Submitted:
20 February 2023
Posted:
21 February 2023
Read the latest preprint version here
Abstract
Keywords:
1. Introduction
1.1. Interferon Introduction
- (i).
- Overview
- (ii).
- Interferons Context
1.2. Epidemiological context
2. Methodology
3. Discussion
3.1. Important highlights of innate immune evasion
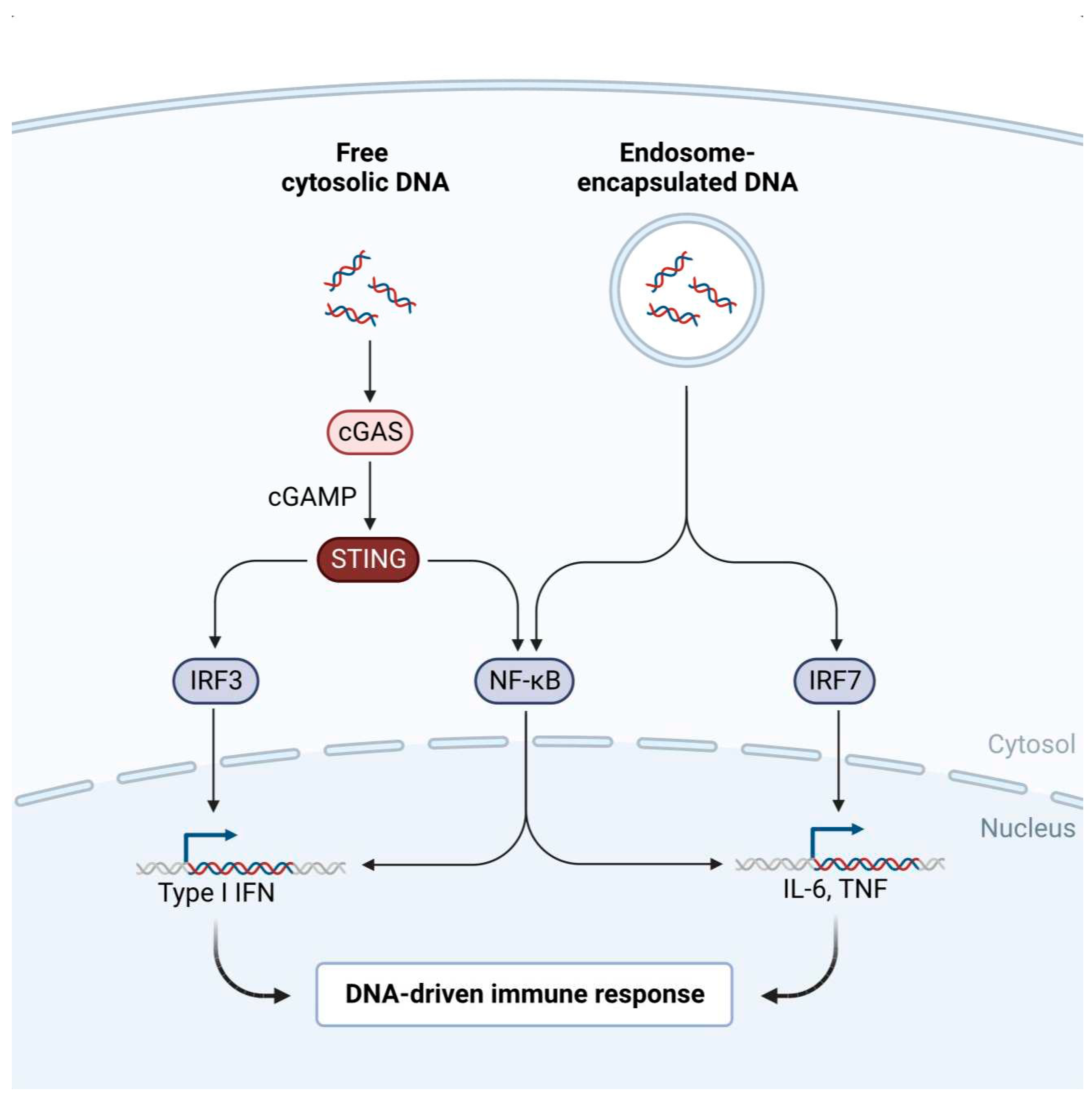
3.2. SARS-CoV-2 Mechanism of Cellular Action in Disease
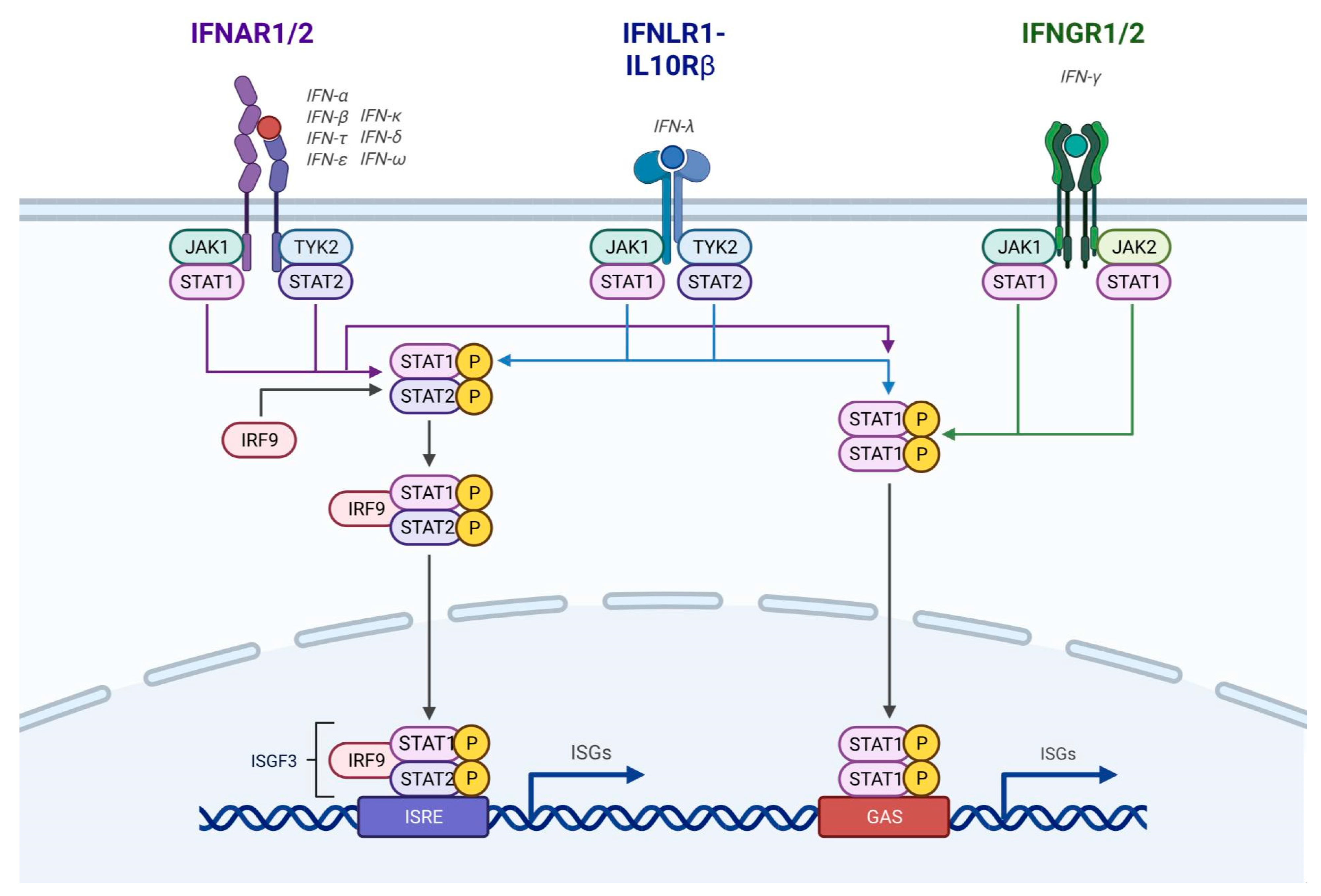
3.3. Cellular Mechanism of IFN Action in Disease
4. Conclusion
Funding
Informed Consent Statement
Data Availability Statement
List of Abbreviations
| SARS-CoV-2 | Severe Acute Respiratory Syndrome-Coronavirus 2 |
| COVID-19 | Coronavirus Infectious Disease 2019 |
| ECM | Extracellular Matrix |
| ACE2 | Angiotensin Convertase Enzyme 2 |
| IAV | Influenza A Virus |
| RSV | Respiratory Syncytial Virus |
| ICM | Intracellular Matrix |
| ORF | Open Reading Frame |
| NSP | Non-Structural Protein |
| IFN | Interferon |
| PAMP | Pathogen-Associated Molecular Pattern |
| PRR | Pattern Recognition Receptor |
| TLR | Toll-Like Receptor |
| MDA-5 | Melanoma-Differentiation Associated protein 5 |
| RIG-I | Retinoic acid-Inducible Gene I |
| ADAR | Adenosine Deaminase Acting on RNA / RNA-specific Adenosine Deaminase |
| ADAR1 | Adenosine Deaminase 1 |
| MyD88 | Myeloid-Differentiation primary response 88 |
| IKK | IkB Kinase |
| IFNAR | Interferon alpha/beta Receptor |
| IFNLR1 | Interferon lambda Receptor 1 |
| IL10R1 | Interleukin-10 Receptor 1 |
| STAT | Signal Transducer and Activator of Transcription |
| IRF | Interferon Regulatory Factor |
| NLRC | Nucleotide-binding oligomerization domain-Like Receptor C |
| NF-kB | Nuclear Factor-kappa-light-chain-enhancer of B-Lymphocytes |
| ISG | Interferon-Stimulated Gene |
| IFITM3 | Interferon-Induced Transmembrane Protein 3 |
| cGAS-STING | cyclic AMP-GMP Synthase-Stimulator of Interferon Genes |
| IL | Interleukin |
| TNF | Tumor Necrosis Factor |
| TGF-α | Tumor Growth Factor-alpha |
| TGF-β | Transforming Growth Factor-beta |
| CRP | C-Reactive Protein |
| CCL | Chemokine (C-C motif) Ligand |
| CXCL | Chemokine (C-X-C motif) Ligand |
| CCR | Chemokine (C-C motif) Receptor |
| CXCR | Chemokine (C-X-C motif) Receptor |
| NK Cell | Natural Killer Cell |
| IMM | Inflammatory Monocyte and Macrophage |
| pDC | plasmacytoid Dendritic Cell |
| HLA | Human Leukocyte Antigen |
| MHC | Major Histocompatibility Complex |
| Th Lymphocyte | T-helper Lymphocyte |
| CTL | Cytotoxic T-Lymphocyte |
| CD | Cluster of Differentiation |
| LINE-1 | Long Interspersed Nuclear Element 1 |
| AIDS | Acute Immunodeficiency Syndrome |
| Ab | Antibody |
| Ig | Immunoglobulin |
| C5a | Complement component 5a |
| GRP78 | 78 kDa-Glucose-Regulated Protein |
| HSP | Heat-Shock Protein |
| ARS | Acute Radiation Syndrome |
| +ssRNA | positive-sense single-stranded Ribonucleic Acid |
| BMI | Body-Mass Index |
| IU | International Unit |
References
- Murray, C.J.L. The Global Burden of Disease Study at 30 years. Nat Med. 2022, 28, 2019–2026. [Google Scholar] [CrossRef]
- Brisse, M.; Ly, H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef]
- Schmidt, A.; Rothenfusser, S.; Hopfner, K.P. Sensing of viral nucleic acids by RIG-I: From translocation to translation. Eur. J. Cell Biol. 2012, 91, 78–85. [Google Scholar] [CrossRef]
- Yao, H.; Dittmann, M.; Peisley, A.; Hoffmann, H.H.; Gilmore, R.H.; Schmidt, T.; Schmidt-Burgk, J.; Hornung, V.; Rice, C.M.; Hur, S. ATP-dependent effector-like functions of RIG-I-like receptors. Mol. Cell 2015, 58, 541–548. [Google Scholar] [CrossRef]
- Cervantes, J.; Weinerman, B.; Basole, C.; et al. TLR8: The forgotten relative revindicated. Cell Mol Immunol 2012, 9, 434–438. [Google Scholar] [CrossRef]
- Bowie, A.G.; Unterholzner, L. Viral evasion and subversion of pattern-recognition receptor signalling. Nat. Rev. Immunol. 2008, 8, 911–922. [Google Scholar] [CrossRef]
- Biron, C.A. (2016, February 12). Innate immunity: Recognizing and responding to foreign invaders-no training needed. Viral Pathogenesis (Third Edition). Retrieved October 22, 2022, from https://www.sciencedirect.com/science/article/pii/B9780128009642000045.
- De Maeyer, E.; De Maeyer-Guignard, J. Type I interferons. Int. Rev. Immunol. 1998, 17, 53–73. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. Type I interferons: Diversity of sources, production pathways and effects on immune responses. Curr. Opin. Virol. 2011, 1, 463–475. [Google Scholar] [CrossRef]
- Lazear, H.M.; Schoggins, J.W.; Diamond, M.S. Shared and Distinct Functions of Type I and Type III Interferons. Immunity 2019, 50, 907–923. [Google Scholar] [CrossRef]
- Stanifer, M.L.; Guo, C.; Doldan, P.; Boulant, S. Importance of Type I and III Interferons at Respiratory and Intestinal Barrier Surfaces. Front. Immunol. 2020, 11, 608645. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Shin, E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp Mol Med 2021, 53, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Glennon-Alty, L.; Moots, R.J.; Edwards, S.W.; Wright, H.L. Type I interferon regulates cytokine-delayed neutrophil apoptosis, reactive oxygen species production and chemokine expression. Clin. Exp. Immunol. 2021, 203, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Pierangeli, A.; Gentile, M.; Oliveto, G.; Frasca, F.; Sorrentino, L.; Matera, L.; Nenna, R.; Viscido, A.; Fracella, M.; Petrarca, L.; et al. Comparison by Age of the Local Interferon Response to SARS-CoV-2 Suggests a Role for IFN-ε and -ω. Front. Immunol. 2022, 13, 873232. [Google Scholar] [CrossRef] [PubMed]
- Chieux, V.; Hober, D.; Chehadeh, W.; Wattré, P. Interféron alpha, protéines antivirales et applications médicales [Alpha interferon, antiviral proteins and their value in clinical medicine]. Ann. De Biol. Clin. 1999, 57, 659–666. [Google Scholar]
- David Vremec, Meredith O’Keeffe, Hubertus Hochrein, Martina Fuchsberger, Irina Caminschi, Mireille Lahoud, Ken Shortman; Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood 2007, 109, 1165–1173. [CrossRef] [PubMed]
- Odendall, C.; Kagan, J.C. The unique regulation and functions of type III interferons in antiviral immunity. Curr. Opin. Virol. 2015, 12, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Hefti, H.P.; Frese, M.; Landis, H.; Di Paolo, C.; Aguzzi, A.; Haller, O.; Pavlovic, J. Human MxA protein protects mice lacking a functional alpha/beta interferon system against La crosse virus and other lethal viral infections. J. Virol. 1999, 73, 6984–6991. [Google Scholar] [CrossRef] [PubMed]
- Haller, O.; Kochs, G. Human MxA protein: An interferon-induced dynamin-like GTPase with broad antiviral activity. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011, 31, 79–87. [Google Scholar] [CrossRef]
- Haller, O.; Gao, S.; von der Malsburg, A.; Daumke, O.; Kochs, G. Dynamin-like MxA GTPase: Structural insights into oligomerization and implications for antiviral activity. J. Biol. Chem. 2010, 285, 28419–28424. [Google Scholar] [CrossRef]
- Dick, A.; Graf, L.; Olal, D.; von der Malsburg, A.; Gao, S.; Kochs, G.; Daumke, O. Role of nucleotide binding and GTPase domain dimerization in dynamin-like myxovirus resistance protein A for GTPase activation and antiviral activity. J. Biol. Chem. 2015, 290, 12779–12792. [Google Scholar] [CrossRef] [PubMed]
- Malmgaard, L. Induction and regulation of IFNs during viral infections. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2004, 24, 439–454. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.Y.; Suthar, M.S. Mechanisms of innate immune evasion in re-emerging RNA viruses. Curr. Opin. Virol. 2015, 12, 26–37. [Google Scholar] [CrossRef]
- Munir S, Hillyer P, Le Nouën C, Buchholz UJ, Rabin RL; et al. Respiratory Syncytial Virus Interferon Antagonist NS1 Protein Suppresses and Skews the Human T Lymphocyte Response. PLOS Pathogens 2011, 7, e1001336. [Google Scholar] [CrossRef]
- Mo, S.; Tang, W.; Xie, J.; Chen, S.; Ren, L.; Zang, N.; Xie, X.; Deng, Y.; Gao, L.; Liu, E. Respiratory syncytial virus activates rab5a to suppress IRF1-dependent lambda interferon production, subverting the antiviral defense of airway epithelial cells. J. Virol. 2021, 95. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, J.; Cao, X. Regulation of type I interferon signaling in immunity and inflammation: A comprehensive review. J. Autoimmun. 2017, 83, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Ivashkiv, L.B.; Donlin, L.T. Regulation of type I interferon responses. Nat. Rev. Immunol. 2014, 14, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Qian, C.; Cao, X. Post-Translational Modification Control of Innate Immunity. Immunity 2016, 45, 15–30. [Google Scholar] [CrossRef]
- Kosciuczuk, E.M.; Mehrotra, S.; Saleiro, D.; Kroczynska, B.; Majchrzak-Kita, B.; Lisowski, P.; Driehaus, C.; Rogalska, A.; Turner, A.; Lienhoop, T.; et al. Sirtuin 2-mediated deacetylation of cyclin-dependent kinase 9 promotes STAT1 signaling in type I interferon responses. J. Biol. Chem. 2019, 294, 827–837. [Google Scholar] [CrossRef]
- Xing, F.; Matsumiya, T.; Shiba, Y.; Hayakari, R.; Yoshida, H.; Imaizumi, T. Non-Canonical Role of IKKα in the Regulation of STAT1 Phosphorylation in Antiviral Signaling. PLoS ONE 2016, 11, e0168696. [Google Scholar] [CrossRef]
- Perwitasari, O.; Cho, H.; Diamond, M.S.; Gale, M.; Jr Inhibitor of κB kinase epsilon (IKK(epsilon)), STAT1, and IFIT2 proteins define novel innate immune effector pathway against West Nile virus infection. J. Biol. Chem. 2011, 286, 44412–44423. [CrossRef] [PubMed]
- Dempoya, J.; Matsumiya, T.; Imaizumi, T.; Hayakari, R.; Xing, F.; Yoshida, H.; Okumura, K.; Satoh, K. Double-stranded RNA induces biphasic STAT1 phosphorylation by both type I interferon (IFN)-dependent and type I IFN-independent pathways. J. Virol. 2012, 86, 12760–12769. [Google Scholar] [CrossRef] [PubMed]
- Dong, L.W.; Kong, X.N.; Yan, H.X.; Yu, L.X.; Chen, L.; Yang, W.; Liu, Q.; Huang, D.D.; Wu, M.C.; Wang, H.Y. Signal regulatory protein alpha negatively regulates both TLR3 and cytoplasmic pathways in type I interferon induction. Mol. Immunol. 2008, 45, 3025–3035. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; An, H.; Yao, M.; Hou, J.; Yu, Y.; Feng, G.; Cao, X. Scaffolding adaptor protein Gab1 is required for TLR3/4- and RIG-I-mediated production of proinflammatory cytokines and type I IFN in macrophages. J. Immunol. 2010, 184, 6447–6456. [Google Scholar] [CrossRef]
- Oshiumi, H.; Matsumoto, M.; Funami, K.; Akazawa, T.; Seya, T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003, 4, 161–167. [Google Scholar] [CrossRef]
- Seya, T.; Matsumoto, M.; Ebihara, T.; Oshiumi, H. Functional evolution of the TICAM-1 pathway for extrinsic RNA sensing. Immunol. Rev. 2009, 227, 44–53. [Google Scholar] [CrossRef]
- Takaki, H.; Oshiumi, H.; Sasai, M.; Kawanishi, T.; Matsumoto, M.; Seya, T. Oligomerized TICAM-1 (TRIF) in the cytoplasm recruits nuclear BS69 to enhance NF-kappaB activation and type I IFN induction. Eur. J. Immunol. 2009, 39, 3469–3476. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, C.; Xue, P.; Zhong, B.; Mao, A.P.; Ran, Y.; Chen, H.; Wang, Y.Y.; Yang, F.; Shu, H.B. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA 2009, 106, 7945–7950. [Google Scholar] [CrossRef] [PubMed]
- Bressy, C.; Droby, G.N.; Maldonado, B.D.; Steuerwald, N.; Grdzelishvili, V.Z. Cell Cycle Arrest in G2/M Phase Enhances Replication of Interferon-Sensitive Cytoplasmic RNA Viruses via Inhibition of Antiviral Gene Expression. J. Virol. 2019, 93, e01885-18. [Google Scholar] [CrossRef]
- Moerdyk-Schauwecker, M.; Shah, N.R.; Murphy, A.M.; Hastie, E.; Mukherjee, P.; Grdzelishvili, V.Z. Resistance of pancreatic cancer cells to oncolytic vesicular stomatitis virus: Role of type I interferon signaling. Virology 2013, 436, 221–234. [Google Scholar] [CrossRef]
- Cataldi, M.; Shah, N.R.; Felt, S.A.; Grdzelishvili, V.Z. Breaking resistance of pancreatic cancer cells to an attenuated vesicular stomatitis virus through a novel activity of IKK inhibitor TPCA-1. Virology 2015, 485, 340–354. [Google Scholar] [CrossRef]
- Aliyari, S.R.; Quanquin, N.; Pernet, O.; Zhang, S.; Wang, L.; Cheng, G. The Evolutionary Dance between Innate Host Antiviral Pathways and SARS-CoV-2. Pathogens 2022, 11, 538. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, F.; Guillomot, M.; D’Andréa, S.; Battegay, S.; La Bonnardière, C. Interferon-delta: The first member of a novel type I interferon family. Biochimie 1998, 80, 779–788. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, J.; Feng, H.; Wei, Q.; Zhao, S.; Yang, S.; Ma, H.; Liu, D.; Zhang, G. Antiviral activity of porcine interferon delta 8 against pseudorabies virus in vitro. Int. J. Biol. Macromol. 2021, 177, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Nagai, M.; Bando, S.; Dobashi, H.; Takahara, J. Emergence of acute interstitial pneumonia following high dose interferon delta treatment in a case of chronic myelogenous leukemia. Intern. Med. 1993, 32, 716–718. [Google Scholar] [CrossRef] [PubMed]
- La Bonnardière, C.; Lefèvre, F.; Charley, B. Interferon response in pigs: Molecular and biological aspects. Vet. Immunol. Immunopathol. 1994, 43, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Mesev, E.V.; LeDesma, R.A.; Ploss, A. Decoding type I and III interferon signalling during viral infection. Nat. Microbiol. 2019, 4, 914–924. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Mian, F.; Poznanski, S.M.; Stackaruk, M.; Chan, T.; Chew, M.V.; Ashkar, A.A. Type I Interferon Receptor on NK Cells Negatively Regulates Interferon-γ Production. Front. Immunol. 2019, 10, 1261. [Google Scholar] [CrossRef] [PubMed]
- Khalil, B.A.; Elemam, N.M.; Maghazachi, A.A. Chemokines and chemokine receptors during COVID-19 infection. Comput. Struct. Biotechnol. J. 2021, 19, 976–988. [Google Scholar] [CrossRef]
