mutation with other diseases. The first one was a case report of a 2.5-year-old Saudi boy from Al-Ahsa diagnosed with ambiguous genitalia and intrauterine growth restriction (IUGR). This study found that ambiguous genitalia in Saudi male infants could be related to c.2605 + 1G>A variant resulting from a homozygous mutation in the FANCD2 gene. The second study revealed that the FANCD2 expression level is significantly increased in Glioblastoma patients. Moreover, it also plays a role in drug resistance and the progression of the disease (Xu et al., 2023). The impact of FANCD2 mutation on CML progression and drug resistance was evident as discussed above. This highlights the significance of understanding the relationship between FANCD2 and CML (Leung et al., 2023; Valeri net al., 2023).
5. Conclusions
We discovered that the FANCD2 gene was mutated in every AP-CML patient. FANCD2 is member of Fanconi anemia (FA-) pathway gene involved in DNA repair and genomic instability. Therefore, our studies show that FANCD2 (c. 2022-5C>T) mutation as a very specific molecular biomarker for early CML progression. We recommend to clinical validate this biomarker is prospective clinical trials.
Funding
This work was funded by the National Plan for Science, Technology and Innovation (MAARIFAH), King Abdul-Aziz City for Science and Technology, Kingdom of Saudi Arabia, Grant Number 14-Med-2817-02.
Acknowledgements
The study was approved by King Abdullah International Medical Research Centre (KAIMRC), National Guard Health Affairs, Saudi Arabia, although no research funding was provided (project # RA17/002/A). This study was partially supported by the College of Medicine, Research Centre, Deanship of Scientific Research, King Saud University, Riyadh, Saudi Arabia. All authors have read and have consented to the acknowledgement.
Research Ethics’ Statements and Approval of the Study
The regulations of the Declaration of Helsinki were followed throughout the study. All patients included in the study provided written informed consent (World Medical Association, 2007; Goodyear, 2007). The approval of study protocols was obtained from Scientific Committees and Ethical Review Boards (ERBs) of King Abdullah International Medical Research Center (KAIMRC); King Saud bin Abdulaziz University for Health Sciences (KSAU-HS), Hayatabad Medical Complex (HMC), Peshawar, Pakistan; and University of the Punjab, Lahore, Pakistan.
Conflicts of Interest
Honoraria and travel grants from Novartis and Roche have been given to Abid Jameel (AJ). The other authors acknowledge no financial or other conflicts of interest.
References
- D’Andrea, A.D.; Grompe, M. The Fanconi anaemia/BRCA pathway. Nat. Rev. Cancer 2003, 3, 23–34. [Google Scholar] [CrossRef]
- Absar, M.; Mahmood, A.; Akhtar, T.; Basit, S.; Ramzan, K.; Jameel, A.; Afzal, S.; Ullah, A.; Qureshi, K.; Alanazi, N.; et al. Whole exome sequencing identifies a novel FANCD2 gene splice site mutation associated with disease progression in chronic myeloid leukemia: Implication in targeted therapy of advanced phase CML. Pak. J. Pharm. Sci. 2020, 33, 1419–1426. [Google Scholar] [CrossRef]
- AlAsiri, S.; Basit, S.; Wood-Trageser, M.A.; Yatsenko, S.A.; Jeffries, E.P.; Surti, U.; Ketterer, D.M.; Afzal, S.; Ramzan, K.; Haque, M.F.-U.; et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J. Clin. Investig. 2015, 125, 258–262. [Google Scholar] [CrossRef]
