3.1. Numerical verification and the sensitivity profile.
In this section, we will use pairs of multivariate independent variables, obtained from a multinomial distribution with a uniform Dirichlet prior, to investigate and verify numerically the behavior of the uncertainty penalty term obtained in the previous section.
An example of the data obtained from a sequence of
8-variate categorical pairs of independent variables with sample size
is shown in
Table 1. For brevity, we only include the results for 5 pairs (this is sufficiently representative given that the statistical behavior across all pairwise comparisons is summarized in
Table 2 below). The negative sign retained in the table arises due to the effect the penalty terms play in the evaluation of both
where
and
represent the MDL complexity penalty and the uncertainty penalty (obtained in this work), respectively. More importantly, the negative sign is an indicator that the update should be rejected due to near-independence in the case of
, and due to high storage requirements in the case of
. As we shall see further, the update rejection, equivalent to the detection of near-independence within the framework developed in this study, is not guaranteed for all independent variables, at least for the MDL score (see
Table 4).
Table 2 summarizes the statistical behavior across the same sequence of
pairs. Note that the
is constant, while
reacts to the local properties of every pair of variables under consideration, and is a tighter bound for the deviation from independence, given by
.
Table 3 summarizes the results of
pairwise comparisons of 4-variate independent variables and
. Note that the lower category count resulted in a decrease in the deviation from perfect independence, and that this effect is also accounted by the drop in both penalty terms, although at vastly different rates.
Table 4, on the other hand, indicates a failure of MDL to properly detect near-independent pair, as can be seen in the last row of the
column. Here, the pairwise comparisons are carried out for 2-variate independent pairs with sample size
, and MDL misclassifies 920 independent pairs out of
, approximately
.
Table 4.
Statistical summary for independent variable pairs with 2 categories and .
Table 4.
Statistical summary for independent variable pairs with 2 categories and .
| |
|
|
|
|
|
| mean |
0.00050636 |
0.01663084 |
0.00345388 |
-0.01612448 |
-0.00294752 |
| median |
0.00023039 |
0.01696400 |
0.00345388 |
-0.01647059 |
-0.00322349 |
|
0.00071399 |
0.00087238 |
0.00000000 |
0.00112515 |
0.00071399 |
| max |
0.00953119 |
0.01725168 |
0.00345388 |
-0.00693925 |
0.00607731 |
Note that the failure of
to identify independent variables is not mitigated by an order of magnitude increase in the sample size, i.e.
, as can be observed in
Table 5. But the total number of misclassifications of independent pairs falls to 227, which is approximately
. These observations are consistent with the well-known observation that the MDL complexity term tends to under-penalize low category count variable pairs, causing at times severe overfitting in the context of BN recovery from data. Observed sample size dependence in MDL’s ability to classify independent variables correctly, however, fails to explain why
needs to be sensitive to sample size, given the relatively well-behaved, well-represented profile of conditional and joint events of the scenario presented here. Clearly, this undesirable under-penalizing property of MDL complexity term cannot be easily dismissed as the shortcoming of the data (see
Figure 1), particularly given the fact that
depends only on
N and reflects nothing else about the variable pairs in question.