- Sidahmed, A.M.; León, A.J.; Bosinger, S.E.; Banner, D.; Danesh, A.; Cameron, M.J.; Kelvin, D.J. CXCL10 contributes to p38-mediated apoptosis in primary T lymphocytes in vitro. Cytokine 2012, 59, 433–441. [Google Scholar] [CrossRef]
- David Vremec, Meredith O’Keeffe, Hubertus Hochrein, Martina Fuchsberger, Irina Caminschi, Mireille Lahoud, Ken Shortman; Production of interferons by dendritic cells, plasmacytoid cells, natural killer cells, and interferon-producing killer dendritic cells. Blood 2007, 109, 1165–1173. [CrossRef] [PubMed]
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. SARS-CoV-2: A Master of Immune Evasion. Biomedicines 2022, 10, 1339. [Google Scholar] [CrossRef] [PubMed]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V. High IL-6/IFN-γ ratio could be associated with severe disease in COVID-19 patients. J. Med. Virol. 2020, 92, 1789–1790. [Google Scholar] [CrossRef]
- Kwon, J.S.; Kim, J.Y.; Kim, M.C.; Park, S.Y.; Kim, B.N.; Bae, S.; Cha, H.H.; Jung, J.; Kim, M.J.; Lee, M.J.; Choi, S.H.; Chung, J.W.; Shin, E.C.; Kim, S.H. Factors of Severity in Patients with COVID-19: Cytokine/Chemokine Concentrations, Viral Load, and Antibody Responses. Am. J. Trop. Med. Hyg. 2020, 103, 2412–2418. [Google Scholar] [CrossRef]
- Sun, J.; Tang, X.; Bai, R.; Liang, C.; Zeng, L.; Lin, H.; Yuan, R.; Zhou, P.; Huang, X.; Xiong, Q.; Peng, J.; Cui, F.; Ke, B.; Su, J.; Liu, Z.; Lu, J.; Tian, J.; Sun, R.; Ke, C. The kinetics of viral load and antibodies to SARS-CoV-2. Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 2020, 26, 1690.e1–1690.e4. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, S.; Li, H.; Wang, Y.; Lu, Z.; Liu, Z.; Lai, Q.; Ji, Y.; Huang, X.; Li, Y.; Sun, J.; Wu, Y.; Xu, X.; Hou, J. Viral and Antibody Kinetics of COVID-19 Patients with Different Disease Severities in Acute and Convalescent Phases: A 6-Month Follow-Up Study. Virol. Sin. 2020, 35, 820–829. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kwon, J.S.; Bae, S.; Cha, H.H.; Lim, J.S.; Kim, M.C.; Chung, J.W.; Park, S.Y.; Lee, M.J.; Kim, B.N.; Jung, J.; Kim, M.J.; Shin, E.C.; Kim, S.H. SARS-CoV-2-Specific Antibody and T Cell Response Kinetics According to Symptom Severity. Am. J. Trop. Med. Hyg. 2021, 105, 395–400. [Google Scholar] [CrossRef] [PubMed]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Fukuda, Y.; Homma, T.; Inoue, H.; Onitsuka, C.; Ikeda, H.; Goto, Y.; Sato, Y.; Kimura, T.; Hirai, K.; Ohta, S.; et al. Downregulation of type III interferons in patients with severe COVID-19. J. Med. Virol. 2021, 93, 4559–4563. [Google Scholar] [CrossRef]
- Ank, N.; West, H.; Bartholdy, C.; Eriksson, K.; Thomsen, A.R.; Paludan, S.R. Lambda interferon (IFN-lambda), a type III IFN, is induced by viruses and IFNs and displays potent antiviral activity against select virus infections in vivo. J. Virol. 2006, 80, 4501–4509. [Google Scholar] [CrossRef] [PubMed]
- Lasfar, A.; Gogas, H.; Zloza, A.; Kaufman, H.L.; Kirkwood, J.M. IFN-λ cancer immunotherapy: New kid on the block. Immunotherapy 2016, 8, 877–888. [Google Scholar] [CrossRef]
- Lasfar, A.; Abushahba, W.; Balan, M.; Cohen-Solal, K.A. Interferon lambda: A new sword in cancer immunotherapy. Clin. Dev. Immunol. 2011, 2011, 349575. [Google Scholar] [CrossRef] [PubMed]
- de Groen, R.A.; Groothuismink, Z.M.; Liu, B.S.; Boonstra, A. IFN-λ is able to augment TLR-mediated activation and subsequent function of primary human B cells. J. Leukoc. Biol. 2015, 98, 623–630. [Google Scholar] [CrossRef]
- Salerno, F.; Guislain, A.; Cansever, D.; Wolkers, M.C. TLR-Mediated Innate Production of IFN-γ by CD8+ T Cells Is Independent of Glycolysis. J. Immunol. 2016, 196, 3695–3705. [Google Scholar] [CrossRef] [PubMed]
- Lasfar, A.; Zloza, A.; Cohen-Solal, K.A. IFN-lambda therapy: Current status and future perspectives. Drug Discov. Today 2016, 21, 167–171. [Google Scholar] [CrossRef]
- Santer, D.M.; Li, D.; Ghosheh, Y.; et al. Interferon-λ treatment accelerates SARS-CoV-2 clearance despite age-related delays in the induction of T cell immunity. Nat Commun 2022, 13, 6992. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, Y.; Homma, T.; Inoue, H.; Goto, Y.; Sato, Y.; Ikeda, H.; Onitsuka, C.; Sato, H.; Akimoto, K.; Ebato, T.; et al. Serum IL-28A/IFN-λ2 is linked to disease severity of COVID-19. Sci. Rep. 2022, 12, 5458. [Google Scholar] [CrossRef] [PubMed]
- Zanoni, I.; Granucci, F.; Broggi, A. Interferon (IFN)-λ Takes the Helm: Immunomodulatory Roles of Type III IFNs. Front. Immunol. 2017, 8, 1661. [Google Scholar] [CrossRef]
- Mordstein, M.; Michiels, T.; Staeheli, P. What have we learned from the IL28 receptor knockout mouse? Journal of interferon & cytokine research: The official journal of the International Society for Interferon and Cytokine Research 2010, 30, 579–584. [Google Scholar] [CrossRef]
- Gaudieri, S.; Lucas, M.; Lucas, A.; McKinnon, E.; Albloushi, H.; Rauch, A.; di Iulio, J.; Martino, D.; Prescott, S.L.; Tulic, M.K. Genetic variations in IL28B and allergic disease in children. PLoS ONE 2012, 7, e30607. [Google Scholar] [CrossRef] [PubMed]
- Bellanti, F.; Vendemiale, G.; Altomare, E.; Serviddio, G. The impact of interferon lambda 3 gene polymorphism on natural course and treatment of hepatitis C. Clin. Dev. Immunol. 2012, 2012, 849373. [Google Scholar] [CrossRef] [PubMed]
- Mangia, A.; Mottola, L.; Santoro, R. Interleukin 28B polymorphisms as predictor of response in hepatitis C virus genotype 2 and 3 infected patients. World J. Gastroenterol. 2013, 19, 8924–8928. [Google Scholar] [CrossRef] [PubMed]
- Naggie, S.; Osinusi, A.; Katsounas, A.; Lempicki, R.; Herrmann, E.; Thompson, A.J.; Clark, P.J.; Patel, K.; Muir, A.J.; McHutchison, J.G.; et al. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: Impaired viral kinetics and therapeutic response. Hepatology 2012, 56, 444–454. [Google Scholar] [CrossRef] [PubMed]
- Barber, G.N. Host defense, viruses and apoptosis. Cell Death Differ. 2001, 8, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Ezelle, H.J.; Balachandran, S.; Sicheri, F.; Polyak, S.J.; Barber, G.N. Analyzing the mechanisms of interferon-induced apoptosis using CrmA and hepatitis C virus NS5A. Virology 2001, 281, 124–137. [Google Scholar] [CrossRef] [PubMed]
- Gil, J.; Esteban, M. The interferon-induced protein kinase (PKR), triggers apoptosis through FADD-mediated activation of caspase 8 in a manner independent of Fas and TNF-alpha receptors. Oncogene 2000, 19, 3665–3674. [Google Scholar] [CrossRef] [PubMed]
- Bittner, Z.A.; Schrader, M.; George, S.E.; Amann, R. Pyroptosis and Its Role in SARS-CoV-2 Infection. Cells 2022, 11, 1717. [Google Scholar] [CrossRef]
- Mortezaee, K.; Majidpoor, J. Cellular immune states in SARS-CoV-2-induced disease. Front. Immunol. 2022, 13. [Google Scholar] [CrossRef]
- Sträter, J.; Möller, P. TRAIL and viral infection. Vitam. Horm. 2004, 67, 257–274. [Google Scholar] [CrossRef]
- Robbins, M.A.; Maksumova, L.; Pocock, E.; Chantler, J.K. Nuclear factor-kappaB translocation mediates double-stranded ribonucleic acid-induced NIT-1 beta-cell apoptosis and up-regulates caspase-12 and tumor necrosis factor receptor-associated ligand (TRAIL). Endocrinology 2003, 144, 4616–4625. [Google Scholar] [CrossRef] [PubMed]
- Shan, C.; Miao, S.; Liu, C.; et al. Induction of macrophage pyroptosis-related factors by pathogenic E. coli high pathogenicity island (HPI) in Yunnan Saba pigs. BMC Vet Res 2021, 17, 114. [Google Scholar] [CrossRef]
- Tan, X.; Sun, L.; Chen, J.; Chen, Z.J. Detection of Microbial Infections Through Innate Immune Sensing of Nucleic Acids. Annu. Rev. Microbiol. 2018, 72, 447–478. [Google Scholar] [CrossRef] [PubMed]
- Roers, A.; Hiller, B.; Hornung, V. Recognition of Endogenous Nucleic Acids by the Innate Immune System. Immunity 2016, 44, 739–754. [Google Scholar] [CrossRef]
- Luecke, S.; Paludan, S.R. Molecular requirements for sensing of intracellular microbial nucleic acids by the innate immune system. Cytokine 2017, 98, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Ablasser, A.; Hertrich, C.; Waßermann, R.; Hornung, V. Nucleic acid driven sterile inflammation. Clin. Immunol. 2013, 147, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Mathern, D.R.; Heeger, P.S. Molecules Great and Small: The Complement System. Clin. J. Am. Soc. Nephrol. CJASN 2015, 10, 1636–1650. [Google Scholar] [CrossRef] [PubMed]
- Kunz, N.; Kemper, C. Complement Has Brains-Do Intracellular Complement and Immunometabolism Cooperate in Tissue Homeostasis and Behavior? Front. Immunol. 2021, 12, 629986. [Google Scholar] [CrossRef] [PubMed]
- West, E.E.; Kunz, N.; Kemper, C. Complement and human T cell metabolism: Location, location, location. Immunol. Rev. 2020, 295, 68–81. [Google Scholar] [CrossRef]
- Shibabaw, T.; Molla, M.D.; Teferi, B.; Ayelign, B. Role of IFN and Complements System: Innate Immunity in SARS-CoV-2. J. Inflamm. Res. 2020, 13, 507–518. [Google Scholar] [CrossRef]
- Znaidia, M.; Demeret, C.; van der Werf, S.; Komarova, A.V. Characterization of SARS-CoV-2 Evasion: Interferon Pathway and Therapeutic Options. Viruses 2022, 14, 1247. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.R.; Corrales, L.; Gajewski, T.F. Innate immune recognition of cancer. Annu. Rev. Immunol. 2015, 33, 445–474. [Google Scholar] [CrossRef] [PubMed]
- Haller, O. A tribute to Jean Lindenmann, co-discoverer of interferon (1924-2015). Cytokine 2015, 76, 113–115. [Google Scholar] [CrossRef] [PubMed]
- Roumenina, L.T.; Daugan, M.V.; Noé, R.; Petitprez, F.; Vano, Y.A.; Sanchez-Salas, R.; Becht, E.; Meilleroux, J.; Clec’h, B.L.; Giraldo, N.A.; et al. Tumor Cells Hijack Macrophage-Produced Complement C1q to Promote Tumor Growth. Cancer Immunol. Res. 2019, 7, 1091–1105. [Google Scholar] [CrossRef] [PubMed]
- Posch, W.; Bermejo-Jambrina, M.; Steger, M.; Witting, C.; Diem, G.; Hörtnagl, P.; Hackl, H.; Lass-Flörl, C.; Huber, L.A.; Geijtenbeek, T.; Wilflingseder, D. Complement Potentiates Immune Sensing of HIV-1 and Early Type I Interferon Responses. mBio 2021, 12, e0240821. [Google Scholar] [CrossRef]
- Bermejo-Jambrina, M.; Blatzer, M.; Jauregui-Onieva, P.; Yordanov, T.E.; Hörtnagl, P.; Valovka, T.; Huber, L.A.; Wilflingseder, D.; Posch, W. CR4 Signaling Contributes to a DC-Driven Enhanced Immune Response Against Complement-Opsonized HIV-1. Front. Immunol. 2020, 11, 2010. [Google Scholar] [CrossRef] [PubMed]
- Posch, W.; Bermejo-Jambrina, M.; Lass-Flörl, C.; Wilflingseder, D. Role of Complement Receptors (CRs) on DCs in Anti-HIV-1 Immunity. Front. Immunol. 2020, 11, 572114. [Google Scholar] [CrossRef] [PubMed]
- Steinman, R.M.; Granelli-Piperno, A.; Pope, M.; Trumpfheller, C.; Ignatius, R.; Arrode, G.; Racz, P.; Tenner-Racz, K. The interaction of immunodeficiency viruses with dendritic cells. Curr. Top. Microbiol. Immunol. 2003, 276, 1–30. [Google Scholar] [CrossRef]
- Lekkerkerker, A.N.; van Kooyk, Y.; Geijtenbeek, T.B. Viral piracy: HIV-1 targets dendritic cells for transmission. Curr. HIV Res. 2006, 4, 169–176. [Google Scholar] [CrossRef]
- Elkon, K.B.; Santer, D.M. Complement, interferon and lupus. Curr. Opin. Immunol. 2012, 24, 665–670. [Google Scholar] [CrossRef]
- Qi, Y.Y.; Zhou, X.J.; Cheng, F.J.; Hou, P.; Ren, Y.L.; Wang, S.X.; Zhao, M.H.; Yang, L.; Martinez, J.; Zhang, H. Increased autophagy is cytoprotective against podocyte injury induced by antibody and interferon-α in lupus nephritis. Ann. Rheum. Dis. 2018, 77, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Chauss, D.; Freiwald, T.; McGregor, R.; Yan, B.; Wang, L.; Nova-Lamperti, E.; Kumar, D.; Zhang, Z.; Teague, H.; West, E.E.; et al. Autocrine vitamin D signaling switches off pro-inflammatory programs of TH1 cells. Nat. Immunol. 2022, 23, 62–74. [Google Scholar] [CrossRef]
- Schardey, J.; Globig, A.M.; Janssen, C.; Hofmann, M.; Manegold, P.; Thimme, R.; Hasselblatt, P. Vitamin D Inhibits Pro-Inflammatory T Cell Function in Patients With Inflammatory Bowel Disease. J. Crohn’s Colitis 2019, 13, 1546–1557. [Google Scholar] [CrossRef] [PubMed]
- Battistini, C.; Ballan, R.; Herkenhoff, M.E.; Saad, S.; Sun, J. Vitamin D Modulates Intestinal Microbiota in Inflammatory Bowel Diseases. Int. J. Mol. Sci. 2020, 22, 362. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, Y.; Du, J.; Gao, P.; Zhao, K. The Interplay Among HIV, LINE-1, and the Interferon Signaling System. Front. Immunol. 2021, 12, 732775. [Google Scholar] [CrossRef] [PubMed]
- Mustelin, T.; Ukadike, K.C. How Retroviruses and Retrotransposons in Our Genome May Contribute to Autoimmunity in Rheumatological Conditions. Front. Immunol. 2020, 11, 593891. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.; Iijima, K.; Kano, S.; Ishizaka, Y. Viral protein R of HIV type-1 induces retrotransposition and upregulates glutamate synthesis by the signal transducer and activator of transcription 1 signaling pathway. Microbiol. Immunol. 2015, 59, 398–409. [Google Scholar] [CrossRef]
- Harman, A.N.; Nasr, N.; Feetham, A.; Galoyan, A.; Alshehri, A.A.; Rambukwelle, D.; Botting, R.A.; Hiener, B.M.; Diefenbach, E.; Diefenbach, R.J.; et al. HIV Blocks Interferon Induction in Human Dendritic Cells and Macrophages by Dysregulation of TBK1. J. Virol. 2015, 89, 6575–6584. [Google Scholar] [CrossRef] [PubMed]
- Tunbak, H.; Enriquez-Gasca, R.; Tie, C.; Gould, P.A.; Mlcochova, P.; Gupta, R.K.; Fernandes, L.; Holt, J.; van der Veen, A.G.; Giampazolias, E.; et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat. Commun. 2020, 11, 5387. [Google Scholar] [CrossRef]
- Kuriyama, Y.; Shimizu, A.; Kanai, S.; et al. Coordination of retrotransposons and type I interferon with distinct interferon pathways in dermatomyositis, systemic lupus erythematosus and autoimmune blistering disease. Sci Rep 2021, 11, 23146. [Google Scholar] [CrossRef]
- Emiliani, Yuliana and Muzi, Gabriel and Sanchez, Andres and Munera, Marlon, Prediction of Molecular Mimicry between Proteins from Trypanosoma Sp. And Human Antigens Associated with Systemic Lupus Erythematosus. Available at SSRN: https://ssrn.com/abstract=4184590. [CrossRef]