- Ali, M.A.M. Chronic Myeloid Leukemia in the Era of Tyrosine Kinase Inhibitors: An Evolving Paradigm of Molecularly Targeted Therapy. Mol. Diagn. Ther. 2016, 20, 315–333. [Google Scholar] [CrossRef]
- Apperley, J.F. Chronic myeloid leukaemia. Lancet 2015, 385, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Baccarani, M.; Castagnetti, F.; Gugliotta, G.; Rosti, G. A review of the European LeukemiaNet recommendations for the management of CML. Ann. Hematol. 2015, 94, 141–147. [Google Scholar] [CrossRef]
- Baccarani, M.; Deininger, M.W.; Rosti, G.; Hochhaus, A.; Soverini, S.; Apperley, J.F.; Cervantes, F.; Clark, R.E.; Cortes, J.E.; Guilhot, F.; et al. European LeukemiaNet recommendations for the management of chronic myeloid leukemia: 2013. Blood 2013, 122, 872–884. [Google Scholar] [CrossRef] [PubMed]
- Branford, S.; Wang, P.; Yeung, D.T.; Thomson, D.; Purins, A.; Wadham, C.; Shahrin, N.H.; Marum, J.E.; Nataren, N.; Parker, W.T.; et al. Integrative genomic analysis reveals cancer-associated mutations at diagnosis of CML in patients with high-risk disease. Blood 2018, 132, 948–961. [Google Scholar] [CrossRef] [PubMed]
- Carson, A.R.; Smith, E.N.; Matsui, H.; Brækkan, S.K.; Jepsen, K.; Hansen, J.-B.; A Frazer, K. Effective filtering strategies to improve data quality from population-based whole exome sequencing studies. BMC Bioinform. 2014, 15, 125–125. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekharappa, S.C.; Chinn, S.B.; Donovan, F.X.; Chowdhury, N.I.; Kamat, A.; Adeyemo, A.A.; Thomas, J.W.; Vemulapalli, M.; Hussey, C.S.; Reid, H.H.; et al. Assessing the spectrum of germline variation in Fanconi anemia genes among patients with head and neck carcinoma before age 50. Cancer 2017, 123, 3943–3954. [Google Scholar] [CrossRef]
- Chirnomas, D.; Taniguchi, T.; de la Vega, M.; Vaidya, A.P.; Vasserman, M.; Hartman, A.-R.; Kennedy, R.; Foster, R.; Mahoney, J.; Seiden, M.V.; et al. Chemosensitization to cisplatin by inhibitors of the Fanconi anemia/BRCA pathway. Mol. Cancer Ther. 2006, 5, 952–961. [Google Scholar] [CrossRef] [PubMed]
- Chronic granulocytic leukaemia: comparison of radiotherapy and busulphan therapy. Report of the Medical Research Council’s working party for therapeutic trials in leukaemia. BMJ 1968, 1, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Cornwell, M.J.; Thomson, G.J.; Coates, J.; Belotserkovskaya, R.; Waddell, I.D.; Jackson, S.P.; Galanty, Y. Small-Molecule Inhibition of UBE2T/FANCL-Mediated Ubiquitylation in the Fanconi Anemia Pathway. ACS Chem. Biol. 2019, 14, 2148–2154. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.E.; Lipton, J.H.; Miller, C.B.; Ailawadhi, S.; Akard, L.; Pinilla-Ibarz, J.; Lin, F.P.; Ericson, S.G.; Mauro, M.J. Change in Chronic Low-Grade Nonhematologic Adverse Events (AEs) and Quality of Life (QoL) in Adult Patients (pts) with Philadelphia Chromosome–Positive (Ph+) Chronic Myeloid Leukemia in Chronic Phase (CML-CP) Switched From Imatinib (IM) to Nilotinib (NIL). Blood 2012, 120, 3782–3782. [Google Scholar] [CrossRef]
- Cortes, J.E.; Talpaz, M.; O’Brien, S.; Faderl, S.; Garcia-Manero, G.; Ferrajoli, A.; Verstovsek, S.; Rios, M.B.; Shan, J.; Kantarjian, H.M. Staging of chronic myeloid leukemia in the imatinib era: an evaluation of the World Health Organization 172 proposal. Cancer 2006, 106, 1306–1315. [Google Scholar] [CrossRef] [PubMed]
- Dan, C.; Pei, H.; Zhang, B.; Zheng, X.; Ran, D.; Du, C. Fanconi anemia pathway and its relationship with cancer. Genome Instab. Dis. 2021, 2, 175–183. [Google Scholar] [CrossRef]