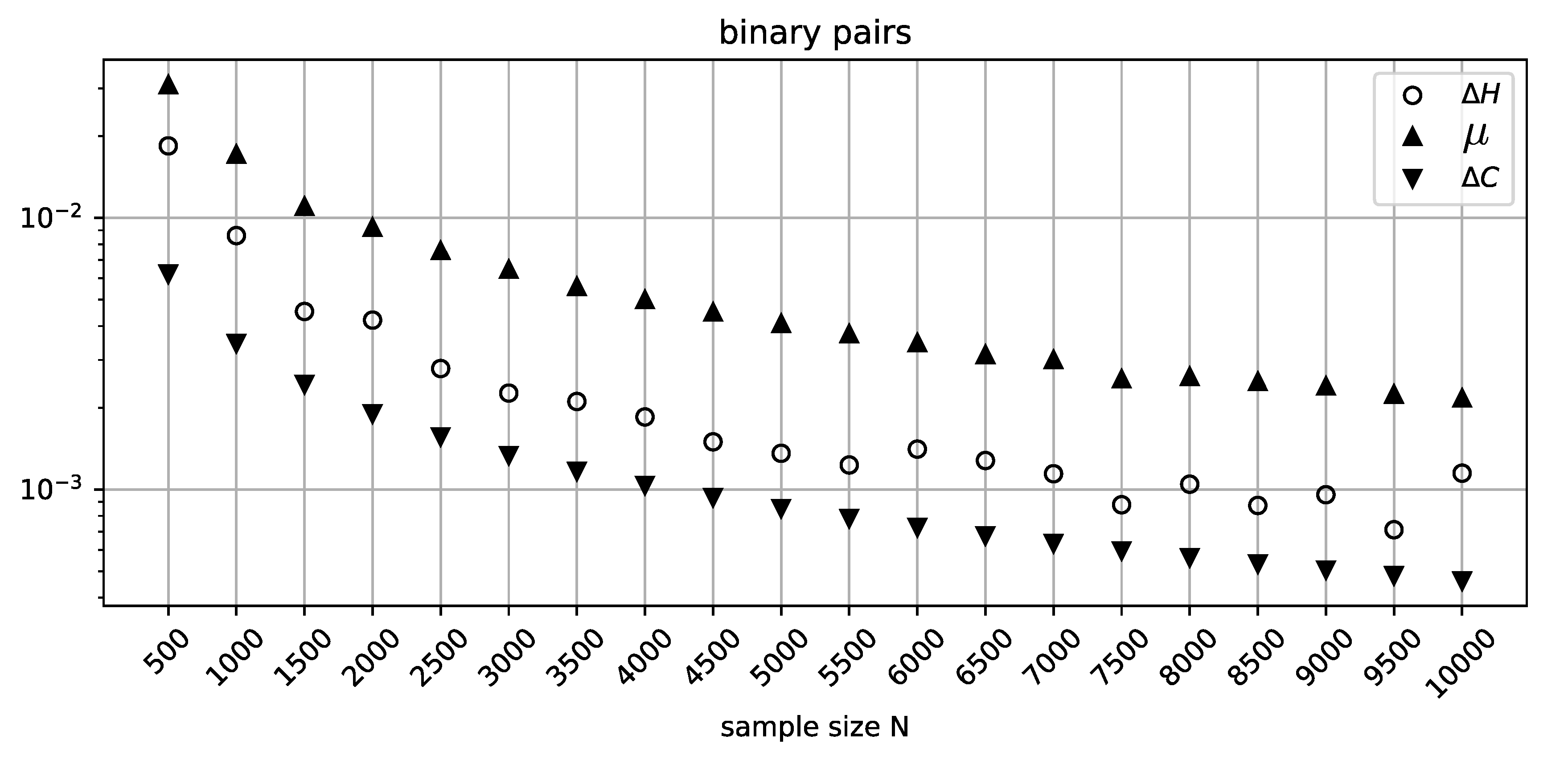
To investigate this misclassification further we consider batches of 10000 randomly generated pairs of binary variables for a range of sample sizes. For every batch, we extract the pair that gives the maximum value of
and evaluate the corresponding values of
and
. These values are then plotted against the increasing sample size in
Figure 1. The MDL penalty term
clearly fails to bound the deviation from independence
across the whole range of sample sizes.