- Yu, Q.; Carbone, C.J.; Katlinskaya, Y.V.; Zheng, H.; Zheng, K.; Luo, M.; Wang, P.J.; Greenberg, R.A.; Fuchs, S.Y. Type I interferon controls propagation of long interspersed element-1. J. Biol. Chem. 2015, 290, 10191–10199. [Google Scholar] [CrossRef] [PubMed]
- Kuriyama, Y.; Shimizu, A.; Kanai, S.; Oikawa, D.; Tokunaga, F.; Tsukagoshi, H.; Ishikawa, O. The synchronized gene expression of retrotransposons and type I interferon in dermatomyositis. J. Am. Acad. Dermatol. 2021, 84, 1103–1105. [Google Scholar] [CrossRef]
- Cassius, C.; Amode, R.; Delord, M.; Battistella, M.; Poirot, J.; How-Kit, A.; Lepelletier, C.; Jachiet, M.; de Masson, A.; Frumholtz, L.; et al. MDA5+ Dermatomyositis Is Associated with Stronger Skin Type I Interferon Transcriptomic Signature with Upregulation of IFN-κ Transcript. J. Investig. Dermatol. 2020, 140, 1276–1279.e7. [Google Scholar] [CrossRef] [PubMed]
- Burns, K.H. Our Conflict with Transposable Elements and Its Implications for Human Disease. Annu. Rev. Pathol. 2020, 15, 51–70. [Google Scholar] [CrossRef]
- Mustelin, T.; Ukadike, K.C. How Retroviruses and Retrotransposons in Our Genome May Contribute to Autoimmunity in Rheumatological Conditions. Front. Immunol. 2020, 11, 593891. [Google Scholar] [CrossRef] [PubMed]
- Crow, M.K. Long interspersed nuclear elements (LINE-1): Potential triggers of systemic autoimmune disease. Autoimmunity 2010, 43, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Cui, G.; Gao, Y. Research progress on inflammatory mechanism of primary Sjögren syndrome. Zhejiang Da Xue Xue Bao. Yi Xue Ban J. Zhejiang University. Med. Sci. 2021, 50, 783–794. [Google Scholar] [CrossRef]
- Gamdzyk, M.; Doycheva, D.M.; Araujo, C.; Ocak, U.; Luo, Y.; Tang, J.; Zhang, J.H. cGAS/STING Pathway Activation Contributes to Delayed Neurodegeneration in Neonatal Hypoxia-Ischemia Rat Model: Possible Involvement of LINE-1. Mol. Neurobiol. 2020, 57, 2600–2619. [Google Scholar] [CrossRef]
- Baechler, E.C.; Bilgic, H.; Reed, A.M. Type I interferon pathway in adult and juvenile dermatomyositis. Arthritis Res. Ther. 2011, 13, 249. [Google Scholar] [CrossRef]
- Hotter, D.; Bosso, M.; Jønsson, K.L.; Krapp, C.; Stürzel, C.M.; Das, A.; Littwitz-Salomon, E.; Berkhout, B.; Russ, A.; Wittmann, S.; et al. IFI16 Targets the Transcription Factor Sp1 to Suppress HIV-1 Transcription and Latency Reactivation. Cell Host Microbe 2019, 25, 858–872.e13. [Google Scholar] [CrossRef]
- Lee-Kirsch, M.A. The Type I Interferonopathies. Annu. Rev. Med. 2017, 68, 297–315. [Google Scholar] [CrossRef]
- Tsuji, R.F.; Geba, G.P.; Wang, Y.; Kawamoto, K.; Matis, L.A.; Askenase, P.W. Required early complement activation in contact sensitivity with generation of local C5-dependent chemotactic activity, and late T cell interferon gamma: A possible initiating role of B cells. J. Exp. Med. 1997, 186, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Laviada-Molina, H.A.; Leal-Berumen, I.; Rodriguez-Ayala, E.; Bastarrachea, R.A. Working Hypothesis for Glucose Metabolism and SARS-CoV-2 Replication: Interplay Between the Hexosamine Pathway and Interferon RF5 Triggering Hyperinflammation. Role of BCG Vaccine? Front. Endocrinol. 2020, 11, 514. [Google Scholar] [CrossRef] [PubMed]
- Narunsky-Haziza, L.; Sepich-Poore, G.D.; Livyatan, I.; Asraf, O.; Martino, C.; Nejman, D.; Gavert, N.; Stajich, J.E.; Amit, G.; González, A.; et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 2022, 185, 3789–3806. [Google Scholar] [CrossRef] [PubMed]
- Dohlman, A.B.; Klug, J.; Mesko, M.; Gao, I.H.; Lipkin, S.M.; Shen, X.; Iliev, I.D. A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell 2022, 185, 3807–3822. [Google Scholar] [CrossRef]
- Di Martino, J.S.; Nobre, A.R.; Mondal, C.; et al. A tumor-derived type III collagen-rich ECM niche regulates tumor cell dormancy. Nat Cancer 2022, 3, 90–107. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, L.; Liu, X. Natural killer cells: The next wave in cancer immunotherapy. Front. Immunol. 2022, 13, 954804. [Google Scholar] [CrossRef]
- Shaver, K.A.; Croom-Perez, T.J.; Copik, A.J. Natural Killer Cells: The Linchpin for Successful Cancer Immunotherapy. Front. Immunol. 2021, 12, 679117. [Google Scholar] [CrossRef]
- Rautela, J.; Huntington, N.D. IL-15 signaling in NK cell cancer immunotherapy. Curr. Opin. Immunol. 2017, 44, 1–6. [Google Scholar] [CrossRef]
- Rah, B.; Rather, R.A.; Bhat, G.R.; Baba, A.B.; Mushtaq, I.; Farooq, M.; Yousuf, T.; Dar, S.B.; Parveen, S.; Hassan, R.; et al. JAK/STAT Signaling: Molecular Targets, Therapeutic Opportunities, and Limitations of Targeted Inhibitions in Solid Malignancies. Front. Pharmacol. 2022, 13, 821344. [Google Scholar] [CrossRef]
- Luo, K.; Li, N.; Ye, W.; Gao, H.; Luo, X.; Cheng, B. Activation of Stimulation of Interferon Genes (STING) Signal and Cancer Immunotherapy. Molecules 2022, 27, 4638. [Google Scholar] [CrossRef] [PubMed]
- Corrales, L.; McWhirter, S.M.; Dubensky, T.W., Jr.; Gajewski, T.F. The host STING pathway at the interface of cancer and immunity. J. Clin. Investig. 2016, 126, 2404–2411. [Google Scholar] [CrossRef] [PubMed]
- Kho, V.M.; Mekers, V.E.; Span, P.N.; Bussink, J.; Adema, G.J. Radiotherapy and cGAS/STING signaling: Impact on MDSCs in the tumor microenvironment. Cell. Immunol. 2021, 362, 104298. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Luo, J.; Alu, A.; Han, X.; Wei, Y.; Wei, X. cGAS-STING pathway in cancer biotherapy. Mol. Cancer 2020, 19, 136. [Google Scholar] [CrossRef]
- Zheng, J.; Mo, J.; Zhu, T.; Zhuo, W.; Yi, Y.; Hu, S.; Yin, J.; Zhang, W.; Zhou, H.; Liu, Z. Comprehensive elaboration of the cGAS-STING signaling axis in cancer development and immunotherapy. Mol. Cancer 2020, 19, 133. [Google Scholar] [CrossRef] [PubMed]
- Zitvogel, L.; Galluzzi, L.; Kepp, O.; Smyth, M.J.; Kroemer, G. Type I interferons in anticancer immunity. Nat. Rev. Immunol. 2015, 15, 405–414. [Google Scholar] [CrossRef]
- Bracci, L.; La Sorsa, V.; Belardelli, F.; Proietti, E. Type I interferons as vaccine adjuvants against infectious diseases and cancer. Expert Rev. Vaccines 2008, 7, 373–381. [Google Scholar] [CrossRef]
- Abushahba, W.; Balan, M.; Castaneda, I.; Yuan, Y.; Reuhl, K.; Raveche, E.; de la Torre, A.; Lasfar, A.; Kotenko, S.V. Antitumor activity of type I and type III interferons in BNL hepatoma model. Cancer Immunol. Immunother. CII 2010, 59, 1059–1071. [Google Scholar] [CrossRef]
- Lasfar, A.; de laTorre, A.; Abushahba, W.; Cohen-Solal, K.A.; Castaneda, I.; Yuan, Y.; Reuhl, K.; Zloza, A.; Raveche, E.; Laskin, D.L.; Kotenko, S.V. Concerted action of IFN-α and IFN-λ induces local NK cell immunity and halts cancer growth. Oncotarget 2016, 7, 49259–49267. [Google Scholar] [CrossRef]
- Yu, R.; Zhu, B.; Chen, D. Type I interferon-mediated tumor immunity and its role in immunotherapy. Cell. Mol. Life Sci. CMLS 2022, 79, 191. [Google Scholar] [CrossRef]
- Saadeldin, M.K.; Abdel-Aziz, A.K.; Abdellatif, A. Dendritic cell vaccine immunotherapy; the beginning of the end of cancer and COVID-19. A hypothesis. Med. Hypotheses 2021, 146, 110365. [Google Scholar] [CrossRef] [PubMed]
- Santini, S.M.; Lapenta, C.; Santodonato, L.; D’Agostino, G.; Belardelli, F.; Ferrantini, M. IFN-alpha in the generation of dendritic cells for cancer immunotherapy. Handb. Exp. Pharmacol. 2009, 188, 295–317. [Google Scholar] [CrossRef]
- Katlinski, K.V.; Gui, J.; Katlinskaya, Y.V.; Ortiz, A.; Chakraborty, R.; Bhattacharya, S.; Carbone, C.J.; Beiting, D.P.; Girondo, M.A.; Peck, A.R.; et al. Inactivation of Interferon Receptor Promotes the Establishment of Immune Privileged Tumor Microenvironment. Cancer Cell 2017, 31, 194–207. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Klement, J.D.; Ibrahim, M.L.; Xiao, W.; Redd, P.S.; Nayak-Kapoor, A.; Zhou, G.; Liu, K. Type I interferon suppresses tumor growth through activating the STAT3-granzyme B pathway in tumor-infiltrating cytotoxic T lymphocytes. J. Immunother. Cancer 2019, 7, 157. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Mukherjee, R.; Peck, A.R.; Sun, Y.; McBrearty, N.; Katlinski, K.V.; Gui, J.; Govindaraju, P.K.; Puré, E.; Rui, H.; Fuchs, S.Y. Cancer-associated fibroblasts downregulate type I interferon receptor to stimulate intratumoral stromagenesis. Oncogene 2020, 39, 6129–6137. [Google Scholar] [CrossRef]
- Alicea-Torres, K.; Sanseviero, E.; Gui, J.; Chen, J.; Veglia, F.; Yu, Q.; Donthireddy, L.; Kossenkov, A.; Lin, C.; Fu, S.; et al. Immune suppressive activity of myeloid-derived suppressor cells in cancer requires inactivation of the type I interferon pathway. Nat. Commun. 2021, 12, 1717. [Google Scholar] [CrossRef] [PubMed]
- Odnokoz, O.; Yu, P.; Peck, A.R.; Sun, Y.; Kovatich, A.J.; Hooke, J.A.; Hu, H.; Mitchell, E.P.; Rui, H.; Fuchs, S.Y. Malignant cell-specific pro-tumorigenic role of type I interferon receptor in breast cancers. Cancer Biol. Ther. 2020, 21, 629–636. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald-Bocarsly, P.; Feng, D. The role of type I interferon production by dendritic cells in host defense. Biochimie 2007, 89, 843–855. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Mann-Nüttel, R.; Schulze, A.; Richter, L.; Alferink, J.; Scheu, S. Sources of Type I Interferons in Infectious Immunity: Plasmacytoid Dendritic Cells Not Always in the Driver’s Seat. Front. Immunol. 2019, 10, 778. [Google Scholar] [CrossRef] [PubMed]
- Matic, S.; Popovic, S.; Djurdjevic, P.; Todorovic, D.; Djordjevic, N.; Mijailovic, Z.; Baskic, D. SARS-CoV-2 infection induces mixed M1/M2 phenotype in circulating monocytes and alterations in both dendritic cell and monocyte subsets. PLoS ONE 2020, 15, e0241097. [Google Scholar] [CrossRef]
- Gigante, M.; Mandic, M.; Wesa, A.K.; Cavalcanti, E.; Dambrosio, M.; Mancini, V.; Battaglia, M.; Gesualdo, L.; Storkus, W.J.; Ranieri, E. Interferon-alpha (IFN-alpha)-conditioned DC preferentially stimulate type-1 and limit Treg-type in vitro T-cell responses from RCC patients. J. Immunother. 2008, 31, 254–262. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Su, X.Z.; Lu, F. The Roles of Type I Interferon in Co-infections with Parasites and Viruses, Bacteria, or Other Parasites. Front. Immunol. 2020, 11, 1805. [Google Scholar] [CrossRef] [PubMed]
- Carrero, J.A. Confounding roles for type I interferons during bacterial and viral pathogenesis. Int. Immunol. 2013, 25, 663–669. [Google Scholar] [CrossRef]
- Ferran, M.C.; Lucas-Lenard, J.M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J. Virol. 1997, 71, 371–377. [Google Scholar] [CrossRef] [PubMed]
- Marquis, K.A.; Becker, R.L.; Weiss, A.N.; Morris, M.C.; Ferran, M.C. The VSV matrix protein inhibits NF-κB and the interferon response independently in mouse L929 cells. Virology 2020, 548, 117–123. [Google Scholar] [CrossRef]
- Varble, A.J.; Ried, C.D.; Hammond, W.J.; Marquis, K.A.; Woodruff, M.C.; Ferran, M.C. The vesicular stomatitis virus matrix protein inhibits NF-κB activation in mouse L929 cells. Virology 2016, 499, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Dinh, P.X.; Panda, D.; Pattnaik, A.K. Interferon-inducible protein IFI35 negatively regulates RIG-I antiviral signaling and supports vesicular stomatitis virus replication. J. Virol. 2014, 88, 3103–3113. [Google Scholar] [CrossRef] [PubMed]
- Kotenko, S.V.; Rivera, A.; Parker, D.; Durbin, J.E. Type III IFNs: Beyond antiviral protection. Semin. Immunol. 2019, 43, 101303. [Google Scholar] [CrossRef]
- Meng, Z.; Wang, T.; Chen, L.; Chen, X.; Li, L.; Qin, X.; Li, H.; Luo, J. The Effect of Recombinant Human Interferon Alpha Nasal Drops to Prevent COVID-19 Pneumonia for Medical Staff in an Epidemic Area. Curr. Top. Med. Chem. 2021, 21. [Google Scholar] [CrossRef]
- Wang, N.; Zhan, Y.; Zhu, L.; Hou, Z.; Liu, F.; Song, P.; Qiu, F.; Wang, X.; Zou, X.; Wan, D.; et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell Host Microbe 2020, 28, 455–464.e2. [Google Scholar] [CrossRef]
- Sette, A.; Crotty, S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell 2021, 184, 861–880. [Google Scholar] [CrossRef] [PubMed]
- Müller, L.; Aigner, P.; Stoiber, D. Type I Interferons and Natural Killer Cell Regulation in Cancer. Front. Immunol. 2017, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gargan, S.; Roche, F.M.; Frieman, M.; Stevenson, N.J. Inhibition of the IFN-α JAK/STAT Pathway by MERS-CoV and SARS-CoV-1 Proteins in Human Epithelial Cells. Viruses 2022, 14, 667. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Cao, Z.; Xie, X.; Zhang, X.; Chen, J.Y.; Wang, H.; Menachery, V.D.; Rajsbaum, R.; Shi, P.Y. Evasion of Type I Interferon by SARS-CoV-2. Cell Rep. 2020, 33, 108234. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, C.; Lefeuvre, C.; Preisser, L.; Pivert, A.; Soleti, R.; Blanchard, S.; Delneste, Y.; Ducancelle, A.; Couez, D.; Jeannin, P. Age-Related Expression of IFN-λ1 Versus IFN-I and Beta-Defensins in the Nasopharynx of SARS-CoV-2-Infected Individuals. Front. Immunol. 2021, 12, 750279. [Google Scholar] [CrossRef]
- Mestecky, J. The common mucosal immune system and current strategies for induction of immune responses in external secretions. J. Clin. Immunol. 1987, 7, 265–276. [Google Scholar] [CrossRef]
- Mestecky, J.; McGhee, J.R. Prospects for human mucosal vaccines. Adv. Exp. Med. Biol. 1992, 327, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Chan, R.; Chan, K.; Lui, G.; Tsun, J.; Chan, K.; Yip, J.; Liu, S.; Yu, M.; Ng, R.; Chong, K.; Wang, M.H.; Chan, P.; Li, A.M.; Lam, H.S. Mucosal Antibody Response to SARS-CoV-2 in Paediatric and Adult Patients: A Longitudinal Study. Pathogens 2022, 11, 397. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Humphries, F.; Girardin, R.C.; Wallace, A.; Ejemel, M.; Amcheslavsky, A.; McMahon, C.T.; Schiller, Z.A.; Ma, Z.; Cruz, J.; et al. Mucosal nanobody IgA as inhalable and affordable prophylactic and therapeutic treatment against SARS-CoV-2 and emerging variants. Front. Immunol. 2022, 13, 995412. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Low, Z.Y.; Zabidi, N.Z.; Yip, A.; Puniyamurti, A.; Chow, V.; Lal, S.K. SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion. Viruses 2022, 14, 1991. [Google Scholar] [CrossRef] [PubMed]