- Dong, H.; Nebert, D.W.; Bruford, E.A.; Thompson, D.C.; Joenje, H.; Vasiliou, V. Update of the human and mouse Fanconi anemia genes. Hum. Genom. 2015, 9, 32. [Google Scholar] [CrossRef]
- Feng, L.; Jin, F. Expression and prognostic significance of Fanconi anemia group D2 protein and breast cancer type 1 susceptibility protein in familial and sporadic breast cancer. Oncol. Lett. 2019, 17, 3687–3700. [Google Scholar] [CrossRef]
- Gnirke, A.; Melnikov, A.; Maguire, J.; Rogov, P.; LeProust, E.M.; Brockman, W.; Fennell, T.; Giannoukos, G.; Fisher, S.; Russ, C.; et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009, 27, 182–189. [Google Scholar] [CrossRef]
- Goodyear, E.; Krleza-Jeric, M.D.; Lemmens, K. The Declaration of Helsinki. BMJ 2007, 335, 624–625. [Google Scholar] [CrossRef]
- Hasford, J.; Baccarani, M.; Hoffmann, V.; Guilhot, J.; Saussele, S.; Rosti, G.; Guilhot, F.; Porkka, K.; Ossenkoppele, G.; Lindoerfer, D.; et al. Predicting complete cytogenetic response and subsequent progression-free survival in 2060 patients with CML on imatinib treatment: the EUTOS score. Blood 2011, 118, 686–692. [Google Scholar] [CrossRef] [PubMed]
- Hasford, J.; Pfirrmann, M.; Hehlmann, R.; Allan, N.C.; Baccarani, M.; Kluin-Nelemans, J.C.; Alimena, G.; Steegmann, J.L.; Ansari, H. A New Prognostic Score for Survival of Patients With Chronic Myeloid Leukemia Treated With Interferon Alfa Writing Committee for the Collaborative CML Prognostic Factors Project Group. JNCI J. Natl. Cancer Inst. 1998, 90, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Jabbour, E.; Kantarjian, H. Chronic myeloid leukemia: 2020 update on diagnosis, therapy and monitoring. Am. J. Hematol. 2020, 95, 691–709. [Google Scholar] [CrossRef] [PubMed]
- Joo, W.; Xu, G.; Persky, N.S.; Smogorzewska, A.; Rudge, D.G.; Buzovetsky, O.; Elledge, S.J.; Pavletich, N.P. Structure of the FANCI-FANCD2 Complex: Insights into the Fanconi Anemia DNA Repair Pathway. Science 2011, 333, 312–316. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Campbell, S.; Lim, J.Y.; McWeeney, S.; Krieg, A.; Bean, Y.; Pejovic, N.; Mhawech-Fauceglia, P.; Pejovic, T. Subcellular localization of FANCD2 is associated with survival in ovarian carcinoma. Oncotarget 2020, 11, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Kalb, R.; Neveling, K.; Hoehn, H.; Schneider, H.; Linka, Y.; Batish, S.D.; Hunt, C.; Berwick, M.; Callén, E.; Surrallés, J.; et al. Hypomorphic Mutations in the Gene Encoding a Key Fanconi Anemia Protein, FANCD2, Sustain a Significant Group of FA-D2 Patients with Severe Phenotype. Am. J. Hum. Genet. 2007, 80, 895–910. [Google Scholar] [CrossRef] [PubMed]
- Landais, I.; Hiddingh, S.; McCarroll, M.; Yang, C.; Sun, A.; Turker, M.S.; Snyder, J.P.; E Hoatlin, M. Monoketone analogs of curcumin, a new class of Fanconi anemia pathway inhibitors. Mol. Cancer 2009, 8, 133–133. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.C.; Yu, V.Z.; Ko, J.M.Y.; Ning, L.; Lung, M.L. FANCD2 Confers a Malignant Phenotype in Esophageal Squamous Cell Carcinoma by Regulating Cell Cycle Progression. Cancers 2020, 12, 2545. [Google Scholar] [CrossRef]
- Lewis, A.G.; the Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer; Flanagan, J.; Marsh, A.; Pupo, G.M.; Mann, G.; Spurdle, A.B.; Lindeman, G.J.; E Visvader, J.; A Brown, M.; et al. Mutation analysis of FANCD2, BRIP1/BACH1, LMO4 and SFN in familial breast cancer. Breast Cancer Res. 2005, 7, R1005–R1016. [Google Scholar] [CrossRef]