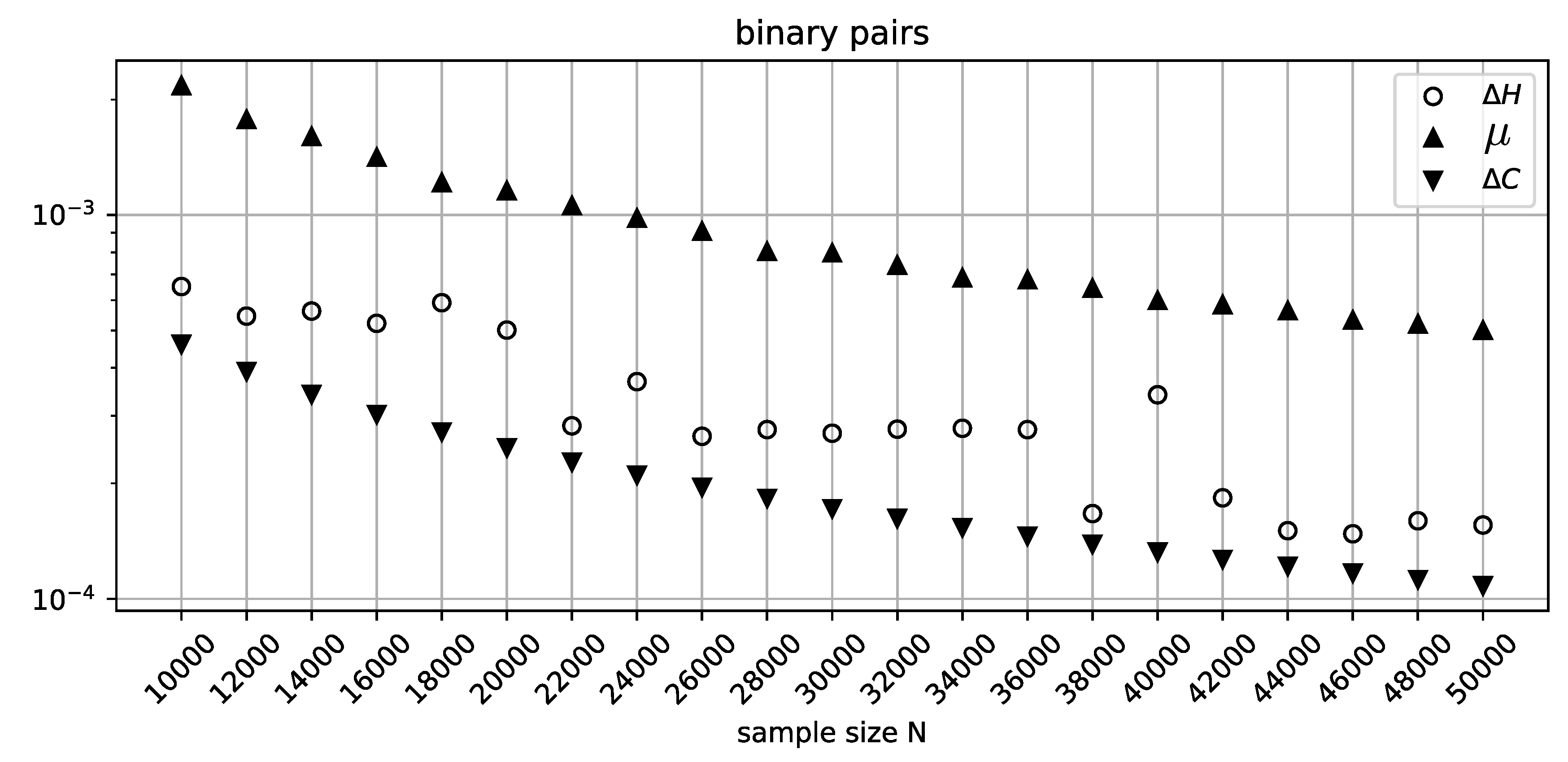
In
Figure 2, the range of sample sizes is extended to 50000 with a coarser increment to show that the misclassification rate of
sees general improvement as
N increases, although at
the MDL penalty term once again fails to identify an independent pair. As expected,
has no trouble in this range of parameters, and its stricter penalization profile is justified by the general volatility exhibited by
.
To continue, we return to our previous setup generating
independent pairs and consider the scenario with 4-variate variables and
.
Table 6 reveals the behavior consistent with the expectations, where both updates identify near-independent pairs equally well. In this range of data parameters the terms are very close in their magnitude, so it is not surprising that the behavior is almost identical.
In
Table 7 (8-variate pairs,
), the MDL penalty term is on average several times greater than
. Both scores, however, perform equally well in this range of parameters, identifying all independent pairs correctly.
Table 8 reveals a misclassification on the part of
, as can be seen in the last row of the
column. Further investigation reveals approximately
of misclassified pairs and sample size dependence of the misclassification rate which is completely resolved by increasing the sample size by one order of magnitude (
Table 9). This observation is fully consistent with the general understanding of the effect that limited sample size may have on conditional or joint events.