- Khanmohammadi, S.; Rezaei, N.; Khazaei, M.; et al. A Case of Autosomal Recessive Interferon Alpha/Beta Receptor Alpha Chain (IFNAR1) Deficiency with Severe COVID-19. J Clin Immunol 2022, 42, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Shibabaw, T.; Molla, M.D.; Teferi, B.; Ayelign, B. Role of IFN and Complements System: Innate Immunity in SARS-CoV-2. J Inflamm Res. 2020, 13, 507–518. [Google Scholar] [CrossRef]
- Alunno, A.; Najm, A.; Mariette, X.; De Marco, G.; Emmel, J.; Mason, L.; McGonagle, D.G.; Machado, P.M. Immunomodulatory therapies for SARS-CoV-2 infection: A systematic literature review to inform EULAR points to consider. Ann. Rheum. Dis. 2021, 80, 803–815. [Google Scholar] [CrossRef]
- Setaro, A.C.; Gaglia, M.M. All hands on deck: SARS-CoV-2 proteins that block early anti-viral interferon responses. Curr. Res. Virol. Sci. 2021, 2, 100015. [Google Scholar] [CrossRef]
- Yuen, C.K.; Lam, J.Y.; Wong, W.M.; Mak, L.F.; Wang, X.; Chu, H.; Cai, J.P.; Jin, D.Y.; To, K.K.; Chan, J.F.; Yuen, K.Y.; Kok, K.H. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg. Microbes Infect. 2020, 9, 1418–1428. [Google Scholar] [CrossRef] [PubMed]
- Santerre, M.; Arjona, S.P.; Allen, C.N.; Shcherbik, N.; Sawaya, B.E. Why do SARS-CoV-2 NSPs rush to the ER? J. Neurol. 2021, 268, 2013–2022. [Google Scholar] [CrossRef]
- Hackstadt, T.; Chiramel, A.I.; Hoyt, F.H.; Williamson, B.N.; Dooley, C.A.; Beare, P.A.; de Wit, E.; Best, S.M.; Fischer, E.R. Disruption of the Golgi Apparatus and Contribution of the Endoplasmic Reticulum to the SARS-CoV-2 Replication Complex. Viruses 2021, 13, 1798. [Google Scholar] [CrossRef] [PubMed]
- Hui, K.; Cheung, M.C.; Perera, R.; Ng, K.C.; Bui, C.; Ho, J.; Ng, M.; Kuok, D.; Shih, K.C.; Tsao, S.W.; et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: An analysis in ex-vivo and in-vitro cultures. Lancet. Respir. Med. 2020, 8, 687–695. [Google Scholar] [CrossRef]
- Hossain, A.; Akter, S.; Rashid, A.A.; Khair, S.; Alam, A. Unique mutations in SARS-CoV-2 Omicron subvariants’ non-spike proteins: Potential impacts on viral pathogenesis and host immune evasion. Microb. Pathog. 2022, 170, 105699. [Google Scholar] [CrossRef]
- Grant, A.H.; Estrada, A., 3rd; Ayala-Marin, Y.M.; Alvidrez-Camacho, A.Y.; Rodriguez, G.; Robles-Escajeda, E.; Cadena-Medina, D.A.; Rodriguez, A.C.; Kirken, R.A. The Many Faces of JAKs and STATs Within the COVID-19 Storm. Front. Immunol. 2021, 12, 690477. [Google Scholar] [CrossRef] [PubMed]
- Minich, D.M.; Henning, M.; Darley, C.; Fahoum, M.; Schuler, C.B.; Frame, J. Is Melatonin the “Next Vitamin D”?: A Review of Emerging Science, Clinical Uses, Safety, and Dietary Supplements. Nutrients 2022, 14, 3934. [Google Scholar] [CrossRef] [PubMed]
- Smolensky, M.H.; Sackett-Lundeen, L.L.; Portaluppi, F. Nocturnal light pollution and underexposure to daytime sunlight: Complementary mechanisms of circadian disruption and related diseases. Chronobiol. Int. 2015, 32, 1029–1048. [Google Scholar] [CrossRef] [PubMed]
- Richardson, G.; Tate, B. Hormonal and pharmacological manipulation of the circadian clock: Recent developments and future strategies. Sleep 2000, 23 Suppl 3, S77–S85. [Google Scholar]
- Skene, D.J.; Deacon, S.; Arendt, J. Use of melatonin in circadian rhythm disorders and following phase shifts. Acta Neurobiol. Exp. 1996, 56, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Chattree, V.; Singh, K.; Singh, K.; Goel, A.; Maity, A.; Lone, A. A comprehensive review on modulation of SIRT1 signaling pathways in the immune system of COVID-19 patients by phytotherapeutic melatonin and epigallocatechin-3-gallate. Journal of food biochemistry 2022, e14259. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Quinones, Q.J.; de Ridder, G.G.; Pizzo, S.V. GRP78: A chaperone with diverse roles beyond the endoplasmic reticulum. Histol. Histopathol. 2008, 23, 1409–1416. [Google Scholar] [CrossRef]
- Carlos, A.J.; Ha, D.P.; Yeh, D.W.; Van Krieken, R.; Tseng, C.C.; Zhang, P.; Gill, P.; Machida, K.; Lee, A.S. The chaperone GRP78 is a host auxiliary factor for SARS-CoV-2 and GRP78 depleting antibody blocks viral entry and infection. J. Biol. Chem. 2021, 296, 100759. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Toyoda, S.; Fukuhara, A.; Shimomura, I. GRP78, a Novel Host Factor for SARS-CoV-2: The Emerging Roles in COVID-19 Related to Metabolic Risk Factors. Biomedicines 2022, 10, 1995. [Google Scholar] [CrossRef]
- Shin, J.; Toyoda, S.; Nishitani, S.; Fukuhara, A.; Kita, S.; Otsuki, M.; Shimomura, I. Possible Involvement of Adipose Tissue in Patients With Older Age, Obesity, and Diabetes With SARS-CoV-2 Infection (COVID-19) via GRP78 (BIP/HSPA5): Significance of Hyperinsulinemia Management in COVID-19. Diabetes 2021, 70, 2745–2755. [Google Scholar] [CrossRef]
- Ghasemitarei, M.; Privat-Maldonado, A.; Yusupov, M.; Rahnama, S.; Bogaerts, A.; Ejtehadi, M.R. Effect of Cysteine Oxidation in SARS-CoV-2 Receptor-Binding Domain on Its Interaction with Two Cell Receptors: Insights from Atomistic Simulations. J. Chem. Inf. Model. 2022, 62, 129–141. [Google Scholar] [CrossRef] [PubMed]
- Rayner, J.O.; Roberts, R.A.; Kim, J.; Poklepovic, A.; Roberts, J.L.; Booth, L.; Dent, P. AR12 (OSU-03012) suppresses GRP78 expression and inhibits SARS-CoV-2 replication. Biochem. Pharmacol. 2020, 182, 114227. [Google Scholar] [CrossRef] [PubMed]
- Booth, L.; Roberts, J.L.; Ecroyd, H.; Tritsch, S.R.; Bavari, S.; Reid, S.P.; Proniuk, S.; Zukiwski, A.; Jacob, A.; Sepúlveda, C.S.; Giovannoni, F.; García, C.C.; Damonte, E.; González-Gallego, J.; Tuñón, M.J.; Dent, P. AR-12 Inhibits Multiple Chaperones Concomitant With Stimulating Autophagosome Formation Collectively Preventing Virus Replication. J. Cell. Physiol. 2016, 231, 2286–2302. [Google Scholar] [CrossRef] [PubMed]
- Bayati, A.; Kumar, R.; Francis, V.; McPherson, P.S. SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis. J. Biol. Chem. 2021, 296, 100306. [Google Scholar] [CrossRef] [PubMed]
- Prabhakara, C.; Godbole, R.; Sil, P.; Jahnavi, S.; Gulzar, S.E.; van Zanten, T.S.; Sheth, D.; Subhash, N.; Chandra, A.; Shivaraj, A.; et al. Strategies to target SARS-CoV-2 entry and infection using dual mechanisms of inhibition by acidification inhibitors. PLoS Pathog. 2021, 17, e1009706. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, F.A. Hyperinflammation, apoptosis, and organ damage. Exp. Biol. Med. 2022, 247, 1112–1123. [Google Scholar] [CrossRef] [PubMed]
- Puhl, A.C.; Gomes, G.F.; Damasceno, S.; Fritch, E.J.; Levi, J.A.; Johnson, N.J.; Scholle, F.; Premkumar, L.; Hurst, B.L.; Lee-Montiel, F.; Veras, F.P.; Batah, S.S.; Fabro, A.T.; Moorman, N.J.; Yount, B.L.; Dickmander, R.J.; Baric, R.S.; Pearce, K.H.; Cunha, F.Q.; Alves-Filho, J.C.; Ekins, S. Vandetanib Blocks the Cytokine Storm in SARS-CoV-2-Infected Mice. ACS Omega 2022, 7, 31935–31944. [Google Scholar] [CrossRef] [PubMed]
- Romano, M.; Ruggiero, A.; Squeglia, F.; Maga, G.; Berisio, R. A Structural View of SARS-CoV-2 RNA Replication Machinery: RNA Synthesis, Proofreading and Final Capping. Cells 2020, 9, 1267. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.S.; Xiang, X.; Wang, X.; Wang, Z.H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020, 11, 1061. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, V.; Shannon, C.P.; Wei, X.S.; Xiang, X.; Wang, X.; Wang, Z.H.; Tebbutt, S.J.; Kollmann, T.R.; Fish, E.N. Corrigendum: Interferon-α2b Treatment for COVID-19. Front. Immunol. 2020, 11, 615275. [Google Scholar] [CrossRef]
- Zhou, Q.; MacArthur, M.R.; He, X.; Wei, X.; Zarin, P.; Hanna, B.S.; Wang, Z.H.; Xiang, X.; Fish, E.N. Interferon-α2b Treatment for COVID-19 Is Associated with Improvements in Lung Abnormalities. Viruses 2020, 13, 44. [Google Scholar] [CrossRef]
- Pereda, R.; González, D.; Rivero, H.B.; Rivero, J.C.; Pérez, A.; López, L.; Mezquia, N.; Venegas, R.; Betancourt, J.R.; Domínguez, R.E. Therapeutic Effectiveness of Interferon-α2b Against COVID-19: The Cuban Experience. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2020, 40, 438–442. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Yu, S.; Chen, Q.; Duan, Z.; Zhou, J.; Mao, C.; Yu, D.; Zhu, W.; Nie, J.; Hou, Y. A randomized controlled trial of low-dose recombinant human interferons alpha-2b nasal spray to prevent acute viral respiratory infections in military recruits. Vaccine 2010, 28, 4445–4451. [Google Scholar] [CrossRef] [PubMed]
- Bennett, A.L.; Smith, D.W.; Cummins, M.J.; Jacoby, P.A.; Cummins, J.M.; Beilharz, M.W. Low-dose oral interferon alpha as prophylaxis against viral respiratory illness: A double-blind, parallel controlled trial during an influenza pandemic year. Influenza Other Respir. Viruses 2013, 7, 854–862. [Google Scholar] [CrossRef]
- Lee, A.C.; Jeong, Y.; Lee, S.; Jang, H.; Zheng, A.; Kwon, S.; Repine, J.E. Repine (19/05/2021), Nasopharyngeal Type I-Interferon for Immediately Available Prophylaxis Against Emerging Respiratory Viral Infections, Front. Immunol. 2021, 12, 660298. [Google Scholar] [CrossRef]
- Alibek, K.; Tskhay, A. Ahead of a vaccine: A safe method of protection against COVID-19 exists. Res. Ideas Outcomes 2020, 6, e61709. [Google Scholar] [CrossRef]
- Sodeifian, F.; Nikfarjam, M.; Kian, N.; Mohamed, K.; Rezaei, N. The role of type I interferon in the treatment of COVID-19. J Med Virol. 2021, 94, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Qiong Zhou, Virginia Chen; et al. Interferon-a2b Treatment for COVID-19. Front. Immunol. 2020, 11, 1601. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Wu, A.; Xu, S.; Pan, R.; Zeng, C.; Jin, X.; Ge, X.; Shi, Z.; Ahola, T.; Chen, Y.; Guo, D. Coronavirus nsp10/nsp16 Methyltransferase Can Be Targeted by nsp10-Derived Peptide In Vitro and In Vivo To Reduce Replication and Pathogenesis. J. Virol. 2015, 89, 8416–8427. [Google Scholar] [CrossRef]
- Mahalapbutr, P.; Kongtaworn, N.; Rungrotmongkol, T. Structural insight into the recognition of S-adenosyl-L-homocysteine and sinefungin in SARS-CoV-2 Nsp16/Nsp10 RNA cap 2′-O-Methyltransferase. Comput. Struct. Biotechnol. J. 2020, 18, 2757–2765. [Google Scholar] [CrossRef]
- Nencka, R.; Silhan, J.; Klima, M.; Otava, T.; Kocek, H.; Krafcikova, P.; Boura, E. Coronaviral RNA-methyltransferases: Function, structure and inhibition. Nucleic Acids Res. 2022, 50, 635–650. [Google Scholar] [CrossRef] [PubMed]
- Saliu, T.P.; Umar, H.I.; Ogunsile, O.J.; Okpara, M.O.; Yanaka, N.; Elekofehinti, O.O. Molecular docking and pharmacokinetic studies of phytocompounds from nigerian medicinal plants as promising inhibitory agents against SARS-CoV-2 methyltransferase (nsp16). J. Genet. Eng. Biotechnol. 2021, 19, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Ehrhardt, C.; Wolff, T.; Pleschka, S.; Planz, O.; Beermann, W.; Bode, J.G.; Schmolke, M.; Ludwig, S. Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J. Virol. 2007, 81, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Choi, J.H.; Dai, D.L.; Luo, J.; Ladak, R.J.; Li, Q.; Wang, Y.; Zhang, C.; Wiebe, S.; Liu, A.C.H.; et al. SARS-CoV-2 impairs interferon production via NSP2-induced repression of mRNA translation. Proc. Natl. Acad. Sci. USA 2022, 119, e2204539119. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, T.; Fukushi, S.; Saijo, M.; Kurane, I.; Morikawa, S. Importance of Akt signaling pathway for apoptosis in SARS-CoV-infected Vero E6 cells. Virology 2004, 327, 169–174. [Google Scholar] [CrossRef] [PubMed]
- Yoo, J.S.; Sasaki, M.; Cho, S.X.; et al. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat Commun 2021, 12, 6602. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Cao, Y.; Shah, A.U.; Zuo, J.; Zhang, S.; Yu, Q.; Chong, M.; Yang, Q. Inhibition of the antigen-presenting ability of dendritic cells by non-structural protein 2 of influenza A virus. Vet. Microbiol. 2022, 267, 109392. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Han, Q. Oral Methioninase for Covid-19 Methionine-restriction Therapy. Vivo 2020, 34 (Suppl. S3), 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- McCullough, P.A.; Kelly, R.J.; Ruocco, G.; Lerma, E.; Tumlin, J.; Wheelan, K.R.; Katz, N.; Lepor, N.E.; Vijay, K.; Carter, H.; et al. Pathophysiological basis and rationale for early outpatient treatment of SARS-CoV-2 (COVID-19) Infection. Am J Med 2021, 134, 16–22. [Google Scholar] [CrossRef]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11, e01928-20. [Google Scholar] [CrossRef]
- Aiman, A.; Basir, S.F.; Islam, A. (2022). Interferons Horizon Therapeutics. In (Ed.), Basic and Clinical Aspects of Interferon Gamma. IntechOpen. [CrossRef]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020, 94, e00985-20. [Google Scholar] [CrossRef] [PubMed]
- Felgenhauer, U.; Schoen, A.; Gad, H.H.; Hartmann, R.; Schaubmar, A.R.; Failing, K.; Drosten, C.; Weber, F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020, 295, 13958–13964. [Google Scholar] [CrossRef] [PubMed]
- Sodeifian, F.; Nikfarjam, M.; Kian, N.; Mohamed, K.; Rezaei, N. The role of type I interferon in the treatment of COVID-19. J. Med. Virol. 2022, 94, 63–81. [Google Scholar] [CrossRef] [PubMed]
- Barros-Martins, J.; Förster, R.; Bošnjak, B. NK cell dysfunction in severe COVID-19: TGF-β-induced downregulation of integrin beta-2 restricts NK cell cytotoxicity. Sig Transduct Target Ther 2022, 7, 32. [Google Scholar] [CrossRef] [PubMed]
- Fazeli, M.R.; Hezarjaribi, N. A Simplified Process for Purification and Refolding of Recombinant Human Interferon-α2b. Iran. Biomed. J. 2022, 26, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Ma, L.L.; Jiang, J.D.; Pang, R.; Chen, Y.J.; Li, Y.H. Yao Xue Xue Bao = Acta Pharm. Sin. 2014, 49, 1547–1553, Available online: https://pubmed.ncbi.nlm.nih.gov/25757280/.
