Innovations and Achievements of the Magnified ROC (mROC) and the AUC-mROC
The adjustment of the AUC-ROC for early retrieval evaluation can be thought as an engineering problem, hence there is not a unique way to solve it, and different designing principles can be followed. In this study we aim to propose a solution that, following the legacy of Clark et al. [
11], embraces a designing strategy inspired by simplicity. This means that our approach will not require elaborated choices of transformation functions and parameters. Furthermore, we aim to progress current knowledge by addressing the two main limitations discussed above about the previous adjusted ROC solutions: (i) we wish that in the proposed mROC plot the random predictor is always a straight diagonal line y=x between the points (0,0) and (1,1); (ii) as a consequence of the first point, we wish that the AUC-mROC of a random predictor is always equal to 0.5.
We define the un-normalized magnified TPR (umTPR) and un-normalized magnified FPR (umFPR) at each
, where
is the number of samples, as:
The un-normalized mROC curve is composed of the points at coordinates
for
. The un-normalized AUC-mROC is obtained by computing the area under the un-normalized mROC curve (for example using the trapezoidal rule), which is between 0 and 1. We borrow from Clark et al. [
11] the basic rationale to adopt the logarithm function to decrease the influence of late hits in adjusting the AUC-ROC, which is a solution previously supported by Järvelin et al. in the NDCG measure [
32,
33]. However, the main difference is that Clark et al. [
11] apply a fixed log10 transformation directly and only to the FPR (in practice a semilogarithmic ROC plot), whereas we apply an adaptive logarithm-based transformation to both TP and FP and, in our case, the attenuation of late hits is varying with the number of P and N respectively. This means that in our adaptive logarithm-based adjustment if P << N (as in unbalanced early retrieval problems) then the attenuation of the logarithm function on FP will be stronger than on TP, and viceversa. This adaptive mechanism is fundamental to automatically adjust the ROC curve to diverse unbalanced prediction evaluation scenarios such as P << N or P >> N. In addition, in the next section of the study we will provide computational evidences that the tactic to apply the transformation to both TP and FP is necessary for an appropriate evaluation of the random predictor performance when the number of samples grows (compare
Figure 3b,d).
For the random predictor, the umTPR and umFPR at each
can be computed analytically:
Therefore, the un-normalized mROC curve of the random predictor (
Figure 3c, grey dashed line) is composed of the points at coordinates
for
. Differently from AUC-ROC, the un-normalized AUC-mROC of the random predictor is not 0.5 and, as for the CROC framework proposed by Swamidass et al. [
12], it is dependent on the proportion of positive and negative samples in the dataset (see
Figure 3d). For this reason, we propose the final mROC curve with a normalization such that the random predictor curve follows the bisector line, and the associated AUC-mROC for the random predictor is 0.5. The procedure is as follows.
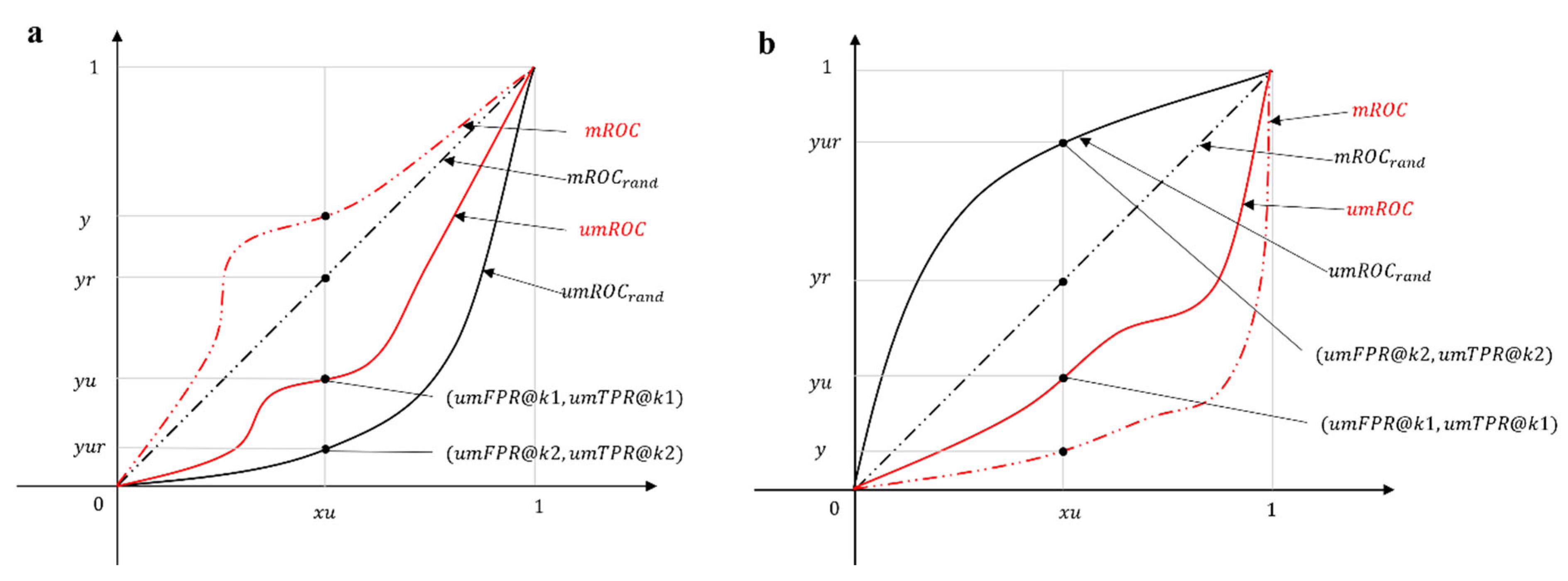
For each point
of a predictor’s curve in the un-normalized mROC plot, we define the respective point
of the random predictor’s curve. The crucial concept to understand is that the same
value on x-axis of the ROC plot can be achieved at two different k values: k1 for the predictor and k2 for the random predictor. Since we already know that
, our goal is to analytically compute (by using the equations reported some lines above) the value of
as a function of
:
. This is achievable by combining the following two equations:
From which it is simple to derive that:
The goal of the normalization is to map these curves in a new mROC plot where the random predictor curve - as for the classical ROC curve - is the bisector of the first quadrant with coordinates
and the new coordinates of the predictor are
. To achieve this, we consider two cases separately, as illustrated in
Figure 1.