In the scenario presented here, a 16-variate random variable can be expected to have unconditional events of the size
. Therefore, any joint event will necessarily be smaller, in the order of the square of the unconditional events due to independence, i.e.
This corresponds to only roughly 40 samples per joint event in the case of , on average. Clearly, for such small probabilities the sample size should be larger to be adequately representative, otherwise the unaccounted-for effects of sampling error may dominate the landscape.
It is not surprising that MDL falters in these circumstances, given how much it over-penalizes . This comes at the cost of specificity, i.e. MDL would clearly fail to classify dependent pairs as such for all values of that would fall between, say, the values and presented in the table.
Table 9 reveals the ability of
to recover its sensitivity under the condition of sufficient sample size. This is to be expected, since for
a joint event will correspond to roughly 400 samples, on average. Note that while the MDL complexity term continues to over-penalize
significantly even when provided data of ample size, the sensitivity of
attains a new degree of refinement.
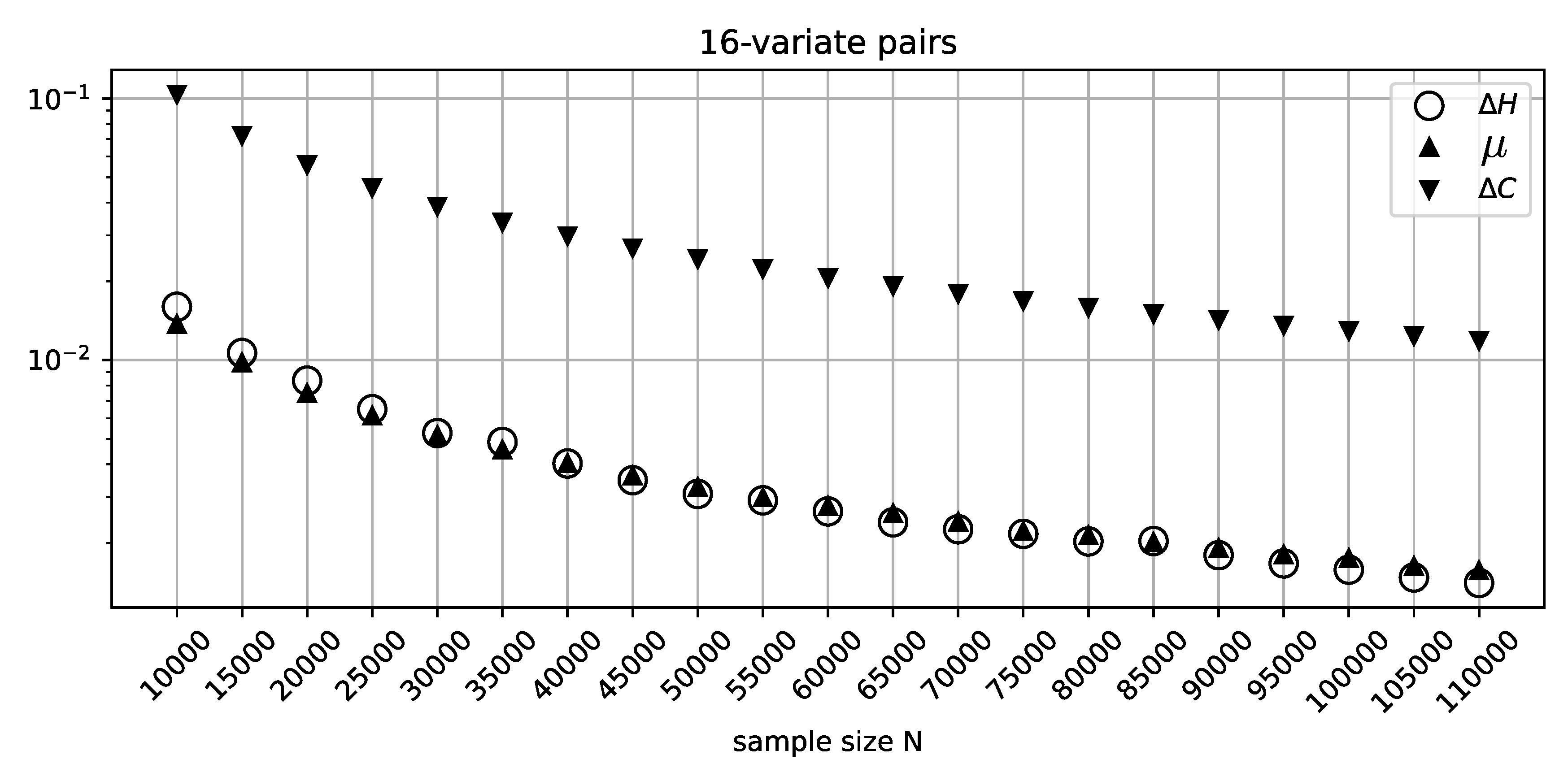
Figure 3 recapitulates the misclassification analysis (that was performed for the binary variables above) for the batches of 10000 random 16-variate independent variable pairs for every value of
N. The figure shows consistently improving classification precision of
, with a somewhat elevated sensitivity profile for smaller sample sizes, as expected due to the unaccounted-for effect of sampling error. On the other hand, the excessive over-penalization imposed by
, clearly visible in this figure, is difficult to justify, given the abundant sample size and very consistent behavior on the part of
.