- Malik, G.; Zhou, Y. Innate Immune Sensing of Influenza A Virus. Viruses 2020, 12, 755. [Google Scholar] [CrossRef] [PubMed]
- Saito, H.; Takenaka, H.; Yoshida, S.; Tsubokawa, T.; Ogata, A.; Imanishi, F.; Imanishi, J. Prevention from naturally acquired viral respiratory infection by interferon nasal spray. Rhinology 1985, 23, 291–295. [Google Scholar]
- Brune, J.E.; Chang, M.Y.; Altemeier, W.A.; Frevert, C.W. Type I Interferon Signaling Increases Versican Expression and Synthesis in Lung Stromal Cells During Influenza Infection. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2021, 69, 691–709. [Google Scholar] [CrossRef]
- Mordstein, M.; Neugebauer, E.; Ditt, V.; Jessen, B.; Rieger, T.; Falcone, V.; Sorgeloos, F.; Ehl, S.; Mayer, D.; Kochs, G.; et al. Lambda interferon renders epithelial cells of the respiratory and gastrointestinal tracts resistant to viral infections. J. Virol. 2010, 84, 5670–5677. [Google Scholar] [CrossRef]
- Klinkhammer, J.; Schnepf, D.; Ye, L.; Schwaderlapp, M.; Gad, H.H.; Hartmann, R.; Garcin, D.; Mahlakõiv, T.; Staeheli, P. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 2018, 7, e33354. [Google Scholar] [CrossRef] [PubMed]
- Mordstein, M.; Kochs, G.; Dumoutier, L.; Renauld, J.C.; Paludan, S.R.; Klucher, K.; Staeheli, P. Interferon-lambda contributes to innate immunity of mice against influenza A virus but not against hepatotropic viruses. PLoS Pathog. 2008, 4, e1000151. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.Y.; Boisson-Dupuis, S.; Chapgier, A.; Yang, K.; Bustamante, J.; Puel, A.; Picard, C.; Abel, L.; Jouanguy, E.; Casanova, J.L. Inborn errors of interferon (IFN)-mediated immunity in humans: Insights into the respective roles of IFN-alpha/beta, IFN-gamma, and IFN-lambda in host defense. Immunol. Rev. 2008, 226, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Daza-Cajigal, V.; Albuquerque, A.S.; Young, D.F.; Ciancanelli, M.J.; Moulding, D.; Angulo, I.; Jeanne-Julien, V.; Rosain, J.; Minskaia, E.; Casanova, J.L.; et al. Partial human Janus kinase 1 deficiency predominantly impairs responses to interferon gamma and intracellular control of mycobacteria. Front. Immunol. 2022, 13, 888427. [Google Scholar] [CrossRef] [PubMed]
- Gill, N.; Deacon, P.M.; Lichty, B.; Mossman, K.L.; Ashkar, A.A. Induction of innate immunity against herpes simplex virus type 2 infection via local delivery of Toll-like receptor ligands correlates with beta interferon production. J. Virol. 2006, 80, 9943–9950. [Google Scholar] [CrossRef] [PubMed]
- Harandi, A.M. The potential of immunostimulatory CpG DNA for inducing immunity against genital herpes: Opportunities and challenges. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2004, 30, 207–210. [Google Scholar] [CrossRef] [PubMed]
- McKellar, J.; Rebendenne, A.; Wencker, M.; Moncorgé, O.; Goujon, C. Mammalian and Avian Host Cell Influenza A Restriction Factors. Viruses 2021, 13, 522. [Google Scholar] [CrossRef] [PubMed]
- Giotis, E.S.; Robey, R.C.; Skinner, N.G.; Tomlinson, C.D.; Goodbourn, S.; Skinner, M.A. Chicken interferome: Avian interferon-stimulated genes identified by microarray and RNA-seq of primary chick embryo fibroblasts treated with a chicken type I interferon (IFN-α). Vet. Res. 2016, 47, 75. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jiang, Y.; Jiao, P.; Wang, A.; Zhao, F.; Tian, G.; Wang, X.; Yu, K.; Bu, Z.; Chen, H. The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J. Virol. 2006, 80, 11115–11123. [Google Scholar] [CrossRef]
- Safadi, D.E.; Lebeau, G.; Lagrave, A.; Mélade, J.; Grondin, L.; Rosanaly, S.; Begue, F.; Hoareau, M.; Veeren, B.; Roche, M.; et al. Extracellular Vesicles Are Conveyors of the NS1 Toxin during Dengue Virus and Zika Virus Infection. Viruses 2023, 15, 364. [Google Scholar] [CrossRef]
- Nogales, A.; Villamayor, L.; Utrilla-Trigo, S.; Ortego, J.; Martinez-Sobrido, L.; DeDiego, M.L. Natural Selection of H5N1 Avian Influenza A Viruses with Increased PA-X and NS1 Shutoff Activity. Viruses 2021, 13, 1760. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, G.; Zhang, H.; Xin, G.; Zhang, D.; Zeng, J.; Chen, X.; Xu, Y.; Cui, Y.; Li, K. Effects of NS1 variants of H5N1 influenza virus on interferon induction, TNFalpha response and p53 activity. Cell. Mol. Immunol. 2010, 7, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Tian, G.; Li, Y.; Deng, G.; Jiang, Y.; Liu, C.; Liu, W.; Bu, Z.; Kawaoka, Y.; Chen, H. A single-amino-acid substitution in the NS1 protein changes the pathogenicity of H5N1 avian influenza viruses in mice. J. Virol. 2008, 82, 1146–1154. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Zhu, J.; Tu, J.; Zou, W.; Hu, Y.; Yu, Z.; Yin, W.; Li, Y.; Zhang, A.; Wu, Y.; et al. Effect on virulence and pathogenicity of H5N1 influenza A virus through truncations of NS1 eIF4GI binding domain. J. Infect. Dis. 2010, 202, 1338–1346. [Google Scholar] [CrossRef] [PubMed]
- Haasbach, E.; Droebner, K.; Vogel, A.B.; Planz, O. Low-dose interferon Type I treatment is effective against H5N1 and swine-origin H1N1 influenza A viruses in vitro and in vivo. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2011, 31, 515–525. [Google Scholar] [CrossRef] [PubMed]
- DeDiego, M.L.; Nogales, A.; Lambert-Emo, K.; Martinez-Sobrido, L.; Topham, D.J. NS1 Protein Mutation I64T Affects Interferon Responses and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2016, 90, 9693–9711. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; Martinez-Sobrido, L.; Topham, D.J.; DeDiego, M.L. NS1 Protein Amino Acid Changes D189N and V194I Affect Interferon Responses, Thermosensitivity, and Virulence of Circulating H3N2 Human Influenza A Viruses. J. Virol. 2017, 91, e01930-16. [Google Scholar] [CrossRef] [PubMed]
- Vandoorn, E.; Stadejek, W.; Parys, A.; Chepkwony, S.; Chiers, K.; Van Reeth, K. Pathobiology of an NS1-Truncated H3N2 Swine Influenza Virus Strain in Pigs. J. Virol. 2022, 96, e0051922. [Google Scholar] [CrossRef]
- Iverson, E.; Griswold, K.; Song, D.; Gagliardi, T.B.; Hamidzadeh, K.; Kesimer, M.; Sinha, S.; Perry, M.; Duncan, G.A.; Scull, M.A. Membrane-Tethered Mucin 1 Is Stimulated by Interferon and Virus Infection in Multiple Cell Types and Inhibits Influenza A Virus Infection in Human Airway Epithelium. mBio 2022, 13, e0105522. [Google Scholar] [CrossRef]
- Malur, M.; Gale, M., Jr.; Krug, R.M. LGP2 downregulates interferon production during infection with seasonal human influenza A viruses that activate interferon regulatory factor 3. J. Virol. 2012, 86, 10733–10738. [Google Scholar] [CrossRef]
- Smith, B.L.; Chen, G.; Wilke, C.O.; Krug, R.M. Avian Influenza Virus PB1 Gene in H3N2 Viruses Evolved in Humans To Reduce Interferon Inhibition by Skewing Codon Usage toward Interferon-Altered tRNA Pools. mBio 2018, 9, e01222-18. [Google Scholar] [CrossRef] [PubMed]
- Solov’ev, V.D. The results of controlled observations on the prophylaxis of influenza with interferon. Bull. World Health Organ. 1969, 41, 683–688. [Google Scholar] [PubMed]
- Jordan, W.S.; Hopps, H.E.; Merigan, T.C. Influenza and Interferon Research in the Soviet Union: January 1973. J. Infect. Dis. 1973, 128, 261–264. [Google Scholar] [CrossRef]
- Sun, X.; Belser, J.A.; Maines, T.R. Adaptation of H9N2 Influenza Viruses to Mammalian Hosts: A Review of Molecular Markers. Viruses 2020, 12, 541. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E.; Neyts, J. Avian influenza A (H5N1) infection: Targets and strategies for chemotherapeutic intervention. Trends Pharmacol. Sci. 2007, 28, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Robinson, W.A.; Mughal, T.I.; Thomas, M.R.; Johnson, M.; Spiegel, R.J. Treatment of Metastatic Malignant Melanoma with Recombinant Interferon Alpha 2. Immunobiology 1986, 172, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Sheng, L.; Chen, X.; Wang, Q.; Lyu, S.; Li, P. Interferon-α2b enhances survival and modulates transcriptional profiles and the immune response in melanoma patients treated with dendritic cell vaccines. Biomed. Pharmacother. Biomed. Pharmacother. 2020, 125, 109966. [Google Scholar] [CrossRef] [PubMed]
- Di Trolio, R.; Simeone, E.; Di Lorenzo, G.; Grimaldi, A.M.; Romano, A.; Ayala, F.; Caracò, C.; Mozzillo, N.; Ascierto, P.A. Update on PEG-interferon α-2b as adjuvant therapy in melanoma. Anticancer Res. 2012, 32, 3901–3909. [Google Scholar] [PubMed]
- Kim, Y.M.; Shin, E.C. Type I and III interferon responses in SARS-CoV-2 infection. Exp. Mol. Med. 2021, 53, 750–760. [Google Scholar] [CrossRef]
- Lee, J.S.; Shin, E.C. The type I interferon response in COVID-19: Implications for treatment. Nat Rev Immunol 2020, 20, 585–586. [Google Scholar] [CrossRef]
- Lyudmila Shalamova, Ulrike Felgenhauer, Andreas R. Schaubmar, Kathrin Buettner, Marek Widera, Sandra Ciesek, Friedemann Weber (2022), Omicron variant of SARS-CoV-2 exhibits an increased resilience to the antiviral type I interferon response. [CrossRef]
- Daniel, B. Stetson and Rusian Medzhitov. Type I Interferons Host Defense. Immun. 2006, 25, 373–381. [Google Scholar] [CrossRef]
- Emily Mantlo, Natalya Bukreyeva, Junki Maruyama; et al. Antiviral Activities of Type I interferons to SARS-CoV-2 Infection, J. Antivir. Resist. 2020, 178, 104811. [CrossRef]
- Zhao, J.; Zhu, F.C.; Shu, Y.L.; Zhou, R.; Liu, L.Q.; Zhang, L.L.; Shi, Z.Y.; Tang, Z.; Lin, L.Z.; Yu, Z.A.; Zhang, L.P.; Zhang, B.; Hou, Y.D. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi = Zhonghua Shiyan He Linchuang Bingduxue Zazhi Chin. J. Exp. Clin. Virol. 2005, 19, 220–222.
- Yu, D.X.; Chen, Q.; Zhang, L.L.; Liu, Y.; Yu, Z.A.; Li, Z.F.; Zhang, L.P.; Hu, G.F.; Duan, Z.J.; Chu, X.W.; et al. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi = Zhonghua Shiyan He Linchuang Bingduxue Zazhi = Chin. J. Exp. Clin. Virol. 2005, 19, 216–219.
- Tannock, G.A.; Gillett, S.M.; Gillett, R.S.; Barry, R.D.; Hensley, M.J.; Herd, R.; Reid, A.L.; Saunders, N.A. A study of intranasally administered interferon A (rIFN-alpha 2A) for the seasonal prophylaxis of natural viral infections of the upper respiratory tract in healthy volunteers. Epidemiol. Infect. 1988, 101, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Birkhoff, M.; Leitz, M.; Marx, D. Advantages of Intranasal Vaccination and Considerations on Device Selection. Indian J. Pharm. Sci. 2009, 71, 729–731. [Google Scholar]
- Islam, A.; Khan, M.A.; Ahmed, R.; Hossain, M.S.; Kabir, S.; Islam, M.S.; Siddiki, A. Transcriptome of nasopharyngeal samples from COVID-19 patients and a comparative analysis with other SARS-CoV-2 infection models reveal disparate host responses against SARS-CoV-2. Journal of translational medicine 2021, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- Alexander, M.R.; Brice, A.M.; Jansen van Vuren, P.; Rootes, C.L.; Tribolet, L.; Cowled, C.; Bean, A.; Stewart, C.R. Ribosome-Profiling Reveals Restricted Post Transcriptional Expression of Antiviral Cytokines and Transcription Factors during SARS-CoV-2 Infection. Int. J. Mol. Sci. 2021, 22, 3392. [Google Scholar] [CrossRef] [PubMed]
- Yang, Q.; Lin, F.; Wang, Y.; Zeng, M.; Luo, M. Long Noncoding RNAs as Emerging Regulators of COVID-19. Frontiers in immunology 2021, 12, 700184. [Google Scholar] [CrossRef]
- Vishnubalaji, R.; Shaath, H.; Alajez, N.M. Protein Coding and Long Noncoding RNA (lncRNA) Transcriptional Landscape in SARS-CoV-2 Infected Bronchial Epithelial Cells Highlight a Role for Interferon and Inflammatory Response. Genes 2020, 11, 760. [Google Scholar] [CrossRef]
- Herder, V.; Dee, K.; Wojtus, J.K.; Epifano, I.; Goldfarb, D.; Rozario, C.; Gu, Q.; Da Silva Filipe, A.; Nomikou, K.; Nichols, J.; et al. Elevated temperature inhibits SARS-CoV-2 replication in respiratory epithelium independently of IFN-mediated innate immune defenses. PLoS Biol. 2021, 19, e3001065. [Google Scholar] [CrossRef] [PubMed]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020, 94, e00985-20. [Google Scholar] [CrossRef] [PubMed]
- Alfi, O.; Yakirevitch, A.; Wald, O.; Wandel, O.; Izhar, U.; Oiknine-Djian, E.; Nevo, Y.; Elgavish, S.; Dagan, E.; Madgar, O.; et al. Human Nasal and Lung Tissues Infected Ex Vivo with SARS-CoV-2 Provide Insights into Differential Tissue-Specific and Virus-Specific Innate Immune Responses in the Upper and Lower Respiratory Tract. J. Virol. 2021, 95, e0013021. [Google Scholar] [CrossRef] [PubMed]
- Kouwaki, T.; Nishimura, T.; Wang, G.; Oshiumi, H. RIG-I-Like Receptor-Mediated Recognition of Viral Genomic RNA of Severe Acute Respiratory Syndrome Coronavirus-2 and Viral Escape From the Host Innate Immune Responses. Front. Immunol. 2021, 12, 700926. [Google Scholar] [CrossRef] [PubMed]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11, e01928-20. [Google Scholar] [CrossRef] [PubMed]
- Park, A.; Iwasaki, A. Type I and Type III Interferons—Induction, Signaling, Evasion, and Application to Combat COVID-19. Cell Host Microbe 2020, 27, 870–878. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, C.; Zhao, W. Virus Caused Imbalance of Type I IFN Responses and Inflammation in COVID-19. Front. Immunol. 2021, 12, 633769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, S.; Jin, Y.; Ji, W.; Zhang, W.; Duan, G. An Update on Innate Immune Responses during SARS-CoV-2 Infection. Viruses 2021, 13, 2060. [Google Scholar] [CrossRef] [PubMed]
- Frumholtz, L.; Bouaziz, J.D.; Battistella, M.; Hadjadj, J.; Chocron, R.; Bengoufa, D.; Le Buanec, H.; Barnabei, L.; Meynier, S.; Schwartz, O.; et al. Saint-Louis CORE (COVID Research) Type I interferon response and vascular alteration in chilblain-like lesions during the COVID-19 outbreak. Br. J. Dermatol. 2021, 185, 1176–1185. [Google Scholar] [CrossRef]
- Yamada, T.; Sato, S.; Sotoyama, Y.; Orba, Y.; Sawa, H.; Yamauchi, H.; Sasaki, M.; Takaoka, A. RIG-I triggers a signaling-abortive anti-SARS-CoV-2 defense in human lung cells. Nat. Immunol. 2021, 22, 820–828. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Nemati, M.; Saha, B.; Bansode, Y.D.; Jafarzadeh, S. Protective Potentials of Type III Interferons in COVID-19 Patients: Lessons from Differential Properties of Type I- and III Interferons. Viral Immunol. 2021, 34, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Tannock, G.A.; Gillett, S.M.; Gillett, R.S.; Barry, R.D.; Hensley, M.J.; Herd, R.; Reid, A.L.; Saunders, N.A. A study of intranasally administered interferon A (rIFN-alpha 2A) for the seasonal prophylaxis of natural viral infections of the upper respiratory tract in healthy volunteers. Epidemiol. Infect. 1988, 101, 611–621. [Google Scholar] [CrossRef]
- Birkhoff, M.; Leitz, M.; Marx, D. Advantages of Intranasal Vaccination and Considerations on Device Selection. Indian J. Pharm. Sci. 2009, 71, 729–731. [Google Scholar]
- Amanda, J. Lee and Ali A. Ashkar. The Dual Nature of Type I and Type II Interferons. Frontline Immunol. 2018. [Google Scholar] [CrossRef]
- Lv, L.; Cao, M.; Bai, J.; Jin, L.; Wang, X.; Gao, Y.; Liu, X.; Jiang, P. PRV-encoded UL13 protein kinase acts as an antagonist of innate immunity by targeting IRF3-signaling pathways. Vet. Microbiol. 2020, 250, 108860. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Romero, N.; Favoreel, H.W. Pseudorabies Virus Inhibits Type I and Type III Interferon-Induced Signaling via Proteasomal Degradation of Janus Kinases. J. Virol. 2021, 95, e0079321. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, Y.; Qin, C.; Lang, Y.; Xu, A.; Yu, C.; Zhao, Z.; Zhang, R.; Yang, J.; Tang, J. Pseudorabies Virus EP0 Antagonizes the Type I Interferon Response via Inhibiting IRF9 Transcription. J. Virol. 2022, 96, e0217121. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; Jiang, Z.; Tang, J. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef]
- Schreiber, G. The Role of Type I Interferons in the Pathogenesis and Treatment of COVID-19. Front. Immunol. 2020, 11, 595739. [Google Scholar] [CrossRef]
- Tonutti, A.; Motta, F.; Ceribelli, A.; Isailovic, N.; Selmi, C.; De Santis, M. Anti-MDA5 Antibody Linking COVID-19, Type I Interferon, and Autoimmunity: A Case Report and Systematic Literature Review. Front. Immunol. 2022, 13, 937667. [Google Scholar] [CrossRef]