The first case is visualized in
Figure 1a, where
. The un-normalized mROC curve of the actual predictor lies below the corresponding curve of the random predictor. In order to implement this mapping to the new coordinates, we perform a rescaling such that:
The rescaling preserves the ratio of the y-axis coordinates of the two predictors with respect to the upper bound (
), ensuring that
always holds. The second case, where
, as visualized in
Figure 1b, requires a different rescaling such that:
Similarly, the rescaling preserves the ratio of the y-axis coordinates of the two predictors with respect to the lower bound (
), ensuring that
always holds. Finally, by considering both cases, we arrive to the unified formula
in which
is the Heaviside step function. This implies that, since
:
And, by substituting all the terms with their actual values, we obtain the final formula:
The normalization is only applied to the y-axis, so that .
From Unbalanced to Balanced: A Generalized ROC (gROC) and AUC-gROC
Although the link prediction problem, like many other early retrieval problems, is often characterized by extremely unbalanced classes within the dataset, a border range of early retrieval problems encompass all possible proportions of positive and negative samples.
From the previous analysis, we know that mROC applies the same logarithm transformation on the both axes, regardless of the ratio of P and N. It defaults to the presumption that
, such that the top-most region of the rankings is attached with more importance, while the other parts, such as the middle and bottom, are considered less relevant. This behavior is not desired for a meticulous control of the evaluation in face of less unbalanced or balanced datasets. Thus, we propose to use the mROC for extremely unbalanced dataset with few positives, while switching back to the standard ROC for balanced and inversely unbalanced cases. To allow for the automatic transition from unbalanced to balanced scenario, we proposed the generalized ROC (gROC) as follows:
Indeed, gROC is a proportional mixing of mROC and ROC, such that when it is equal to mROC and when it is equal to ROC. One crucial merit of this adjustment is that the curve of a random predictor is again the bisector of the first quadrant, and the AUC-gROC of the random predictor remains at 0.5.
We claim that gROC is a class-proportion-aware metric that is apt for evaluating all types of classification and early retrieval problems, including link prediction. In the following sections, we will provide more computational evidence to demonstrate the adaptability of gROC.
Computational Experiments to Assess the Validity of AUC-gROC and AUC-mROC
As we briefly sketched in the introduction, in 2007 Truchon and Bayly [
13] proposed a ‘quintessential’ argument about the inadequateness of AUC-ROC measure for evaluation of early recognition problems. They hypothesized three basic cases in which, regardless of their fundamental differences, the AUC-ROC is equal to 1/2. The first is that half of the items are retrieved at the very beginning of the rank-ordered list and the other half at the end; the second is that the items are randomly distributed all across the ranks; and the third is that all of the items are retrieved in the middle of the list. Truchon and Bayly noted that, in terms of the ‘early recognition’, the first case is clearly better than second one, which is also significantly better than the third one.
We analysed the results of a large-scale experimental study [
18,
19] on link prediction, which reports AUC-ROC and AUC-PR evaluations of two landmark link prediction methods - Cannistraci-Hebb adaptive network automata (CHA) and stochastic block model (SBM) - tested over 5500 simulations (550 networks x 10 repetitions). One of the interesting finding is that in 31% cases CHA has higher AUC-PR and SBM has higher AUC-ROC. In our opinion, this incongruency represents a new crucial scenario to investigate for improving our understanding about the inadequateness of AUC-ROC in evaluation of early recognition.