These results demonstrate that not only does the uncertainty penalty term have an edge in interpretability, but that it is also far more balanced and consistent in its sensitivity profile, a matter of direct relevance to practical performance.
3.2. Application to the structural biology of tRNA molecules: Sample size invariance study.
In our prior work [
17], we used BN modeling (with conventional scoring criteria) to study and dissect the structure of intra-tRNA-molecule residue/position relationships across the three domains of life, with an eye toward identifying informative positions and sections of the tRNA molecule in different tRNA subclasses. In this study, we have recapitulated this analysis with the current version of our BN modeling software BNOmics [
18] using the "standard" MDL, and evaluated the resulting BN across different sample sizes using the standard MDL and the novel criterion, MU. The tRNA sequences and alignment were assembled as in [
17] with slight modifications, amounting to 9378 tRNA sequences.
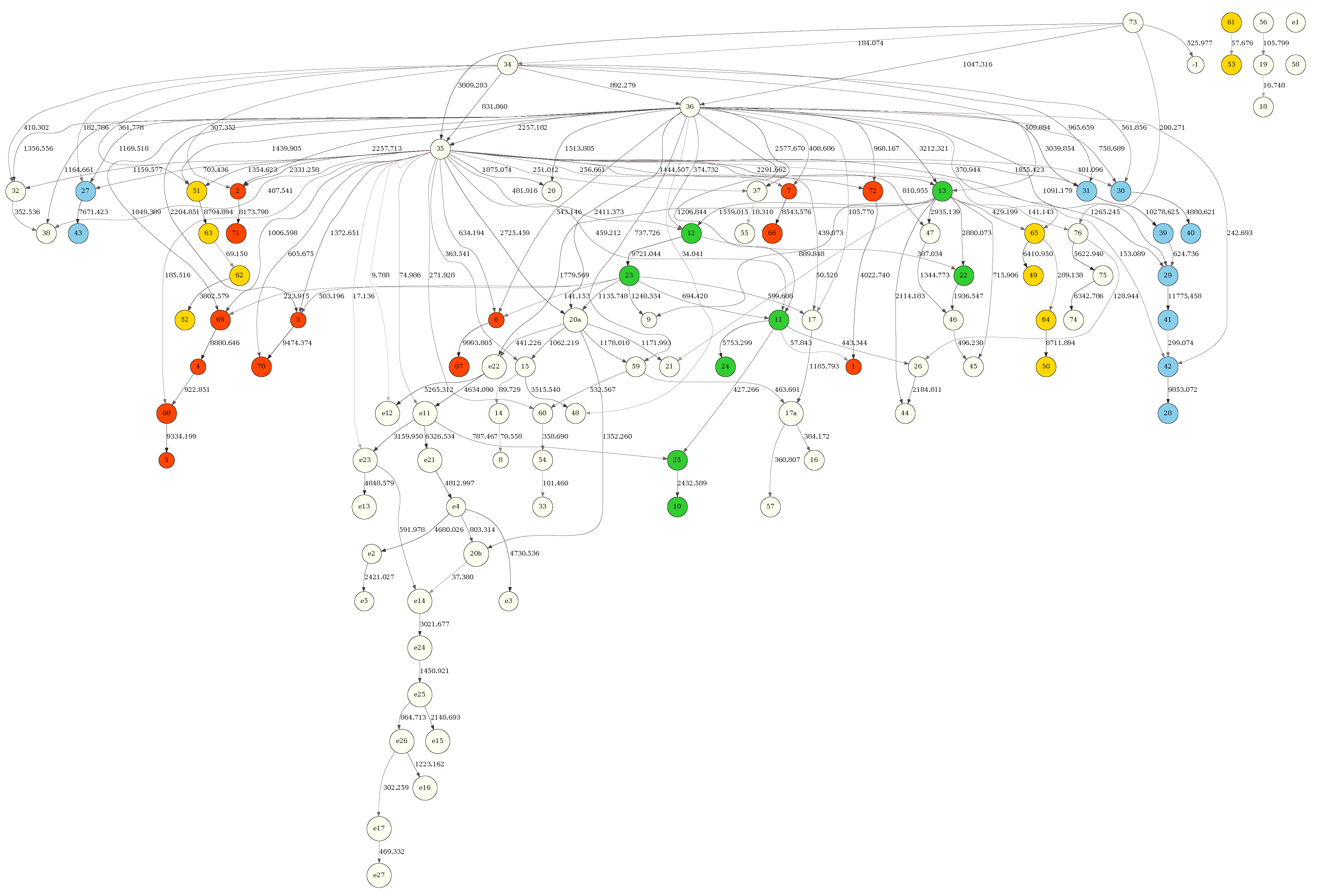
Figure 4 depicts the BN obtained from the full (9378 tRNAs) sample with the MDL criterion. The tRNA residues in the network were colored according to the tRNA molecule structural domains — red (acceptor stem), green (D-arm), blue (anticodon 131 arm) and yellow (T-arm). The tRNA residue positions, shown inside the network node labels, followed the universally accepted tRNA position numbering standard [
22]. Figure S1 depicts the same structure but scored, also with MDL, against a random subsample of 4689 tRNAs (50 percent of the full sample). Figure S2 depicts the same structure scored with the MU criterion against the full (9378 tRNAs) sample. Finally, Figure S3 depicts the same structure scored with the MU criterion against a random subsample of 4689 tRNAs (50 percent of the full sample). It is clear that edge strengths obtained with the MDL scoring criterion strongly depend on sample size (