- Li, L.; Tan, W.; Deans, A.J. Structural insight into FANCI–FANCD2 monoubiquitination. Essays Biochem. 2020, 64, 807–817. [Google Scholar] [CrossRef]
- Li, N.; Ding, L.; Li, B.; Wang, J.; D’Andrea, A.D.; Chen, J. Functional analysis of Fanconi anemia mutations in China. Exp. Hematol. 2018, 66, 32–41. [Google Scholar] [CrossRef]
- Liang, C.-C.; Li, Z.; Lopez-Martinez, D.; Nicholson, W.V.; Vénien-Bryan, C.; Cohn, M.A. The FANCD2–FANCI complex is recruited to DNA interstrand crosslinks before monoubiquitination of FANCD2. Nat. Commun. 2016, 7, 12124. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Palovcak, A.; Li, F.; Zafar, A.; Yuan, F.; Zhang, Y. Fanconi anemia pathway as a prospective target for cancer intervention. Cell Biosci. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- MacKay, C.; Déclais, A.-C.; Lundin, C.; Agostinho, A.; Deans, A.J.; MacArtney, T.J.; Hofmann, K.; Gartner, A.; West, S.C.; Helleday, T.; et al. Identification of KIAA1018/FAN1, a DNA Repair Nuclease Recruited to DNA Damage by Monoubiquitinated FANCD2. Cell 2010, 142, 65–76. [Google Scholar] [CrossRef]
- Mahon, F.-X.; Etienne, G. Deep Molecular Response in Chronic Myeloid Leukemia: The New Goal of Therapy? Clin. Cancer Res. 2014, 20, 310–322. [Google Scholar] [CrossRef]
- Mani, C.; Tripathi, K.; Chaudhary, S.; Somasagara, R.R.; Rocconi, R.P.; Crasto, C.; Reedy, M.; Athar, M.; Palle, K. Hedgehog/GLI1 Transcriptionally Regulates FANCD2 in Ovarian Tumor Cells: Its Inhibition Induces HR-Deficiency and Synergistic Lethality with PARP Inhibition. Neoplasia 2021, 23, 1002–1015. [Google Scholar] [CrossRef]
- Mantere, T.; Tervasmäki, A.; Nurmi, A.; Rapakko, K.; Kauppila, S.; Tang, J.; Schleutker, J.; Kallioniemi, A.; Hartikainen, J.M.; Mannermaa, A.; et al. Case-control analysis of truncating mutations in DNA damage response genes connects TEX15 and FANCD2 with hereditary breast cancer susceptibility. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Meetei, A.R.; de Winter, J.P.; Medhurst, A.L.; Wallisch, M.; Waisfisz, Q.; van de Vrugt, H.J.; Oostra, A.B.; Yan, Z.; Ling, C.; E Bishop, C.; et al. A novel ubiquitin ligase is deficient in Fanconi anemia. Nat. Genet. 2003, 35, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Miao, H.; Ren, Q.; Li, H.; Zeng, M.; Chen, D.; Xu, C.; Chen, Y.; Wen, Z. Comprehensive analysis of the autophagy-dependent ferroptosis-related gene FANCD2 in lung adenocarcinoma. BMC Cancer 2022, 22, 1–18. [Google Scholar] [CrossRef]
- Moes-Sosnowska, J.; Rzepecka, I.K.; Chodzynska, J.; Dansonka-Mieszkowska, A.; Szafron, L.M.; Balabas, A.; Lotocka, R.; Sobiczewski, P.; Kupryjanczyk, J. Clinical importance of FANCD2, BRIP1, BRCA1, BRCA2 and FANCF expression in ovarian carcinomas. Cancer Biol. Ther. 2019, 20, 843–854. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information. ClinVar; [VCV000218824.22]. Available online: https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000218824.22 (accessed on 25 October 2022).
- Niraj, J.; Färkkilä, A.; D’Andrea, A.D. The Fanconi Anemia Pathway in Cancer. Annu. Rev. Cancer Biol. 2019, 3, 457–478. [Google Scholar] [CrossRef]
- Osman, A.E.; Deininger, M.W. Chronic Myeloid Leukemia: Modern therapies, current challenges and future directions. Blood Rev. 2021, 49, 100825. [Google Scholar] [CrossRef]
- Quintás-Cardama, A.; Cortes, J. Molecular biology of bcr-abl1–positive chronic myeloid leukemia. Blood 2009, 113, 1619–1630. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2012; Available online: http://www.R-project.org/ISBN 3-900051-07-0.