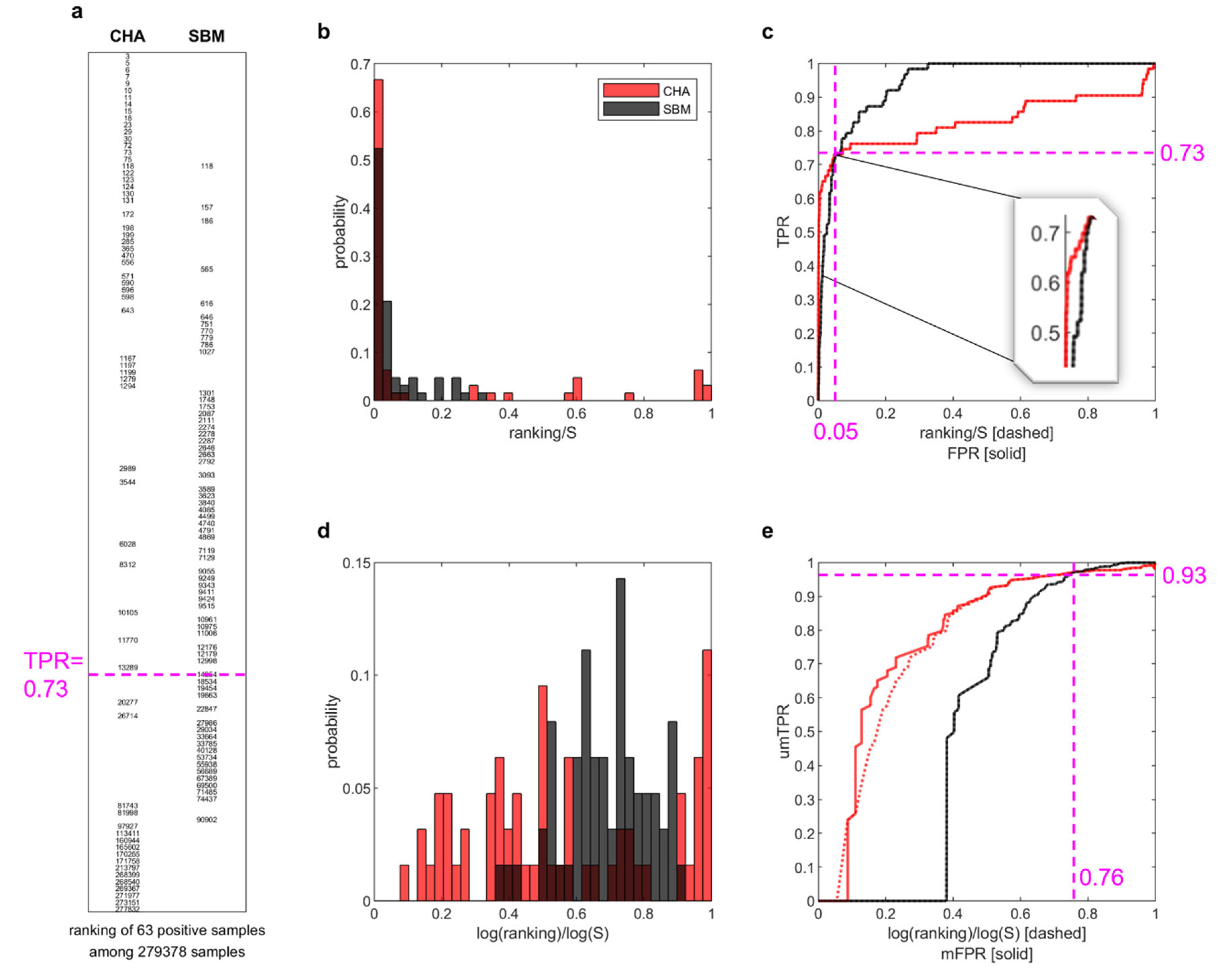
To this aim, (
Figure 2 reports a paradigmatic example to investigate the AUC-ROC problem in this novel scenario. Specifically,
Figure 2a displays, for both CHA and SBM, a table with the ranking positions of the 63 positive samples (i.e. the links removed in one of the tested simulations) in the ranking of 279378 non-observed links. For the first 46/63 positive samples retrieved both by CHA and SBM (i.e. up to recall 0.73, see items above magenta dashed line in
Figure 2a), the ranks assigned by CHA are overall much lower than the ones assigned by SBM. Instead, the last 17/63 retrieved positive samples are better ranked by SBM (see items below the magenta dashed line in
Figure 2a). Looking at this table, it is visually evident that CHA is performing better than SBM at top-ranking relevant links. This emerges, although not patently, also in
Figure 2b that reports the probability of an item to occur at the different levels (ratios) of the ranking. Indeed, in this plot CHA achieves probability close to 0.7 to rank items at the very beginning, whereas SBM achieves probability slightly above 0.5. Nevertheless, the same plot might be visually misleading because the top-ranking zone is compressed very close to the y-axis and a matching with the information reported in the table of
Figure 2a is not visually evident. This issue is solved in
Figure 2d where the proposed adaptive logarithmic magnification of the x-axis is applied. Indeed,
Figure 2d is matching the same visual information of
Figure 2a, and the supremacy of CHA on SBM in top-ranking relevant links is evident.
Figure 2c displays the ROC plot associated to this example and here also, as for
Figure 2a, the relevant early retrieval information is visually hidden because it is compressed very close to the y-axis. However, the fact that CHA is better then SBM in early retrieval is emerging, although not patently, also in ROC plot of
Figure 2c. Indeed, if we give a closer look at the area before the crossing point (the point in which the ROCs of CHA and SBM cross each other), we can notice that (see inset in
Figure 2c) the ROC of CHA clearly dominates the one of SBM. This means that in the ROC plot there is a clear early retrieval information about the fact that CHA is able to top-rank relevant links better than SBM with a 0.73 sensitivity (also termed: recall or true positive rate) and with a false positive rate of 0.05, which is low with respect to the sensitivity achieved. Practically CHA is better than SBM for the top ~15000 links of the ranking, which is the most relevant part at the application level, but this relevant part is constrained within a very small area, because the top 15000 links are only ~5% of the whole ranking (and correspond to FPR ~0.05), and ROC is giving similar importance to the entire ranking. In conclusion SBM gets a higher AUC-ROC because it performs better in the zone of the ranking that is less relevant from the application point of view. This information is not visually conveyed in the ROC plot that is misleading when we refer to the area under the curve (AUC-ROC). Indeed, the number of positive links (P = 63) is small in comparison to the non-observed ones (N = 279378 - 63), therefore the AUC-ROC ‘tells’ us more about the most abundant items that are the N, and visually neglects (confining in a small early area, see inset in
Figure 2c) the information on the positive items that are in reality the one to which we are interested. Consequently, if used to evaluate early retrieval, in this scenario the AUC-ROC provides the misleading information that the performance of CHA (AUC-ROC = 0.83) is worse than SBM (AUC-ROC = 0.94).