Figure 4 vs Figure S1), just as expected. In contrast, the edge strengths obtained with the MU score are practically independent of sample size (Figure S2
vs Figure S3).
A "naive" alternative, described above in the introduction, would be to rescale MDL scores by , thereby eliminating sample size dependence in the likelihood term (converting it to conditional entropy). However, this transformation fails to eliminate sample size dependence from the complexity term, demanding a separate interpretation for its contribution. The results of the "naive" rescaling can be observed in Figure S4 and Figure S5 which exhibit significant instability in the strengths of many weaker edges across the two sample sizes. For reference, Figures S6 and S7 show the scores as calculated by alone, i.e. with the penalty term omitted, where we can see a much more predictable and relatively stable behavior across the two considered sample sizes. Notably, a more stable score behavior is demonstrated in Figures S2 and S3, suggesting that the MU penalty derived in this study is in congruence with .
Note that the effect of the penalty term on the score can be one of the primary deciding factors in network configuration preference, and play the role of a termination criterion for seeking dependencies. Since MDL penalizes the arity of ancestor variable bundles, i.e. complexity, one has to account for description efficiency when interrogating MDL score fluctuations. On the contrary, the MU score derived in this study seamlessly integrates its penalty term as an acceptable evaluation uncertainty. This allows the interpretation of score fluctuations directly in terms of satisfiability of the independence criterion.
In summary, the MU principle overcomes the limitations of the MDL principle in that it (i) naturally handles data of varying sample size, (ii) seamlessly integrates an adaptive penalty term commensurate with into edge scores, thereby simplifying interpretation, (iii) implicitly penalizes model complexity with higher regularity.