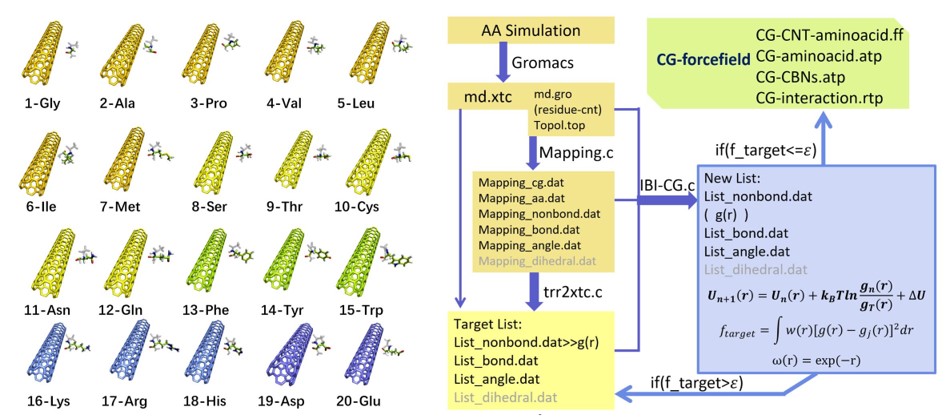
Our work uses Iterative Boltzmann Inversion (IBI) to study the coarse-grained interaction between 20 amino acids and the representative carbon nanotube CNT55L3. IBI is a multi-scale simulation method that has attracted the attention of many researchers in recent years. It can effectively modify the coarse-grained model derived from the Potential of Mean Force (PMF). IBI is based on the distribution result obtained by All-Atom molecular dynamics simulation, that is, the target distribution function, the PMF potential energy is extracted, and then the initial potential energy extracted by the PMF is used to perform simulation iterations using IBI. Our research results have gone through more than 100 iterations, and finally, the distribution obtained by coarse-grained molecular simulation (CGMD) can effectively overlap with the results of all-atom molecular dynamics simulation (AAMD). In addition, our work lays the foundation for the study of force fields for the simulation of the coarse-graining of super-large proteins and other important nanoparticles.