Figure 2e shows that, by applying the log adaptive magnification (proposed in this study) to both axes of the ROC plot, we can adjust the ROC plot in a way that is matching the same visual information of
Figure 2a, and the supremacy of CHA on SBM in top-ranking relevant links is evident. Indeed, now the crossing point is moved to umTPR = 0.93 and mFPR = 0.76, and the merit of this adjustment is the adaptive mechanism that automatically tunes the logarithm function with respect to the number of items considered on the respective axis. However, this adjustment does not guarantee that the random predictor is always associated to a ROC that is the bisector of the first quadrant (
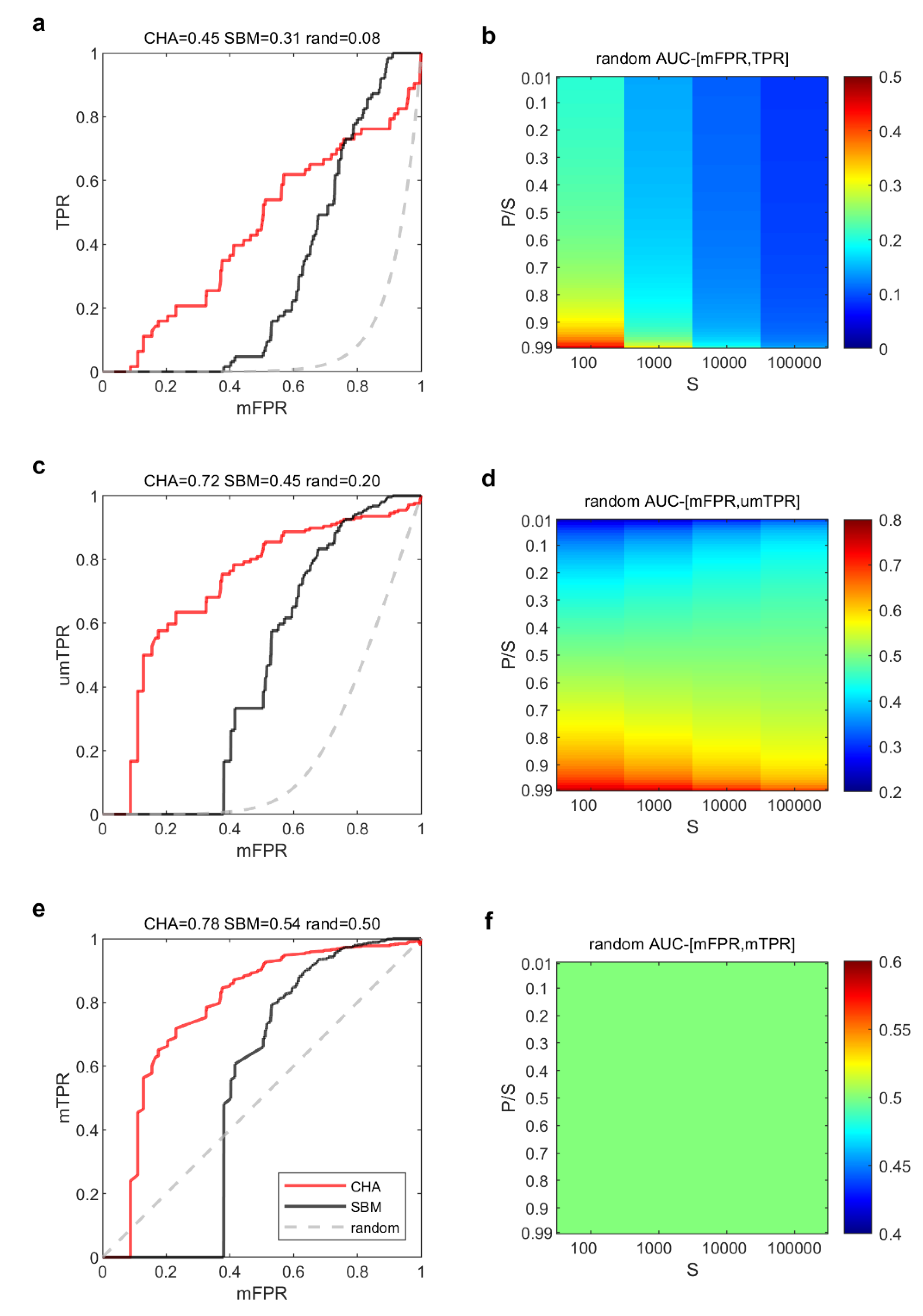
) and whose AUC-ROC is 0.5. This is a fundamental feature of the ROC theory and, in this study, we achieve two main findings. The first is that the log adaptive magnification of the ROC x-axis only (
Figure 3a) is not enough to ensure that the AUC-ROC of the random predictor behaves symmetrically when the ration of P/N is varying, and the number of samples is growing (
Figure 3b). Therefore, the log adaptive magnification of both the ROC axes is necessary (
Figure 3c) in order to guarantee that this symmetry is obtained (
Figure 3d). The second achievement is a consequence of the first finding, indeed once we fixed the issue of being able to analytically (see the previous section for the mathematical formula) compute the performance of the random predictor in our magnified ROC plot, now the last obstacle is to design a mathematical normalization that adjust the magnified ROC plot in a way that the random predictor has always AUC-mROC = 0.5. In the previous section we derived a mathematical theory to design this normalization. Consequently, in
Figure 3e we show that the result of this normalization is able to effectively achieve our aim to provide a magnified ROC plot that respects the basic ROC theory according to which the random predictor has a mROC that follows the bisector of the first quadrant and the AUC-mROC = 0.5. Meanwhile, the magnified ROC plot in Figure3e is able to match the same visual information of
Figure 2a, and the supremacy of CHA on SBM in top-ranking relevant links is explained also in terms of AUC-mROC. Indeed, the AUC-mROC provides the appropriate information that the performance of CHA (AUC-mROC = 0.78) is better than SBM (AUC-mROC = 0.54).
In all these scenarios we compared the evaluations of AUC-ROC with AUC-mROC, AUC-gROC and other baseline early retrieval evaluation measures that we commented in the introduction: precision, AUC-precision, AUC-PR, NDCG. Furthermore, we included in the comparison also the MCC as sanity check that AUC-ROC evaluation is misleading. MCC is a binary classification rate that generates a high score only if the binary predictor is able to correctly predict most of positive data instances and most of negative data instances [
25,
26]. Differently from AUC-ROC, MCC provides a fair estimate of the predictor performance in class unbalanced datasets such as the one in link prediction problem. However, differently from the other early retrieval evaluation measures, MCC does not attribute more importance to the positive class and it fairly and balanced considers the position in the ranking of positive and negative (in our case nonobserved links) instances. In
Figure 4, on the left side, each plot reports the probability (both for CHA and SBM separately) of a positive link to occur at the different levels (ratios) of the ranking, the x-axis is transformed according to the proposed adaptive logarithm magnification function. On the right, each plot reports the performance of CHA and SBM according to the different evaluation measures.