- Anjum, F.R.; Anam, S.; Abbas, G.; Mahmood, M.S.; Rahman, S.U.; Goraya, M.U.; Abdullah, R.M.; Luqman, M.; Ali, A.; Akram, M.K.; Chaudhry, T.H. Type I IFNs: A Blessing in Disguise or Partner in Crime in MERS-CoV-, SARS-CoV-, and SARS-CoV-2-Induced Pathology and Potential Use of Type I IFNs in Synergism with IFN-γ as a Novel Antiviral Approach Against COVID-19. Viral Immunol. 2021, 34, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Karlowitz, R.; Stanifer, M.L.; Roedig, J.; Andrieux, G.; Bojkova, D.; Bechtel, M.; Smith, S.; Kowald, L.; Schubert, R.; Boerries, M.; et al. USP22 controls type III interferon signaling and SARS-CoV-2 infection through activation of STING. Cell Death Dis. 2022, 13, 684. [Google Scholar] [CrossRef] [PubMed]
- Felgenhauer, U.; Schoen, A.; Gad, H.H.; Hartmann, R.; Schaubmar, A.R.; Failing, K.; Drosten, C.; Weber, F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020, 295, 13958–13964. [Google Scholar] [CrossRef] [PubMed]
- Metz-Zumaran, C.; Kee, C.; Doldan, P.; Guo, C.; Stanifer, M.L.; Boulant, S. Increased Sensitivity of SARS-CoV-2 to Type III Interferon in Human Intestinal Epithelial Cells. J. Virol. 2022, 96, e0170521. [Google Scholar] [CrossRef]
- Sampaio, N.G.; Chauveau, L.; Hertzog, J.; Bridgeman, A.; Fowler, G.; Moonen, J.P.; Dupont, M.; Russell, R.A.; Noerenberg, M.; Rehwinkel, J. The RNA sensor MDA5 detects SARS-CoV-2 infection. Sci. Rep. 2021, 11, 13638. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Riva, L.; Pu, Y.; Martin-Sancho, L.; Kanamune, J.; Yamamoto, Y.; Sakai, K.; Gotoh, S.; Miorin, L.; De Jesus, P.D.; et al. MDA5 Governs the Innate Immune Response to SARS-CoV-2 in Lung Epithelial Cells. Cell Rep. 2021, 34, 108628. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zhuang, M.W.; Deng, J.; Zheng, Y.; Zhang, J.; Nan, M.L.; Zhang, X.J.; Gao, C.; Wang, P.H. SARS-CoV-2 ORF9b antagonizes type I and III interferons by targeting multiple components of the RIG-I/MDA-5-MAVS, TLR3-TRIF, and cGAS-STING signaling pathways. J. Med. Virol. 2021, 93, 5376–5389. [Google Scholar] [CrossRef]
- Zheng, Y.; Zhuang, M.W.; Han, L.; Zhang, J.; Nan, M.L.; Zhan, P.; Kang, D.; Liu, X.; Gao, C.; Wang, P.H. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct. Target. Ther. 2020, 5, 299. [Google Scholar] [CrossRef]
- Oh, S.J.; Shin, O.S. SARS-CoV-2 Nucleocapsid Protein Targets RIG-I-Like Receptor Pathways to Inhibit the Induction of Interferon Response. Cells 2021, 10, 530. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, J.; Han, L.; Zhuang, M.W.; Xu, Y.; Zhang, J.; Nan, M.L.; Xiao, Y.; Zhan, P.; Liu, X.; Gao, C.; Wang, P.H. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 2022, 7, 22. [Google Scholar] [CrossRef]
- Chen, A.; et al. (2021) Chitinase-3-like 1 protein complexes modulate macrophage-mediated immune suppression in glioblastoma. Journal of Clinical Investigation, 131. [CrossRef]
- Fukuda, Y.; Homma, T.; Inoue, H.; Onitsuka, C.; Ikeda, H.; Goto, Y.; Sato, Y.; Kimura, T.; Hirai, K.; Ohta, S.; et al. Downregulation of type III interferons in patients with severe COVID-19. J. Med. Virol. 2021, 93, 4559–4563. [Google Scholar] [CrossRef]
- Jafarzadeh, A.; Nemati, M.; Saha, B.; Bansode, Y.D.; Jafarzadeh, S. Protective Potentials of Type III Interferons in COVID-19 Patients: Lessons from Differential Properties of Type I- and III Interferons. Viral Immunol. 2021, 34, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Shin, E.C. Roles of Type I and III Interferons in COVID-19. Yonsei Med. J. 2021, 62, 381–390. [Google Scholar] [CrossRef] [PubMed]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11, e01928-20. [Google Scholar] [CrossRef]
- Choudhary, A. Immune regulatory function of interferon-gamma in acute leukemia. Indian J. Appl. Res. 2021, 75–77. [Google Scholar] [CrossRef]
- Guo, Y.; Yang, C.; Liu, Y.; Li, T.; Li, H.; Han, J.; Jia, L.; Wang, X.; Zhang, B.; Li, J.; Li, L. High Expression of HERV-K (HML-2) Might Stimulate Interferon in COVID-19 Patients. Viruses 2022, 14, 996. [Google Scholar] [CrossRef]
- Farooq, M.; Khan, A.W.; Ahmad, B.; Kim, M.S.; Choi, S. Therapeutic Targeting of Innate Immune Receptors Against SARS-CoV-2 Infection. Front. Pharmacol. 2022, 13, 915565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, S.; Liang, J.Q. Transcriptional and reverse transcriptional regulation of host genes by human endogenous retroviruses in cancers. Front. Microbiol. 2022, 13, 946296. [Google Scholar] [CrossRef]
- Civril, F.; Deimling, T.; de Oliveira Mann, C.; et al. Structural mechanism of cytosolic DNA sensing by cGAS. Nature 2013, 498, 332–337. [Google Scholar] [CrossRef]
- Ablasser, A.; Bauernfeind, F.; Hartmann, G.; et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III–transcribed RNA intermediate. Nat Immunol 2009, 10, 1065–1072. [Google Scholar] [CrossRef]
- Webb, L.G.; Fernandez-Sesma, A. RNA viruses and the cGAS-STING pathway: Reframing our understanding of innate immune sensing. Curr. Opin. Virol. 2022, 53, 101206. [Google Scholar] [CrossRef] [PubMed]
- Pippig, D.A.; Hellmuth, J.C.; Cui, S.; Kirchhofer, A.; Lammens, K.; Lammens, A.; Schmidt, A.; Rothenfusser, S.; Hopfner, K.-P. The regulatory domain of the RIG-I family ATPase LGP2 senses double-stranded RNA. Nucleic Acids Res. 2009, 37, 2014–2025. [Google Scholar] [CrossRef]
- Zhang, Q.; Yoo, D. Immune evasion of porcine enteric coronaviruses and viral modulation of antiviral innate signaling. Virus Res. 2016, 226, 128–141. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Wang, C.; Xue, M.; Fu, F.; Zhang, X.; Li, L.; Yin, L.; Xu, W.; Feng, L.; Liu, P. The Coronavirus Transmissible Gastroenteritis Virus Evades the Type I Interferon Response through IRE1α-Mediated Manipulation of the MicroRNA miR-30a-5p/SOCS1/3 Axis. J. Virol. 2018, 92, e00728-18. [Google Scholar] [CrossRef] [PubMed]
- Gal-Ben-Ari, S.; Barrera, I.; Ehrlich, M.; Rosenblum, K. PKR: A kinase to remember. Front. Mol. Neurosci. 2019, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Shan, L.; Qu, S.; Xue, M.; Wang, K.; Fu, F.; Wang, L.; Wang, Z.; Feng, L.; Xu, W.; Liu, P. The Coronavirus PEDV Evades Type III Interferon Response Through the miR-30c-5p/SOCS1 Axis. Front. Microbiol. 2020, 11, 1180. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Tian, M.; Fu, X. Reduced expression of miR-30c-5p promotes hepatocellular carcinoma progression by targeting RAB32. Molecular therapy. Nucleic Acids 2021, 26, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Tufan, A.B.; Lazarow, K.; Kolesnichenko, M.; Sporbert, A.; von Kries, J.P.; Scheidereit, C. tsg101 associates with parp1 and is essential for parylation and dna damage-induced nf-κb activation. The EMBO Journal 2022. [CrossRef] [PubMed]
- Zhang, Q.; Ke, H.; Blikslager, A.; Fujita, T.; Yoo, D. Type III Interferon Restriction by Porcine Epidemic Diarrhea Virus and the Role of Viral Protein nsp1 in IRF1 Signaling. J. Virol. 2018, 92, e01677-17. [Google Scholar] [CrossRef]
- Cao, L.; Ge, X.; Gao, Y.; Herrler, G.; Ren, Y.; Ren, X.; Li, G. Porcine epidemic diarrhea virus inhibits dsRNA-induced interferon-β production in porcine intestinal epithelial cells by blockade of the RIG-I-mediated pathway. Virol. J. 2015, 12, 127. [Google Scholar] [CrossRef]
- Lu, Y.; Cai, H.; Lu, M.; Ma, Y.; Li, A.; Gao, Y.; Zhou, J.; Gu, H.; Li, J.; Gu, J. Porcine Epidemic Diarrhea Virus Deficient in RNA Cap Guanine-N-7 Methylation Is Attenuated and Induces Higher Type I and III Interferon Responses. J. Virol. 2020, 94, e00447-20. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Liu, X.; Hu, D.; Luo, Y.; Zhang, G.; Liu, P. Swine Enteric Coronaviruses (PEDV, TGEV, and PDCoV) Induce Divergent Interferon-Stimulated Gene Responses and Antigen Presentation in Porcine Intestinal Enteroids. Front. Immunol. 2022, 12, 826882. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; Buckley, A.C.; Pillatzki, A.; Lager, K.M.; Faaberg, K.S.; Baker, S.C. Inactivating Three Interferon Antagonists Attenuates Pathogenesis of an Enteric Coronavirus. J. Virol. 2020, 94, e00565-20. [Google Scholar] [CrossRef] [PubMed]
- Case, J.B.; Ashbrook, A.W.; Dermody, T.S.; Denison, M.R. Mutagenesis of S-Adenosyl-l-Methionine-Binding Residues in Coronavirus nsp14 N7-Methyltransferase Demonstrates Differing Requirements for Genome Translation and Resistance to Innate Immunity. J. Virol. 2016, 90, 7248–7256. [Google Scholar] [CrossRef] [PubMed]
- Ogando, N.S.; Zevenhoven-Dobbe, J.C.; van der Meer, Y.; Bredenbeek, P.J.; Posthuma, C.C.; Snijder, E.J. The Enzymatic Activity of the nsp14 Exoribonuclease Is Critical for Replication of MERS-CoV and SARS-CoV-2. Journal of virology 2020, 94, e01246-20. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yin, L.; Xue, M.; Chen, J.; Li, L.; Fu, F.; Feng, L.; Liu, P. Coronavirus Porcine Deltacoronavirus Upregulates MHC Class I Expression through RIG-I/IRF1-Mediated NLRC5 Induction. J. Virol. 2022, 96, e0015822. [Google Scholar] [CrossRef]
- Han, L.; Zheng, Y.; Deng, J.; Nan, M.L.; Xiao, Y.; Zhuang, M.W.; Zhang, J.; Wang, W.; Gao, C.; Wang, P.H. SARS-CoV-2 ORF10 antagonizes STING-dependent interferon activation and autophagy. Journal of medical virology 2022, 10.1002/jmv.27965. Advance online publication. [CrossRef]
- van den Berg, D.F.; Te Velde, A.A. Severe COVID-19: NLRP3 Inflammasome Dysregulated. Front. Immunol. 2020, 11, 1580. [Google Scholar] [CrossRef] [PubMed]
- Dutta, D.; Liu, J.; Xiong, H. NLRP3 inflammasome activation and SARS-CoV-2-mediated hyperinflammation, cytokine storm and neurological syndromes. International journal of physiology, pathophysiology and pharmacology 2022, 14, 138–160. [Google Scholar]
- Ratajczak, M.Z.; Kucia, M. SARS-CoV-2 infection and overactivation of Nlrp3 inflammasome as a trigger of cytokine “storm” and risk factor for damage of hematopoietic stem cells. Leukemia 2020, 34, 1726–1729. [Google Scholar] [CrossRef]
- Batiha, G.E.; Al-Gareeb, D.; Qusti, S.; Alshammari, E.M.; Rotimi, D.; Adeyemi, O.S.; Al-Kuraishy, H.M. Common NLRP3 inflammasome inhibitors and Covid-19: Divide and Conquer. Scientific African 2021, e01084. Advance online publication. [CrossRef]
- Nguyen, A.A.; Habiballah, S.B.; Platt, C.D.; Geha, R.S.; Chou, J.S.; McDonald, D.R. Immunoglobulins in the treatment of COVID-19 infection: Proceed with caution! Clin. Immunol. 2020, 216, 108459. [Google Scholar] [CrossRef]
- Xia, X. Extreme Genomic CpG Deficiency in SARS-CoV-2 and Evasion of Host Antiviral Defense. Mol. Biol. Evol. 2020, 37, 2699–2705. [Google Scholar] [CrossRef]
- Nchioua, R.; Kmiec, D.; Müller, J.A.; Conzelmann, C.; Groß, R.; Swanson, C.M.; Neil, S.; Stenger, S.; Sauter, D.; Münch, J.; Sparrer, K.; Kirchhoff, F. SARS-CoV-2 Is Restricted by Zinc Finger Antiviral Protein despite Preadaptation to the Low-CpG Environment in Humans. mBio 2020, 11, e01930-20. [Google Scholar] [CrossRef]
- Alice Barbarin, André Herbelin, Jean-Marc Gombert. [The CD8+ T cell innate function in the war against cancer].. médecine/sciences, EDP Sciences, 2017, 33 (11), pp.927-929. 10.1051/med-sci/20173311004. inserm-02508042 (PDF) [The CD8+ T cell innate function in the war against cancer].. Available from: https://www.researchgate.net/publication/339946901_The_CD8_T_cell_innate_function_in_the_war_against_cancer [accessed Oct 05 2022].
- Planas, D.; Saunders, N.; Maes, P.; Guivel-Benhassine, F.; Planchais, C.; Buchrieser, J.; Bolland, W.H.; Porrot, F.; Staropoli, I.; Lemoine, F.; et al. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature 2022, 602, 671–675. [Google Scholar] [CrossRef]
- Flemming, A. Omicron, the great escape artist. Nat. Rev. Immunol. 2022, 22, 75. [Google Scholar] [CrossRef]
- Durmaz, V.; Köchl, K.; Krassnigg, A.; Parigger, L.; Hetmann, M.; Singh, A.; Nutz, D.; Korsunsky, A.; Kahler, U.; König, C.; et al. Structural bioinformatics analysis of SARS-CoV-2 variants reveals higher hACE2 receptor binding affinity for Omicron B.1.1.529 spike RBD compared to wild type reference. Sci. Rep. 2022, 12, 14534. [Google Scholar] [CrossRef]
- Shafiq, A.; Zubair, F.; Ambreen, A.; Suleman, M.; Yousafi, Q.; Rasul Niazi, Z.; Anwar, Z.; Khan, A.; Mohammad, A.; Wei, D.Q. Investigation of the binding and dynamic features of A.30 variant revealed higher binding of RBD for hACE2 and escapes the neutralizing antibody: A molecular simulation approach. Comput. Biol. Med. 2022, 146, 105574. [Google Scholar] [CrossRef]
- Hakami, A.R. Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach. Biology 2022, 11, 258. [Google Scholar] [CrossRef]
- Radzikowska, U.; Ding, M.; Tan, G.; Zhakparov, D.; Peng, Y.; Wawrzyniak, P.; Wang, M.; Li, S.; Morita, H.; Altunbulakli, C.; et al. Distribution of ACE2, CD147, CD26, and other SARS-CoV-2 associated molecules in tissues and immune cells in health and in asthma, COPD, obesity, hypertension, and COVID-19 risk factors. Allergy 2020, 75, 2829–2845. [Google Scholar] [CrossRef]
- LaSalle, T.J.; Gonye, A.L.K.; Freeman, S.S.; Kaplonek, P.; Gushterova, I.; Kays, K.R.; Manakongtreecheep, K.; Tantivit, J.; Rojas-Lopez, M.; Russo, B.C.; et al. Longitudinal characterization of circulating neutrophils uncovers distinct phenotypes associated with disease severity in hospitalized COVID-19 patients. bioRxiv: The preprint server for biology 2021, 2021.10.04.463121. [CrossRef]
- Manan, A.; Pirzada, R.H.; Haseeb, M.; Choi, S. Toll-like Receptor Mediation in SARS-CoV-2: A Therapeutic Approach. Int. J. Mol. Sci. 2022, 23, 10716. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.; Jin, Y.; Ji, W.; Zhang, W.; Duan, G. An Update on Innate Immune Responses during SARS-CoV-2 Infection. Viruses 2021, 13, 2060. [Google Scholar] [CrossRef]
- Huyton, T.; Göttmann, W.; Bade-Döding, C.; Paine, A.; Blasczyk, R. The T/NK cell co-stimulatory molecule SECTM1 is an IFN “early response gene” that is negatively regulated by LPS in human monocytic cells. Biochim. Et Biophys. Acta (BBA) Gen. Subj. 2011, 1810, 1294–1301. [Google Scholar] [CrossRef]
- Xia, H.; Shi, P.Y. Antagonism of Type I Interferon by Severe Acute Respiratory Syndrome Coronavirus 2. Journal of interferon & cytokine research: The official journal of the International Society for Interferon and Cytokine Research 2020, 40, 543–548. [Google Scholar] [CrossRef]
- Zhang, J.; Zhao, C.; Zhao, W. Virus Caused Imbalance of Type I IFN Responses and Inflammation in COVID-19. Front. Immunol. 2021, 12, 633769. [Google Scholar] [CrossRef]
- Prokop, J.W.; Hartog, N.L.; Chesla, D.; Faber, W.; Love, C.P.; Karam, R.; Abualkheir, N.; Feldmann, B.; Teng, L.; McBride, T.; et al. High-Density Blood Transcriptomics Reveals Precision Immune Signatures of SARS-CoV-2 Infection in Hospitalized Individuals. Front. Immunol. 2021, 12, 694243. [Google Scholar] [CrossRef] [PubMed]
- Gardinassi, L.G.; Souza, C.; Sales-Campos, H.; Fonseca, S.G. Immune and Metabolic Signatures of COVID-19 Revealed by Transcriptomics Data Reuse. Front. Immunol. 2020, 11, 1636. [Google Scholar] [CrossRef]
- Vanderheiden, A.; Ralfs, P.; Chirkova, T.; Upadhyay, A.A.; Zimmerman, M.G.; Bedoya, S.; Aoued, H.; Tharp, G.M.; Pellegrini, K.L.; Manfredi, C.; et al. Type I and Type III Interferons Restrict SARS-CoV-2 Infection of Human Airway Epithelial Cultures. J. Virol. 2020, 94, e00985-20. [Google Scholar] [CrossRef]
- Didangelos, A. COVID-19 Hyperinflammation: What about Neutrophils? mSphere 2020, 5, e00367-20. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.; Lareau, C.A.; Keshishian, H.; Ganskih, S.; Schneider, C.; Hennig, T.; Melanson, R.; Werner, S.; Wei, Y.; Zimmer, M.; et al. The SARS-CoV-2 RNA-protein interactome in infected human cells. Nat. Microbiol. 2021, 6, 339–353. [Google Scholar] [CrossRef]
- Yoo, J.S.; Sasaki, M.; Cho, S.X.; Kasuga, Y.; Zhu, B.; Ouda, R.; Orba, Y.; de Figueiredo, P.; Sawa, H.; Kobayashi, K.S. SARS-CoV-2 inhibits induction of the MHC class I pathway by targeting the STAT1-IRF1-NLRC5 axis. Nat. Commun. 2021, 12, 6602. [Google Scholar] [CrossRef]
