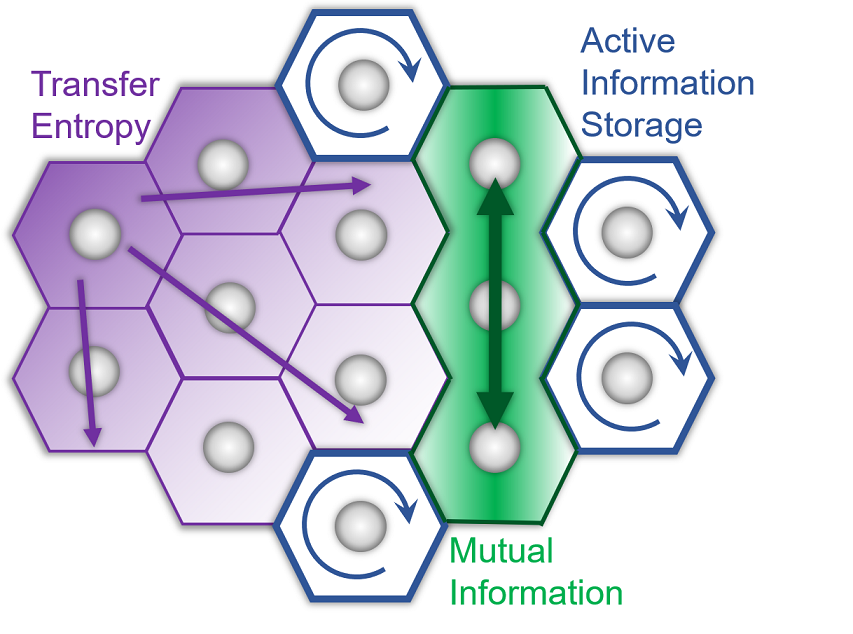
There is a growing appreciation in the fields of Cell and Developmental Biology that cells collectively process information in time and space. While many powerful molecular tools exist to observe biophysical dynamics biologists must find ways to quantitatively understand these phenomena at the systems level. Here, we present a guide for application of well-established information theory metrics to biological datasets and explain these metrics using examples from cell, developmental and regenerative biology. We introduce a novel computational tool (CAIM) for simple, rigorous application of these metrics to timeseries datasets. Finally, we use CAIM to study calcium and cytoskeletal Actin information flow patterns between Xenopus laevis embryonic animal cap stem cells. The tools that we present here will enable biologists to apply information theory to develop systems level understanding of a diverse array of experimental systems.