The first scenario is the one commented till now, indeed
Figure 4a coincides with the
Figure 2d. We already commented above this result explaining that the AUC-ROC provides a misleading evaluation of the early retrieval performance of CHA with respect to SBM (indicating that SBM is better than CHA) because of two reasons. Firstly, in an unbalanced scenario, it gives more importance to the most abundant class that in this case is not the one of interest for the evaluation of the prediction. Secondly, according to its definition the mistakes at the bottom ranking are equally relevant as the correct predictions at the top ranking, which is not matching the purpose of link prediction in real applications. Looking at the full ranking, CHA ranks most of the positives (> 60% positives) in the top 2%, while SBM < 40% positives. In particular, with a zoom in the top 1%, CHA ranks around 16% positives in the top 0.02%, while SBM only 0.8% positives. Therefore, the performance of CHA is remarkably better than SBM at top-ranking positive links. If we look at the bottom-50% ranking, we can notice that CHA positions around 12% positives in the second half of the ranking, while SBM only 0.1%. From an application perspective, having more positives in the top-ranking at the expense of more mistakes in the bottom ranking, is much more valuable than having few positives both in the top and bottom ranking, since often for practical usage only a small fraction of the top predictions is considered, while the bottom predictions are rarely assessed. Therefore, in this scenario we would assess that CHA provides better link recommendations than SBM. This is now confirmed by the fact that in
Figure 4b both AUC-mROC and all the other early retrieval measures agrees that CHA outperforms SBM in link prediction. Most importantly, even MCC - that is designed as AUC-ROC to be a binary classification rate and not an early retrieval measure - disagrees with AUC-ROC and clearly agree with the other early retrieval measures, offering an incontrovertible evidence that AUC-ROC is unreliable for evaluation of this link prediction scenario. MCC, as AUC-ROC, neglects the early retrieval nature of the problem but differently from AUC-ROC is able to adjust for the class unbalance.
The second scenario is commented in
Figure 4c. CHA ranks almost all the positives (99%) in the top 1%. SBM ranks 50% of the positives in the top 1%, with 95% of the positives within the top 10% and only few positives in the bottom ranking. In this scenario, we would argue that the competition between CHA and SBM for link recommendation has a clear winner in CHA. However, looking at the value of the performance measures in
Figure 3d, we can notice that AUC-ROC does not highlight the difference and it is very close for both methods (~1.00 vs 0.97), simply because both methods are good enough at not making many mistakes at the bottom ranking, which is however not the main goal of the application. Therefore, we believe that also in this scenario AUC-ROC provides a misleading assessment. This is confirmed by the results in
Figure 4d, where all the other measures including AUC-mROC and MCC highlight a significant performance gap in favour of CHA with respect to SBM.
The third scenario is commented in