- Frieman, M.; Yount, B.; Heise, M.; Kopecky-Bromberg, S.A.; Palese, P.; Baric, R.S. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J. Virol. 2007, 81, 9812–9824. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, Y.; Li, Y.; Huang, F.; Luo, B.; Yuan, Y.; Xia, B.; Ma, X.; Yang, T.; Yu, F.; et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Ι. Proc. Natl. Acad. Sci. USA 2021, 118, e2024202118. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.Y.; Liu, W.C.; Zheng, J.Q.; Lu, C.L.; Hou, Y.C.; Zheng, C.M.; Song, J.Y.; Lu, K.C.; Chao, Y.C. Immunological Aspects of SARS-CoV-2 Infection and the Putative Beneficial Role of Vitamin-D. Int. J. Mol. Sci. 2021, 22, 5251. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Puyo, C.A. Acetylcysteine to Combat COVID-19: An evidence review. Ther Clin Risk Manag 2020, 16, 1047–1055. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M.; Han, Q. Oral Methioninase for Covid-19 Methionine-restriction Therapy. In vivo 2020, 34(3 Suppl), 1593–1596. [Google Scholar] [CrossRef]
- Hou, W.; Wang, S.; Wu, H.; Xue, L.; Wang, B.; Wang, S.; Wang, H. Small GTPase-A Key Role in Host Cell for Coronavirus Infection and a Potential Target for Coronavirus Vaccine Adjuvant Discovery. Viruses 2022, 14, 2044. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Dominguez, N.; Parnell, C.; Teng, Y. Drugging the Small GTPase Pathways in Cancer Treatment: Promises and Challenges. Cells 2019, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Laskovs, M.; Partridge, L.; Slack, C. Molecular inhibition of RAS signalling to target ageing and age-related health. Disease models & mechanisms 2022, 15, dmm049627. [CrossRef]
- Matos, P.; Pereira, J.; Jordan, P. Targeting Cancer by Using Nanoparticles to Modulate RHO GTPase Signaling. Adv. Exp. Med. Biol. 2022, 1357, 115–127. [Google Scholar] [CrossRef]
- Ding, F.; Yin, Z.; Wang, H.R. Ubiquitination in Rho signaling. Curr. Top. Med. Chem. 2011, 11, 2879–2887. [Google Scholar] [CrossRef] [PubMed]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef]
- Ito, H.; Morishita, R.; Tabata, H.; Nagata, K. Roles of Rho small GTPases in the tangentially migrating neurons. Histol. Histopathol. 2014, 29, 871–879. [Google Scholar] [CrossRef]
- Govek, E.E.; Hatten, M.E.; Van Aelst, L. The role of Rho GTPase proteins in CNS neuronal migration. Dev. Neurobiol. 2011, 71, 528–553. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Chen, Y.; Chen, Y. Spatiotemporal Regulation of Rho GTPases in Neuronal Migration. Cells 2019, 8, 568. [Google Scholar] [CrossRef]
- Pantazi, I.; Al-Qahtani, A.A.; Alhamlan, F.S.; Alothaid, H.; Matou-Nasri, S.; Sourvinos, G.; Vergadi, E.; Tsatsanis, C. SARS-CoV-2/ACE2 Interaction Suppresses IRAK-M Expression and Promotes Pro-Inflammatory Cytokine Production in Macrophages. Front. Immunol. 2021, 12, 683800. [Google Scholar] [CrossRef] [PubMed]
- Pierce, C.A.; Preston-Hurlburt, P.; Dai, Y.; Aschner, C.B.; Cheshenko, N.; Galen, B.; Garforth, S.J.; Herrera, N.G.; Jangra, R.K.; Morano, N.C.; et al. Immune responses to SARS-CoV-2 infection in hospitalized pediatric and adult patients. Science translational medicine 2020, 12, eabd5487. [Google Scholar] [CrossRef]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. [Google Scholar] [CrossRef]
- Letarov, A.V.; Babenko, V.V.; Kulikov, E.E. Free SARS-CoV-2 Spike Protein S1 Particles May Play a Role in the Pathogenesis of COVID-19 Infection. Biochemistry. Biokhimiia 2021, 86, 257–261. [Google Scholar] [CrossRef]
- Meyer, K.; Patra, T.; Vijayamahantesh; Ray, R. SARS-CoV-2 Spike Protein Induces Paracrine Senescence and Leukocyte Adhesion in Endothelial Cells. J. Virol. 2021, 95, e0079421. [CrossRef] [PubMed]
- Xiang, Y.; Wang, M.; Chen, H.; Chen, L. Potential therapeutic approaches for the early entry of SARS-CoV-2 by interrupting the interaction between the spike protein on SARS-CoV-2 and angiotensin-converting enzyme 2 (ACE2). Biochem. Pharmacol. 2021, 192, 114724. [Google Scholar] [CrossRef]
- Paidi, R.K.; Jana, M.; Raha, S.; McKay, M.; Sheinin, M.; Mishra, R.K.; Pahan, K. Eugenol, a Component of Holy Basil (Tulsi) and Common Spice Clove, Inhibits the Interaction Between SARS-CoV-2 Spike S1 and ACE2 to Induce Therapeutic Responses. J. Neuroimmune Pharmacol. Off. J. Soc. NeuroImmune Pharmacol. 2021, 16, 743–755. [Google Scholar] [CrossRef]
- Paidi, R.K.; Jana, M.; Mishra, R.K.; Dutta, D.; Raha, S.; Pahan, K. ACE-2-interacting Domain of SARS-CoV-2 (AIDS) Peptide Suppresses Inflammation to Reduce Fever and Protect Lungs and Heart in Mice: Implications for COVID-19 Therapy. J. Neuroimmune Pharmacol. Off. J. Soc. NeuroImmune Pharmacol. 2021, 16, 59–70. [Google Scholar] [CrossRef]
- Yu, W.; Wu, X.; Zhao, Y.; Chen, C.; Yang, Z.; Zhang, X.; Ren, J.; Wang, Y.; Wu, C.; Li, C.; et al. Computational Simulation of HIV Protease Inhibitors to the Main Protease (Mpro) of SARS-CoV-2: Implications for COVID-19 Drugs Design. Molecules 2021, 26, 7385. [Google Scholar] [CrossRef] [PubMed]
- Paidi, R.K.; Jana, M.; Mishra, R.K.; Dutta, D.; Pahan, K. Selective Inhibition of the Interaction between SARS-CoV-2 Spike S1 and ACE2 by SPIDAR Peptide Induces Anti-Inflammatory Therapeutic Responses. J. Immunol. 2021, 207, 2521–2533. [Google Scholar] [CrossRef] [PubMed]
- Al-Zaidan, L.; Mestiri, S.; Raza, A.; Merhi, M.; Inchakalody, V.P.; Fernandes, Q.; Taib, N.; Uddin, S.; Dermime, S. The expression of hACE2 receptor protein and its involvement in SARS-CoV-2 entry, pathogenesis, and its application as potential therapeutic target. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2021, 43, 177–196. [Google Scholar] [CrossRef] [PubMed]
- Le Coupanec, A.; Desforges, M.; Kaufer, B.; Dubeau, P.; Côté, M.; Talbot, P.J. Potential differences in cleavage of the S protein and type-1 interferon together control human coronavirus infection, propagation, and neuropathology within the central nervous system. J. Virol. 2021, 95, e00140-21. Advance online publication. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Wang, M.; Li, Y.; Wang, P.; Zhao, P.; Yang, Z.; Wang, S.; Zhang, L.; Li, Z.; Jia, K.; et al. SARS-CoV-2 Spike protein enhances ACE2 expression via facilitating Interferon effects in bronchial epithelium. Immunol. Lett. 2021, 237, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Seneff, S.; Nigh, G.; Kyriakopoulos, A.M.; McCullough, P.A. Innate immune suppression by SARS-CoV-2 mRNA vaccinations: The role of G-quadruplexes, exosomes, and MicroRNAs. Food Chem. Toxicol. Int. J. Publ. Br. Ind. Biol. Res. Assoc. 2022, 164, 113008. [Google Scholar] [CrossRef] [PubMed]
- Roskoski, R. ; Jr Janus kinase (JAK) inhibitors in the treatment of inflammatory and neoplastic diseases. Pharmacol. Res. 2016, 111, 784–803. [Google Scholar] [CrossRef] [PubMed]
- Satarker, S.; Tom, A.A.; Shaji, R.A.; Alosious, A.; Luvis, M.; Nampoothiri, M. JAK-STAT Pathway Inhibition and their Implications in COVID-19 Therapy. Postgrad. Med. 2021, 133, 489–507. [Google Scholar] [CrossRef] [PubMed]
- Khaledi, M.; Sameni, F.; Yahyazade, S.; Radandish, M.; Owlia, P.; Bagheri, N.; Afkhami, H.; Mahjoor, M.; Esmaelpour, Z.; Kohansal, M.; Aghaei, F. COVID-19 and the potential of Janus family kinase (JAK) pathway inhibition: A novel treatment strategy. Front. Med. 2022, 9, 961027. [Google Scholar] [CrossRef]
- Barilli, A.; Visigalli, R.; Ferrari, F.; Recchia Luciani, G.; Soli, M.; Dall’Asta, V.; Rotoli, B.M. The JAK1/2 Inhibitor Baricitinib Mitigates the Spike-Induced Inflammatory Response of Immune and Endothelial Cells In Vitro. Biomedicines 2022, 10, 2324. [Google Scholar] [CrossRef]
- Amanda, J. Lee and Ali A. Ashkar The Dual Nature of Type I and Type II Interferons, Frontline Immunology 2018. [CrossRef]
- Felgenhauer, U.; Schoen, A.; Gad, H.H.; Hartmann, R.; Schaubmar, A.R.; Failing, K.; Drosten, C.; Weber, F. Inhibition of SARS-CoV-2 by type I and type III interferons. J. Biol. Chem. 2020, 295, 13958–13964. [Google Scholar] [CrossRef] [PubMed]
- Sung, R.Y.; Yin, J.; Oppenheimer, S.J.; Tam, J.S.; Lau, J. Treatment of respiratory syncytial virus infection with recombinant interferon alfa-2a. Arch. Dis. Child. 1993, 69, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Ma, L.L.; Jiang, J.D.; Pang, R.; Chen, Y.J.; Li, Y.H. Recombinant human interferon alpha 2b broad-spectrum anti-respiratory viruses pharmacodynamics study in vitro. Yao xue xue bao= Acta pharmaceutica Sinica 2014, 49, 1547–1553. [Google Scholar]
- Ma, Y.; Su, X.-Z.; Lu, F. The Roles of Type I Interferon in Co-infections with Parasites and Viruses, Bacteria, or Other Parasites. Front. Immunol. 2020, 11, 1805. [Google Scholar] [CrossRef] [PubMed]
- Sodano, F.; Gazzano, E.; Fruttero, R.; Lazzarato, L. NO in Viral Infections: Role and Development of Antiviral Therapies. Molecules 2022, 27, 2337. [Google Scholar] [CrossRef] [PubMed]
- Pieretti, J.C.; Rubilar, O.; Weller, R.B.; Tortella, G.R.; Seabra, A.B. Nitric oxide (NO) and nanoparticles—Potential small tools for the war against COVID-19 and other human coronavirus infections. Virus Res. 2021, 291, 198202. [Google Scholar] [CrossRef] [PubMed]
- Adusumilli, N.C.; Zhang, D.; Friedman, J.M.; Friedman, A.J. Harnessing nitric oxide for preventing, limiting and treating the severe pulmonary consequences of COVID-19. Nitric Oxide: Biol. Chem. 2020, 103, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zheng, S.X.; Tang, W.; He, P.L.; Li, X.Y.; Yang, Y.F.; Li, Y.C.; Geng, J.G.; Zuo, J.P. Inhibition of inducible nitric-oxide synthase expression by (5R)-5-hydroxytriptolide in interferon-gamma- and bacterial lipopolysaccharide-stimulated macrophages. J. Pharmacol. Exp. Ther. 2006, 316, 121–128. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, D.H.; Baek, S.H.; Lee, H.J.; Kim, M.R.; Kwon, H.J.; Lee, C.H. Rengyolone inhibits inducible nitric oxide synthase expression and nitric oxide production by down-regulation of NF-kappaB and p38 MAP kinase activity in LPS-stimulated RAW 264.7 cells. Biochem. Pharmacol. 2006, 71, 1198–1205. [Google Scholar] [CrossRef]
- Martel, J.; Ko, Y.F.; Young, J.D.; Ojcius, D.M. Could nasal nitric oxide help to mitigate the severity of COVID-19? Microbes Infect. 2020, 22, 168–171. [Google Scholar] [CrossRef] [PubMed]
- Pan, S.; Manabe, N.; Yamaguchi, Y. 3D Structures of IgA, IgM, and Components. Int. J. Mol. Sci. 2021, 22, 12776. [Google Scholar] [CrossRef] [PubMed]
- Naito, Y.; Takagi, T.; Yamamoto, T.; Watanabe, S. Association between selective IgA deficiency and COVID-19. J Clin Biochem Nutr 2020, 67, 122–125. [Google Scholar] [CrossRef] [PubMed]
- Botek, M.; Krejčí, J.; Valenta, M.; McKune, A.; Sládečková, B.; Konečný, P.; Klimešová, I.; Pastucha, D. Molecular Hydrogen Positively Affects Physical and Respiratory Function in Acute Post-COVID-19 Patients: A New Perspective in Rehabilitation. Int. J. Environ. Res. Public Health 2022, 19, 1992. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, P.; Liu, Q.; Cao, L.; Xu, J. Pyroptotic Patterns in Blood Leukocytes Predict Disease Severity and Outcome in COVID-19 Patients. Front. Immunol. 2022, 13, 888661. [Google Scholar] [CrossRef] [PubMed]
- Soy, M.; Keser, G.; Atagündüz, P.; Tabak, F.; Atagündüz, I.; Kayhan, S. Cytokine storm in COVID-19: Pathogenesis and overview of anti-inflammatory agents used in treatment. Clin. Rheumatol. 2020, 39, 2085–2094. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xie, F.; Zhou, X.; Wu, Y.; Yan, H.; Liu, T.; Huang, J.; Wang, F.; Zhou, F.; Zhang, L. Role of pyroptosis in inflammation and cancer. Cell. Mol. Immunol. 2022, 19, 971–992. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhou, J.; Zhang, X.; Hu, W.; Zhao, J.F.; Wang, S.; Wang, F.S.; Zhang, J.Y. Single-Cell Transcriptomic Profiling of MAIT Cells in Patients With COVID-19. Front. Immunol. 2021, 12, 700152. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Ge, H.; Li, S.; Qiu, H.J. Modulation of Macrophage Polarization by Viruses: Turning Off/On Host Antiviral Responses. Front. Microbiol. 2022, 13, 839585. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F. Innate immune evasion by filoviruses. Virology 2015, 479-480, 122–130. [Google Scholar] [CrossRef]
- Kuzmin, I.V.; Schwarz, T.M.; Ilinykh, P.A.; Jordan, I.; Ksiazek, T.G.; Sachidanandam, R.; Basler, C.F.; Bukreyev, A. Innate Immune Responses of Bat and Human Cells to Filoviruses: Commonalities and Distinctions. J. Virol. 2017, 91, e02471-16. [Google Scholar] [CrossRef] [PubMed]
- Arndt, W.D.; Cotsmire, S.; Trainor, K.; Harrington, H.; Hauns, K.; Kibler, K.V.; Huynh, T.P.; Jacobs, B.L. Evasion of the Innate Immune Type I Interferon System by Monkeypox Virus. J. Virol. 2015, 89, 10489–10499. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.L. Vaccinia Virus Protein C6: A Multifunctional Interferon Antagonist. Adv. Exp. Med. Biol. 2018, 1052, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Ogino, T.; Miyajima, N.; Kato, A.; Kohase, M. Dephosphorylation failure of tyrosine-phosphorylated STAT1 in IFN-stimulated Sendai virus C protein-expressing cells. Virology 2002, 293, 205–209. [Google Scholar] [CrossRef] [PubMed]
- Bousse, T.; Chambers, R.L.; Scroggs, R.A.; Portner, A.; Takimoto, T. Human parainfluenza virus type 1 but not Sendai virus replicates in human respiratory cells despite IFN treatment. Virus Res. 2006, 121, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Odkhuu, E.; Komatsu, T.; Naiki, Y.; Koide, N.; Yokochi, T. Sendai virus C protein inhibits lipopolysaccharide-induced nitric oxide production through impairing interferon-β signaling. Int. Immunopharmacol. 2014, 23, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Morita, N.; Tanaka, Y.; Odkhuu, E.; Naiki, Y.; Komatsu, T.; Koide, N. Sendai virus V protein decreases nitric oxide production by inhibiting RIG-I signaling in infected RAW264.7 macrophages. Microbes Infect. 2020, 22, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Kawabata, R.; Honda, T.; Sakai, K.; Ami, Y.; Sakaguchi, T.; Irie, T. A Single Amino Acid Substitution within the Paramyxovirus Sendai Virus Nucleoprotein Is a Critical Determinant for Production of Interferon-Beta-Inducing Copyback-Type Defective Interfering Genomes. J. Virol. 2018, 92, e02094-17. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aparicio, M.T.; Garcin, D.; Rice, C.M.; Kolakofsky, D.; García-Sastre, A.; Baum, A. Loss of Sendai virus C protein leads to accumulation of RIG-I immunostimulatory defective interfering RNA. J. Gen. Virol. 2017, 98, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Kato, A.; Ohnishi, Y.; Kohase, M.; Saito, S.; Tashiro, M.; Nagai, Y. Y2, the smallest of the Sendai virus C proteins, is fully capable of both counteracting the antiviral action of interferons and inhibiting viral RNA synthesis. J. Virol. 2001, 75, 3802–3810. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Mercado-López, X.; Grier, J.T.; Kim, W.K.; Chun, L.F.; Irvine, E.B.; Del Toro Duany, Y.; Kell, A.; Hur, S.; Gale, M., Jr.; Raj, A.; López, C.B. Identification of a Natural Viral RNA Motif That Optimizes Sensing of Viral RNA by RIG-I. mBio 2015, 6, e01265-15. [Google Scholar] [CrossRef] [PubMed]
- Schomacker, H.; Hebner, R.M.; Boonyaratanakornkit, J.; Surman, S.; Amaro-Carambot, E.; Collins, P.L.; Schmidt, A.C. The C proteins of human parainfluenza virus type 1 block IFN signaling by binding and retaining Stat1 in perinuclear aggregates at the late endosome. PLoS ONE 2012, 7, e28382. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, T.; Duan, L.; Chen, H.; Hu, R.; Wang, X.; Jia, Y.; Chu, Z.; Liu, H.; Wang, X.; et al. Evasion of Host Antiviral Innate Immunity by Paramyxovirus Accessory Proteins. Front. Microbiol. 2022, 12, 790191. [Google Scholar] [CrossRef] [PubMed]
- Brzózka, K.; Finke, S.; Conzelmann, K.K. Identification of the rabies virus alpha/beta interferon antagonist: Phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 2005, 79, 7673–7681. [Google Scholar] [CrossRef]
- Vidy, A.; Chelbi-Alix, M.; Blondel, D. Rabies virus P protein interacts with STAT1 and inhibits interferon signal transduction pathways. J. Virol. 2005, 79, 14411–14420. [Google Scholar] [CrossRef] [PubMed]