- Senapati, J.; Jabbour, E.; Kantarjian, H.; Short, N.J. Pathogenesis and management of accelerated and blast phases of chronic myeloid leukemia. Leukemia 2022, 37, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Senapati, J.; Sasaki, K.; Issa, G.C.; Lipton, J.H.; Radich, J.P.; Jabbour, E.; Kantarjian, H.M. Management of chronic myeloid leukemia in 2023 – common ground and common sense. Blood Cancer J. 2023, 13, 1–12. [Google Scholar] [CrossRef]
- Özdemir, Z.N.; Kılıçaslan, N.A.; Yılmaz, M.; Eşkazan, A.E. Guidelines for the treatment of chronic myeloid leukemia from the NCCN and ELN: differences and similarities. Int. J. Hematol. 2022, 117, 3–15. [Google Scholar] [CrossRef]
- Eden, R. E.; Coviello, J. M. Chronic Myelogenous Leukemia; StatPearls Publishing, 2023. [Google Scholar]
- Iurlo, A.; Cattaneo, D.; Consonni, D.; Castagnetti, F.; Miggiano, M.C.; Binotto, G.; Bonifacio, M.; Rege-Cambrin, G.; Tiribelli, M.; Lunghi, F.; et al. Treatment discontinuation following low-dose TKIs in 248 chronic myeloid leukemia patients: Updated results from a campus CML real-life study. Front. Pharmacol. 2023, 14. [Google Scholar] [CrossRef] [PubMed]
- Boucher, L.; Sorel, N.; Desterke, C.; Chollet, M.; Rozalska, L.; Hernanz, M.P.G.; Cayssials, E.; Raimbault, A.; Bennaceur-Griscelli, A.; Turhan, A.G.; et al. Deciphering Potential Molecular Signatures to Differentiate Acute Myeloid Leukemia (AML) with BCR::ABL1 from Chronic Myeloid Leukemia (CML) in Blast Crisis. Int. J. Mol. Sci. 2023, 24, 15441. [Google Scholar] [CrossRef]
- Busch, C.; Mulholland, T.; Zagnoni, M.; Dalby, M.; Berry, C.; Wheadon, H. Overcoming BCR::ABL1 dependent and independent survival mechanisms in chronic myeloid leukaemia using a multi-kinase targeting approach. Cell Commun. Signal. 2023, 21, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N. Rinsho ketsueki. Jpn. J. Clin. Hematol. 2023, 64, 981–987. [Google Scholar]
- Fei, P. Advances in the understanding of Fanconi Anemia Complementation Group D2 Protein (FANCD2) in human cancer. Cancer Cell Microenviron. 2015, 2. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017, 67, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; Desetty, R.; Saito, T.T.; Schlabach, M.; Lach, F.P.; Sowa, M.E.; Clark, A.B.; Kunkel, T.A.; Harper, J.W.; Colaiácovo, M.P.; et al. A Genetic Screen Identifies FAN1, a Fanconi Anemia-Associated Nuclease Necessary for DNA Interstrand Crosslink Repair. Mol. Cell 2010, 39, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Sokal, J.E.; Cox, E.B.; Baccarani, M.; Tura, S.; Gomez, G.A.; Robertson, J.E.; Tao, C.Y.; Braun, T.J.; Clarkson, B.D.; Cervantes, F. Prognostic discrimination in "good-risk" chronic granulocytic leukemia. Blood 1984, 63, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.H.; Hinz, J.M.; Yamada, N.A.; Jones, N.J. How Fanconi anemia proteins promote the four Rs: Replication, recombination, repair, and recovery. Environ. Mol. Mutagen. 2005, 45, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Tsiatis, A.C.; Norris-Kirby, A.; Rich, R.G.; Hafez, M.J.; Gocke, C.D.; Eshleman, J.R.; Murphy, K.M. Comparison of Sanger Sequencing, Pyrosequencing, and Melting Curve Analysis for the Detection of KRAS Mutations: Diagnostic and Clinical Implications. J. Mol. Diagn. 2010, 12, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Valeri, A.; Río, P.; Agirre, X.; Prosper, F.; A Bueren, J. Unraveling the role of FANCD2 in chronic myeloid leukemia. Leukemia 2012, 26, 1447–8. [Google Scholar] [CrossRef]
- van Twest, S.; Murphy, V.J.; Hodson, C.; Tan, W.; Swuec, P.; O’rourke, J.J.; Heierhorst, J.; Crismani, W.; Deans, A.J. Mechanism of Ubiquitination and Deubiquitination in the Fanconi Anemia Pathway. Mol. Cell 2017, 65, 247–259. [Google Scholar] [CrossRef]
- Venkitaraman, A.R. Tracing the network connecting brca and fanconi anaemia proteins. Nat. Rev. Cancer 2004, 4, 266–276. [Google Scholar] [CrossRef]
- Wang, A.T.; Smogorzewska, A. SnapShot: Fanconi Anemia and Associated Proteins. Cell 2015, 160, 354–354. [Google Scholar] [CrossRef]
- Wang, X.; Andreassen, P.R.; D’Andrea, A.D. Functional Interaction of Monoubiquitinated FANCD2 and BRCA2/FANCD1 in Chromatin. Mol. Cell. Biol. 2004, 24, 5850–5862. [Google Scholar] [CrossRef]
- Williams, S.A.; Wilson, J.B.; Clark, A.P.; Mitson-Salazar, A.; Tomashevski, A.; Ananth, S.; Glazer, P.M.; Semmes, O.J.; Bale, A.E.; Jones, N.J.; et al. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Hum. Mol. Genet. 2011, 20, 4395–4410. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association. Declaration of Helsinki 2007. 2007. Available online: www.wma.net/e/ethicsunit/helsinki.htm (accessed on 11 February 2021).