Figure 4e. CHA is better than SBM in the top ranking, and both methods are equally bad at making some mistakes at the bottom. Therefore, in a similar scenario - as already investigated by Tao Zhou in a recent study [
20] – AUC-ROC agrees with all other measures (see
Figure 4f) on the fact that CHA is consistently better than SBM in link prediction. In all three scenarios, the evaluation given by AUC-gROC is almost identical to that given by AUC-mROC due to
.
The above analysis showcases the aptness of mROC and AUC-mROC for unbalanced early retrieval problem, in which the objective is to retrieve a few most relevant samples among the vast majority of irrelevant ones. Moving towards a boarder range of problems, we may encounter cases where the proportion of the positive class is approximately equal to or even higher than that of the negative class. We investigate deeper into the scenarios with various class proportions to provide stronger computational evidence in support of the proposed measure.
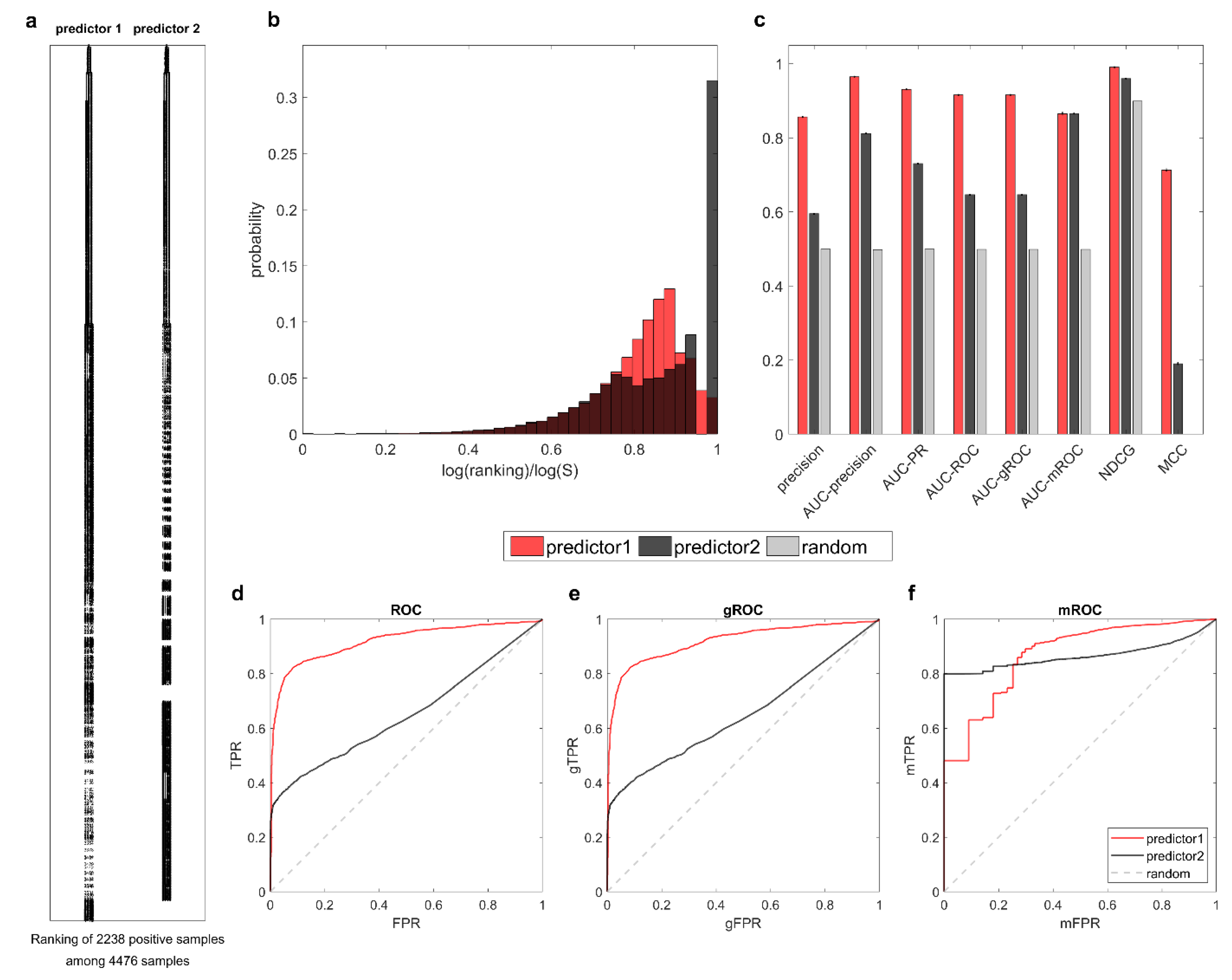
We fist consider the balanced class scenario with
In
Figure 5, the results of predictor1 and predictor2 on the balanced class dataset are plotted, both results are generated artificially to simulate actual cases.
Figure 5a displays for each predictor the ranking positions of the 2238 positive samples among the rankings of 4476 samples, and
Figure 5b reports the probability distribution of the positive samples for the two predictors, in adaptive logarithm magnification scale. Both two predictors correctly rank the positive samples in the top-ranking region. However, predictor1 places more positives in the middle of the rankings than predictor2, and put a few positives at the very bottom. Its visually evident that predictor1 performs better than predictor2 in ranking the most relevant samples, and this is confirmed by all the measures in
Figure 5c except for AUC-mROC. This discrepancy occurs because AUC-mROC always focuses on the top-ranking regions without considering the proportion of the positive class, leading to a misleading evaluation that suggests both predictors have similar performance. This issue is also confirmed in
Figure 5f, which shows that the mTPR of predictor2 rapidly raises to 0.8 at a very low mFPR. The issue is properly resolved by gROC and AUC-gROC. As shown in
Figure 5c and
Figure 5e, the evaluation given by gROC and AUC-gROC is essentially the same as that given by ROC, which is desired for this scenario.
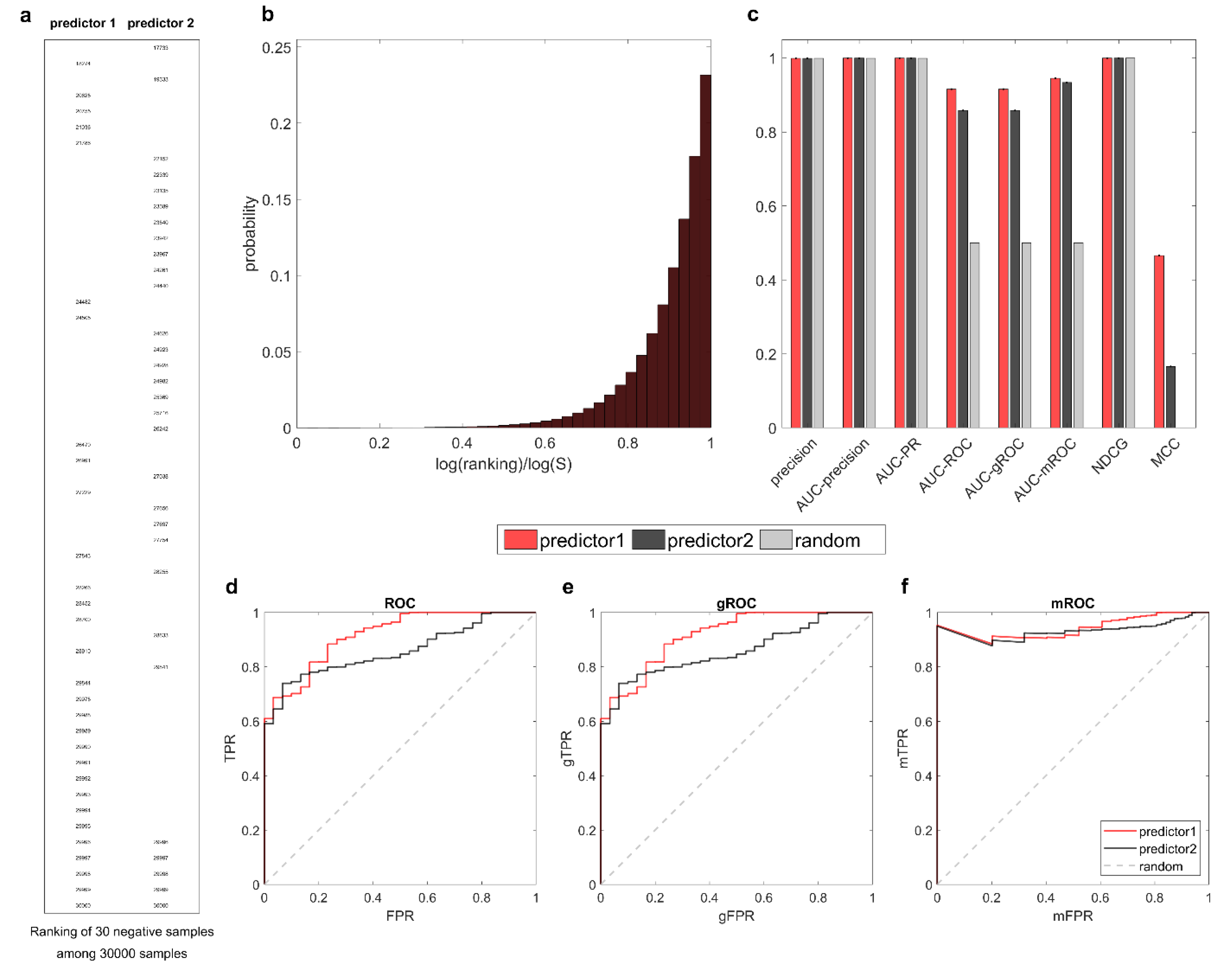
Next, we consider the inversely unbalanced scenario with
Again, we manipulate the results of two predictors on a dataset with 30 negative samples among 30,000 samples. In
Figure 6, the two predictors mistakenly recognize negatives as positives and place them at the top of the rankings. The rankings of negative samples for the two predictors are displayed in
Figure 6a. It is visually evident that predictor1 places fewer negatives at the top than predictor2, and therefore, it performs better than predictor2 under the early retrieval framework. Indeed, this conclusion is agreed upon by all three ROC-based measures: AUC-ROC, AUC-mROC, and AUC-gROC. Note that in this case, AUC-mROC exhibits a stronger discrimination capability owing to its magnification mechanism, but overall, all three measures provide a correct evaluation.
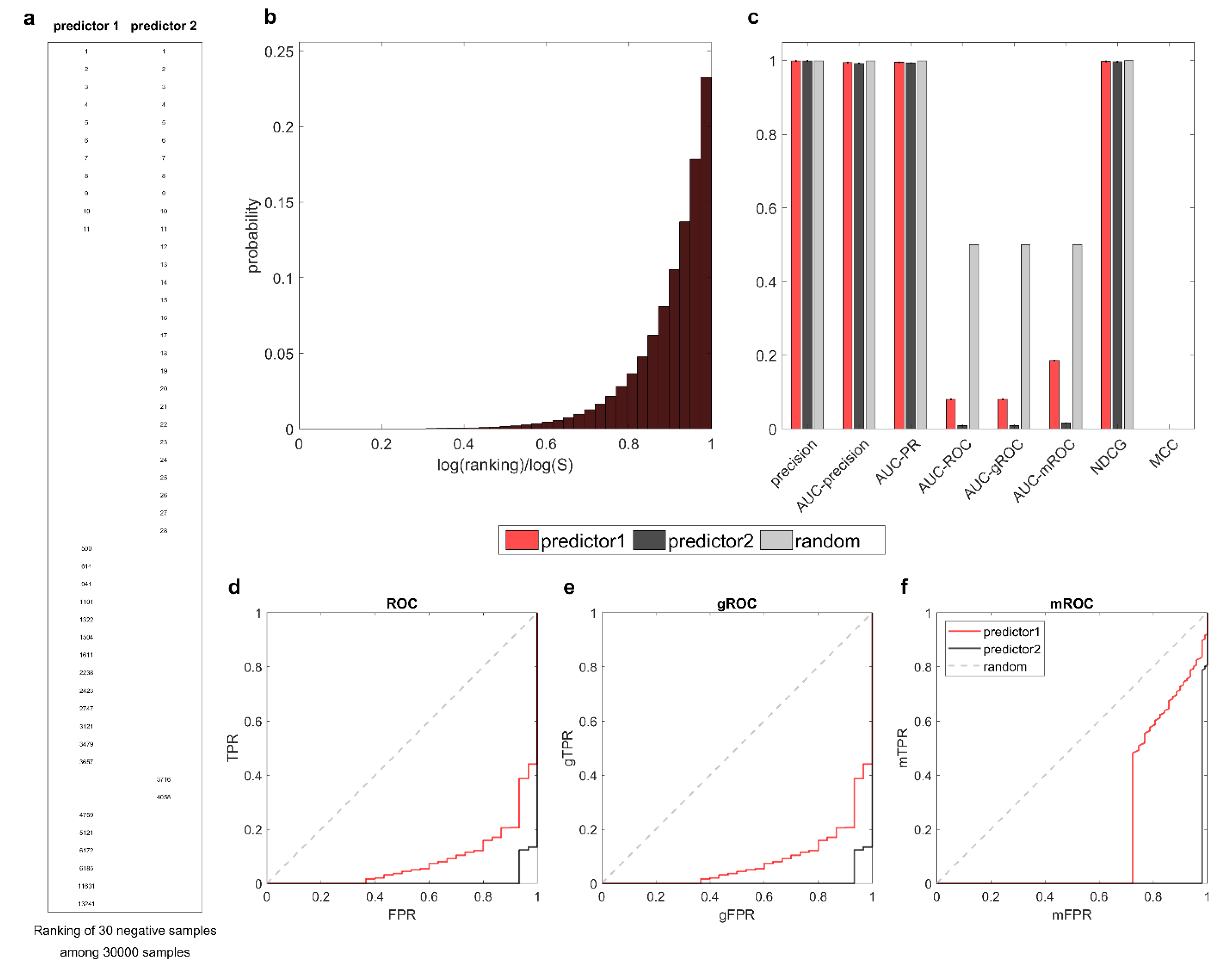
Nevertheless, for a comprehensive study, we provide in
Figure 7 another example of the inversely unbalanced scenario. Analogous to the derivation in
Figure 6, this time the two predictors correctly recognize most of the positives and place the few negatives at the bottom. As observed in
Figure 7a, predictor1 is slightly better than predictor2 by assigning higher rankings to the negatives. Subsequently, in
Figure 7c, both AUC-ROC and AUC-gROC demonstrate the superiority of predictor1, while AUC-mROC provides relatively weak differentiation. Although for the early retrieval problem, the rankings at the bottom are not as important as those at the top, this example shows that even in the worst scenario, AUC-mROC does not ignore the bottom rankings. Finally, through the computational evidence obtained in various scenarios, it is undoubtedly evident that AUC-gROC is a generalized evaluation measure suitable for early retrieval problems with arbitrary class proportions. It is elegant, parameter-free, and highly interpretable.