- Xu, S.; Zhao, F.; Liang, Z.; Feng, H.; Bao, Y.; Xu, W.; Zhao, C.; Qin, G. Expression of FANCD2 is associated with prognosis in patients with nasopharyngeal carcinoma. Int J Clin Exp Pathol. 2019, 12, 3465–3473. [Google Scholar] [PubMed]
- Xu, J.; Wu, M.; Sun, Y.; Zhao, H.; Wang, Y.; Gao, J. Identifying Dysregulated lncRNA-Associated ceRNA Network Biomarkers in CML Based on Dynamical Network Biomarkers. BioMed Res. Int. 2020, 2020, 5189549. [Google Scholar] [CrossRef] [PubMed]
- Yoshimaru, R.; Minami, Y. Genetic Landscape of Chronic Myeloid Leukemia and a Novel Targeted Drug for Overcoming Resistance. Int. J. Mol. Sci. 2023, 24, 13806. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.E.; Kim, S.-H.; Kong, M.; Kim, H.-R.; Yoon, S.; Kee, K.-M.; Kim, J.A.; Kim, D.H.; Park, S.Y.; Park, J.H.; et al. Targeting FLT3-TAZ signaling to suppress drug resistance in blast phase chronic myeloid leukemia. Mol. Cancer 2023, 22, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Zhao, H.F.; Zhang, Y.L.; Song, Y.P. Zhongguo shi yan xue ye xue za zhi. 2023, 31, 649–653. [Google Scholar] [PubMed]
- Telliam, G.; Desterke, C.; Imeri, J.; M’kacher, R.; Oudrhiri, N.; Balducci, E.; Fontaine-Arnoux, M.; Acloque, H.; Bennaceur-Griscelli, A.; Turhan, A.G. Modeling Global Genomic Instability in Chronic Myeloid Leukemia (CML) Using Patient-Derived Induced Pluripotent Stem Cells (iPSCs). Cancers 2023, 15, 2594. [Google Scholar] [CrossRef] [PubMed]
- Leung, W.; Baxley, R.M.; Traband, E.; Chang, Y.-C.; Rogers, C.B.; Wang, L.; Durrett, W.; Bromley, K.S.; Fiedorowicz, L.; Thakar, T.; et al. FANCD2-dependent mitotic DNA synthesis relies on PCNA K164 ubiquitination. Cell Rep. 2023, 42, 113523–113523. [Google Scholar] [CrossRef]
- Yilmaz, U.; Eskazan, A.E. Moving on from 2013 to 2020 European LeukemiaNet recommendations for treating chronic myeloid leukemia: what has changed over the 7 years? Expert Rev. Hematol. 2020, 13, 1035–1038. [Google Scholar] [CrossRef]
- Valeri, A.; Alonso-Ferrero, M.E.; Río, P.; Pujol, M.R.; Casado, J.A.; Pérez, L.; Jacome, A.; Agirre, X.; Calasanz, M.J.; Hanenberg, H.; et al. Bcr/Abl Interferes with the Fanconi Anemia/BRCA Pathway: Implications in the Chromosomal Instability of Chronic Myeloid Leukemia Cells. PLOS ONE 2010, 5, e15525. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).