Table 1 emphasizes that AUC-mROC and AUC-gROC are among the measures with highest correlation to AUC-ROC, along with NDCG. This offers evidence that the AUC-mROC is an appropriate adjustment of the original AUC-ROC, and that AUC-gROC is equivalent to AUC-mROC in unbalanced scenarios. Meanwhile, AUC-mROC and AUC-gROC exhibit the highest mean correlation (Spearman correlation coefficient of 0.915) with the other measures. This evidence suggests that they are the most suitable evaluators for this task, as they demonstrate centrality within the ensemble of all evaluators. Their strong association with all the other measures makes them representative of the entire set.
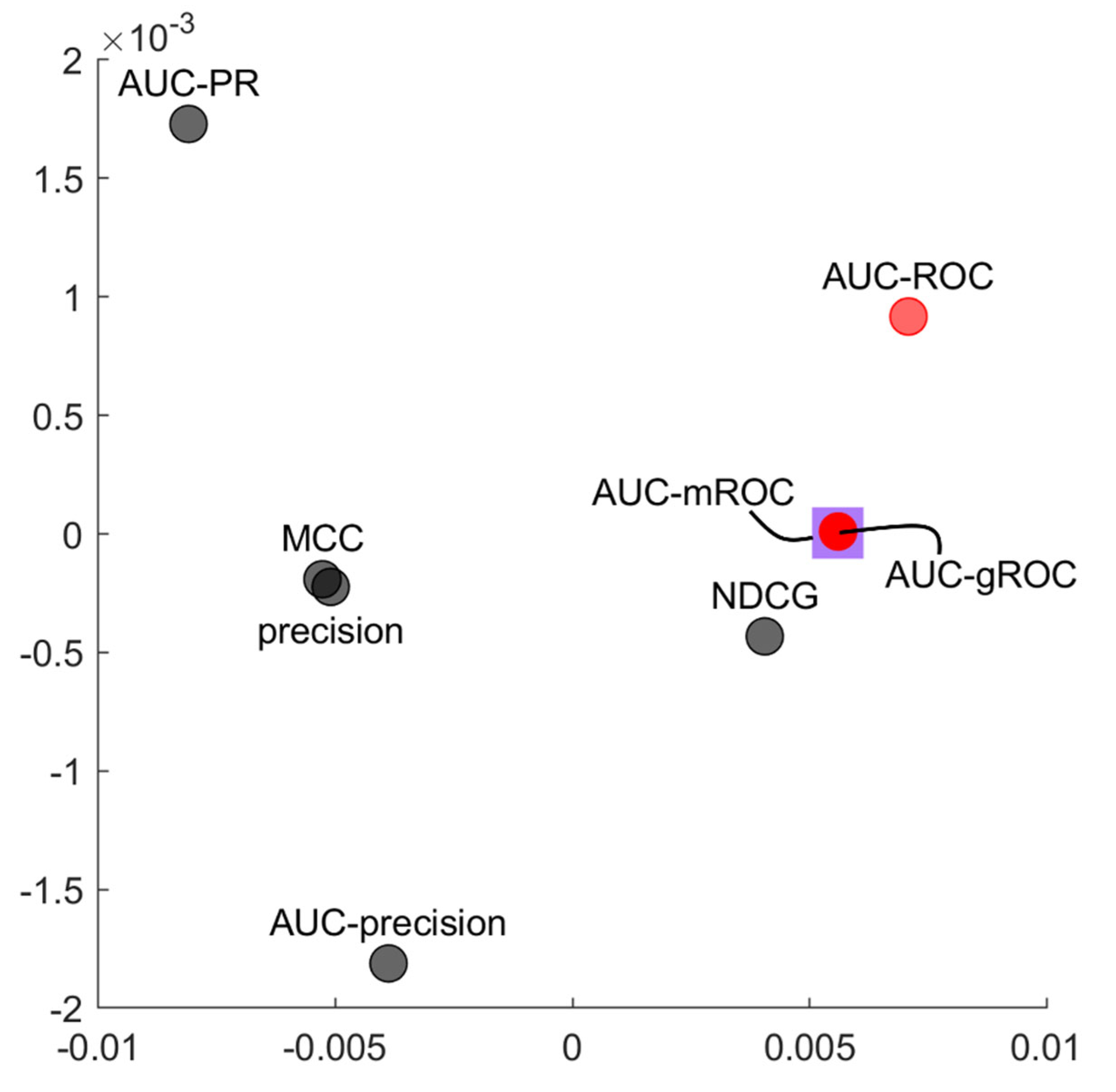
Figure 8 displays an unsupervised multidimensional analysis by means of the principal component analysis of the evaluation measures performance across all networks and link predictors (the details on the way we implemented the analysis are provided in figure legend). This knowledge representation analysis maps in a two-dimensional reduced space the relation of similarities that arise in the multidimensional space between the evaluation measures. Hence, measures that provide a similar evaluation trend (across the networks and link predictors) tend to cluster together in a similar geometrical region of the two-dimensional representation space.
AUC-ROC and AUC-gROC appears very close in the same geometrical neighbourhood and this is a further evidence (a confirmation) that, as we noted in the previous correlation analysis, the AUC-gROC is an appropriate adjustment of the original AUC-ROC. Moreover, since the evaluated networks are mostly composed of , AUC-gROC completely overlaps with AUC-mROC, indicating their high equivalence in this scenario.