- Kong, Z.; Yin, H.; Wang, F.; Liu, Z.; Luan, X.; Sun, L.; Liu, W.; Shang, Y. Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity. PLoS Pathog. 2022, 18, e1010544. [Google Scholar] [CrossRef] [PubMed]
- Bo, Z.; Miao, Y.; Xi, R.; Zhong, Q.; Bao, C.; Chen, H.; Sun, L.; Qian, Y.; Jung, Y.S.; Dai, J. PRV UL13 inhibits cGAS-STING-mediated IFN-β production by phosphorylating IRF3. Vet. Res. 2020, 51, 118. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Xu, A.; Qin, C.; Zhang, Q.; Chen, S.; Lang, Y.; Wang, M.; Li, C.; Feng, W.; Zhang, R.; Jiang, Z.; Tang, J. Pseudorabies Virus dUTPase UL50 Induces Lysosomal Degradation of Type I Interferon Receptor 1 and Antagonizes the Alpha Interferon Response. J. Virol. 2017, 91, e01148-17. [Google Scholar] [CrossRef]
- Lv, L.; Cao, M.; Bai, J.; Jin, L.; Wang, X.; Gao, Y.; Liu, X.; Jiang, P. PRV-encoded UL13 protein kinase acts as an antagonist of innate immunity by targeting IRF3-signaling pathways. Vet. Microbiol. 2020, 250, 108860. [Google Scholar] [CrossRef]
- Li, X.; Xie, J.; Li, D.; Li, H.; Niu, Y.; Wu, B.; Yang, Y.; Yan, Z.; Zhang, X.; Chen, L.; Feng, R. HSP27 Attenuates cGAS-Mediated IFN-β Signaling through Ubiquitination of cGAS and Promotes PRV Infection. Viruses 2022, 14, 1851. [Google Scholar] [CrossRef] [PubMed]
- Margolis, S.R.; Wilson, S.C.; Vance, R.E. Evolutionary Origins of cGAS-STING Signaling. Trends Immunol. 2017, 38, 733–743. [Google Scholar] [CrossRef]
- Zhang, X.; Bai, X.C.; Chen, Z.J. Structures and Mechanisms in the cGAS-STING Innate Immunity Pathway. Immunity 2020, 53, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.; Wang, S.; Gao, P.; Gao, G.; Fan, Z. DNA sensor cGAS-mediated immune recognition. Protein Cell 2016, 7, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Pang, X.; Zhang, H.; Ji, P. The cGAS-STING Pathway in Bacterial Infection and Bacterial Immunity. Front. Immunol. 2022, 12, 814709. [Google Scholar] [CrossRef]
- Kovarik, P.; Castiglia, V.; Ivin, M.; Ebner, F. Type I Interferons in Bacterial Infections: A Balancing Act. Front. Immunol. 2016, 7, 652. [Google Scholar] [CrossRef] [PubMed]
- Adolfo Garcia-Sastre Christine, A. Biron (2006), Type I Interferons and the Virus-Host Relationship: A lesson in Detente, Science, Vol 312, 5775, pp 879-882. [CrossRef]
- Lopez, L.; Sang, P.C.; Tian, Y.; Sang, Y. Dysregulated Interferon Response Underlying Severe COVID-19. Viruses 2020, 12, 1433. [Google Scholar] [CrossRef]
- Habjan, M.; Andersson, I.; Klingström, J.; Schümann, M.; Martin, A.; Zimmermann, P.; Wagner, V.; Pichlmair, A.; Schneider, U.; Mühlberger, E.; Mirazimi, A.; Weber, F. Processing of genome 5’ termini as a strategy of negative-strand RNA viruses to avoid RIG-I-dependent interferon induction. PLoS ONE 2008, 3, e2032. [Google Scholar] [CrossRef] [PubMed]
- Khatamzas, E.; Hipp, M.M.; Gaughan, D.; Pichulik, T.; Leslie, A.; Fernandes, R.A.; Muraro, D.; Booth, S.; Zausmer, K.; Sun, M.Y.; et al. Snapin promotes HIV-1 transmission from dendritic cells by dampening TLR8 signaling. EMBO J. 2017, 36, 2998–3011. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Qian, G.; Zhu, L.; Zhao, Z.; Liu, Y.; Han, W.; Zhang, X.; Zhang, Y.; Xiong, T.; Zeng, H.; et al. HIV-1 Vif suppresses antiviral immunity by targeting STING. Cell. Mol. Immunol. 2022, 19, 108–121. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F.; Amarasinghe, G.K. Evasion of interferon responses by Ebola and Marburg viruses. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2009, 29, 511–520. [Google Scholar] [CrossRef]
- Lubaki, N.M.; Younan, P.; Santos, R.I.; Meyer, M.; Iampietro, M.; Koup, R.A.; Bukreyev, A. The Ebola Interferon Inhibiting Domains Attenuate and Dysregulate Cell-Mediated Immune Responses. PLoS Pathog. 2016, 12, e1006031. [Google Scholar] [CrossRef] [PubMed]
- Paparisto, E.; Hunt, N.R.; Labach, D.S.; Coleman, M.D.; Di Gravio, E.J.; Dodge, M.J.; Friesen, N.J.; Côté, M.; Müller, A.; Hoenen, T.; Barr, S.D. Interferon-Induced HERC5 Inhibits Ebola Virus Particle Production and Is Antagonized by Ebola Glycoprotein. Cells 2021, 10, 2399. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Halfmann, P.J.; Hill-Batorski, L.; Ozawa, M.; Lopes, T.; Neumann, G.; Schoggins, J.W.; Rice, C.M.; Kawaoka, Y. Identification of interferon-stimulated genes that attenuate Ebola virus infection. Nat. Commun. 2020, 11, 2953. [Google Scholar] [CrossRef] [PubMed]
- Di Palma, F.; Daino, G.L.; Ramaswamy, V.K.; Corona, A.; Frau, A.; Fanunza, E.; Vargiu, A.V.; Tramontano, E.; Ruggerone, P. Relevance of Ebola virus VP35 homo-dimerization on the type I interferon cascade inhibition. Antivir. Chem. Chemother. 2019, 27, 2040206619889220. [Google Scholar] [CrossRef] [PubMed]
- Leung, L.W.; Park, M.S.; Martinez, O.; Valmas, C.; López, C.B.; Basler, C.F. Ebolavirus VP35 suppresses IFN production from conventional but not plasmacytoid dendritic cells. Immunol. Cell Biol. 2011, 89, 792–802. [Google Scholar] [CrossRef]
- Ilinykh, P.A.; Lubaki, N.M.; Widen, S.G.; Renn, L.A.; Theisen, T.C.; Rabin, R.L.; Wood, T.G.; Bukreyev, A. Different Temporal Effects of Ebola Virus VP35 and VP24 Proteins on Global Gene Expression in Human Dendritic Cells. J. Virol. 2015, 89, 7567–7583. [Google Scholar] [CrossRef] [PubMed]
- Woolsey, C.; Menicucci, A.R.; Cross, R.W.; Luthra, P.; Agans, K.N.; Borisevich, V.; Geisbert, J.B.; Mire, C.E.; Fenton, K.A.; Jankeel, A.; et al. A VP35 Mutant Ebola Virus Lacks Virulence but Can Elicit Protective Immunity to Wild-Type Virus Challenge. Cell Rep. 2019, 28, 3032–3046.e6. [Google Scholar] [CrossRef] [PubMed]
- Galão, R.P.; Wilson, H.; Schierhorn, K.L.; Debeljak, F.; Bodmer, B.S.; Goldhill, D.; Hoenen, T.; Wilson, S.J.; Swanson, C.M.; Neil, S. TRIM25 and ZAP target the Ebola virus ribonucleoprotein complex to mediate interferon-induced restriction. PLoS Pathog. 2022, 18, e1010530. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.; Lythe, G.; López-García, M.; Muñoz-Fontela, C.; Carroll, M.; Molina-París, C. Quantification of Type I Interferon Inhibition by Viral Proteins: Ebola Virus as a Case Study. Viruses 2021, 13, 2441. [Google Scholar] [CrossRef] [PubMed]
- Fanunza, E.; Frau, A.; Corona, A.; Tramontano, E. Insights into Ebola Virus VP35 and VP24 Interferon Inhibitory Functions and their Initial Exploitation as Drug Targets. Infect. Disord. Drug Targets 2019, 19, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Corona, A.; Fanunza, E.; Salata, C.; Morwitzer, M.J.; Distinto, S.; Zinzula, L.; Sanna, C.; Frau, A.; Daino, G.L.; Quartu, M.; et al. Cynarin blocks Ebola virus replication by counteracting VP35 inhibition of interferon-beta production. Antivir. Res. 2022, 198, 105251. [Google Scholar] [CrossRef] [PubMed]
- Mateo, M.; Reid, S.P.; Leung, L.W.; Basler, C.F.; Volchkov, V.E. Ebolavirus VP24 binding to karyopherins is required for inhibition of interferon signaling. J. Virol. 2010, 84, 1169–1175. [Google Scholar] [CrossRef] [PubMed]
- Le, H.; Spearman, P.; Waggoner, S.N.; Singh, K. Ebola virus protein VP40 stimulates IL-12- and IL-18-dependent activation of human natural killer cells. JCI Insight 2022, 7, e158902. [Google Scholar] [CrossRef] [PubMed]
- Warfield, K.L.; Perkins, J.G.; Swenson, D.L.; Deal, E.M.; Bosio, C.M.; Aman, M.J.; Yokoyama, W.M.; Young, H.A.; Bavari, S. Role of natural killer cells in innate protection against lethal ebola virus infection. J. Exp. Med. 2004, 200, 169–179. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, J.J.; Parisien, J.P.; Horvath, C.M. Nipah virus V protein evades alpha and gamma interferons by preventing STAT1 and STAT2 activation and nuclear accumulation. J. Virol. 2002, 76, 11476–11483. [Google Scholar] [CrossRef] [PubMed]
- Shaw, M.L.; García-Sastre, A.; Palese, P.; Basler, C.F. Nipah virus V and W proteins have a common STAT1-binding domain yet inhibit STAT1 activation from the cytoplasmic and nuclear compartments, respectively. J. Virol. 2004, 78, 5633–5641. [Google Scholar] [CrossRef] [PubMed]
- Bharaj, P.; Wang, Y.E.; Dawes, B.E.; Yun, T.E.; Park, A.; Yen, B.; Basler, C.F.; Freiberg, A.N.; Lee, B.; Rajsbaum, R. The Matrix Protein of Nipah Virus Targets the E3-Ubiquitin Ligase TRIM6 to Inhibit the IKKε Kinase-Mediated Type-I IFN Antiviral Response. PLoS Pathog. 2016, 12, e1005880. [Google Scholar] [CrossRef] [PubMed]
- Marsh, G.A.; Wang, L.F. Hendra and Nipah viruses: Why are they so deadly? Current opinion in virology 2012, 2, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Basler, C.F. Nipah and hendra virus interactions with the innate immune system. Curr. Top. Microbiol. Immunol. 2012, 359, 123–152. [Google Scholar] [CrossRef]
- Sugai, A.; Sato, H.; Takayama, I.; Yoneda, M.; Kai, C. Nipah and Hendra Virus Nucleoproteins Inhibit Nuclear Accumulation of Signal Transducer and Activator of Transcription 1 (STAT1) and STAT2 by Interfering with Their Complex Formation. J. Virol. 2017, 91, e01136-17. [Google Scholar] [CrossRef]
- Glennon, N.B.; Jabado, O.; Lo, M.K.; Shaw, M.L. Transcriptome Profiling of the Virus-Induced Innate Immune Response in Pteropus vampyrus and Its Attenuation by Nipah Virus Interferon Antagonist Functions. J. Virol. 2015, 89, 7550–7566. [Google Scholar] [CrossRef] [PubMed]
- Myasnikov, A.L.; Berns, S.A.; Talyzin, P.A.; Ershov, F.I. Vopr. Virusol. 2021, 66, 47–54. [CrossRef] [PubMed]
- Davoudi-Monfared, E.; Rahmani, H.; Khalili, H.; Hajiabdolbaghi, M.; Salehi, M.; Abbasian, L.; Kazemzadeh, H.; Yekaninejad, M.S. A Randomized Clinical Trial of the Efficacy and Safety of Interferon β-1a in Treatment of Severe COVID-19. Antimicrob. Agents Chemother. 2020, 64, e01061-20. [Google Scholar] [CrossRef] [PubMed]
- Effat Davoudi-Monfared, Hamid Rahmani, Hossein Khalili, Mahboubeh Hajiabdolbaghi, Mohamadreza Salehi, Ladan Abbasian, Hossein Kazemzadeh, Mir Saeed Yekaninejad (2020), A Randomized Clinical Trial of the Efficacy and Safety of Interferon Beta-1a in Treatment of Severe COVID-19, ASM Journals, Vol 64, 9. [CrossRef]
- Higgins, P.G.; Al-Nakib, W.; Willman, J.; Tyrrell, D.A. Interferon-beta ser as prophylaxis against experimental rhinovirus infection in volunteers. J. Interferon Res. 1986, 6, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Sperber, S.J.; Levine, P.A.; Innes, D.J.; Mills, S.E.; Hayden, F.G. Tolerance and efficacy of intranasal administration of recombinant beta serine interferon in healthy adults. J. Infect. Dis. 1988, 158, 166–175. [Google Scholar] [CrossRef]
- Kalyuzhin, O.V.; Ponezheva, L.O.; Turapova, A.N.; Nurtazina, A.Y.; Bykov, A.S.; Karaulov, A.V. Interferons alpha and Gamma, pidotimod, and tilorone in the treatment of acute respiratory infections in patients with allergic rhinitis: A prospective, cohort clinical and immunological study. Bull. Sib. Med. 2022, 21, 48–59. [Google Scholar] [CrossRef]
- Schaefer, A.; Unterberger, C.; Frankenberger, M.; Lohrum, M.; Staples, K.J.; Werner, T.; Stunnenberg, H.; Ziegler-Heitbrock, L. Mechanism of interferon-gamma mediated down-regulation of interleukin-10 gene expression. Mol. Immunol. 2009, 46, 1351–1359. [Google Scholar] [CrossRef] [PubMed]
- Olson, G.S.; Murray, T.A.; Jahn, A.N.; Mai, D.; Diercks, A.H.; Gold, E.S.; Aderem, A. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 2021, 35, 109195. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; You, H.; Su, C.; Li, Y.; Chen, S.; Zheng, C. Herpes Simplex Virus 1 Tegument Protein VP22 Abrogates cGAS/STING-Mediated Antiviral Innate Immunity. J. Virol. 2018, 92, e00841-18. [Google Scholar] [CrossRef] [PubMed]
- Su, C.; Zheng, C. Herpes Simplex Virus 1 Abrogates the cGAS/STING-Mediated Cytosolic DNA-Sensing Pathway via Its Virion Host Shutoff Protein, UL41. J. Virol. 2017, 91, e02414-16. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, M.; Yang, Y.; Zhang, C.; Xie, Z.; Tang, J.; Shi, Z.; Chen, S.; Li, G.; Gu, Y.; Wang, X.; Zhang, F.; Wang, Y.; Shen, X. Salmonella Induces the cGAS-STING-Dependent Type I Interferon Response in Murine Macrophages by Triggering mtDNA Release. mBio 2022, 13, e0363221. [Google Scholar] [CrossRef]
- Bastard, P.; Rosen, L.B.; Zhang, Q.; Michailidis, E.; Hoffmann, H.H.; Zhang, Y.; Dorgham, K.; Philippot, Q.; Rosain, J.; Béziat, V.; et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 2020, 370, eabd4585. [Google Scholar] [CrossRef]
- Makrinioti, H.; Bush, A.; Gern, J.; Johnston, S.L.; Papadopoulos, N.; Feleszko, W.; Camargo, C.A., Jr.; Hasegawa, K.; Jartti, T. The Role of Interferons in Driving Susceptibility to Asthma Following Bronchiolitis: Controversies and Research Gaps. Front. Immunol. 2021, 12, 761660. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Bunn, P.T.; Singh, S.S.; Ng, S.S.; Montes de Oca, M.; De Labastida Rivera, F.; Chauhan, S.B.; Singh, N.; Faleiro, R.J.; Edwards, C.L.; et al. Type I interferons suppress anti-parasitic immunity and can be targeted to improve treatment of visceral leishmaniasis. Cell Reports 2020, 30. [Google Scholar] [CrossRef]
- 477. Faleiro RJ, Kumar R, Bunn PT, Singh N, Chauhan SB; et al. (2016) Combined Immune Therapy for the Treatment of Visceral Leishmaniasis. PLOS Neglected Tropical Diseases 10, e0004415. [CrossRef]
- Kumar R, Singh N, Gautam S, Singh OP, Gidwani K; et al. (2014) Leishmania Specific CD4 T Cells Release IFNγ That Limits Parasite Replication in Patients with Visceral Leishmaniasis. PLOS Neglected Tropical Diseases 8, e3198. [CrossRef]
- Michael, W. Russell, Zina Moldoveanu, Pearay L. Ogra and Jiri Mesteacky, Mucosal Immunity in COVID-19: A Neglected but Critical Aspect of SARS-CoV-2 Infection, Front. Immunol. [CrossRef]
- Muge Cevik, Krutika Kuppalli, Jason Kindrachuk and Malik Peiris (2020), Virology, pathology and pathogenesis of SARS-CoV-2, BMJ, 371:m3862. [CrossRef]
- Padayachee, Y.; Flicker, S.; Linton, S.; Cafferkey, J.; Kon, O.M.; Johnston, S.L.; Ellis, A.K.; Desrosiers, M.; Turner, P.; Valenta, R.; Scadding, G.K. Review: The Nose as a Route for Therapy. Part 2 Immunotherapy. Front. Allergy 2021, 2, 668781. [Google Scholar] [CrossRef] [PubMed]
- Casteleyn, C.; Broos, A.M.; Simoens, P.; Van den Broeck, W. NALT (nasal cavity-associated lymphoid tissue) in the rabbit. Vet. Immunol. Immunopathol. 2010, 133, 212–218. [Google Scholar] [CrossRef]
- Yang, Y.; Jing, Y.; Yang, J.; Yang, Q. Effects of intranasal administration with Bacillus subtilis on immune cells in the nasal mucosa and tonsils of piglets. Exp. Ther. Med. 2018, 15, 5189–5198. [Google Scholar] [CrossRef]
- Pabst, R. Mucosal vaccination by the intranasal route. Nose-associated lymphoid tissue (NALT)-Structure, function and species differences. Vaccine 2015, 33, 4406–4413. [Google Scholar] [CrossRef]
- Acosta, P.L.; Byrne, A.B.; Hijano, D.R.; Talarico, L.B. Human Type I Interferon Antiviral Effects in Respiratory and Re-emerging Viral Infections. J. Immunol. Res. 2020, 2020, 1372494. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Kaiser, D.L.; Albrecht, J.K. Intranasal recombinant alfa-2b interferon treatment of naturally occurring common colds. Antimicrob. Agents Chemother. 1988, 32, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Gwaltney, J.M. ; Jr Intranasal interferon-alpha 2 treatment of experimental rhinoviral colds. J. Infect. Dis. 1984, 150, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Phillpotts, R.J.; Scott, G.M.; Higgins, P.G.; Wallace, J.; Tyrrell, D.A.; Gauci, C.L. An effective dosage regimen for prophylaxis against rhinovirus infection by intranasal administration of HuIFN-alpha 2. Antivir. Res. 1983, 3, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Edahiro, Y.; Yasuda, H.; Gotoh, A.; Morishita, S.; Suzuki, T.; Takeda, J.; Ando, J.; Tsutsui, M.; Itakura, A.; Komatsu, N. Interferon therapy for pregnant patients with essential thrombocythemia in Japan. Int. J. Hematol. 2021, 113, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Sakai, K.; Ueda, A.; Hasegawa, M.; Ueda, Y. Efficacy and safety of interferon alpha for essential thrombocythemia during pregnancy: Two cases and a literature review. Int. J. Hematol. 2018, 108, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Katsurada, T.; Nakabou, Y.; Kawabata, H. Efficacy of interferon-alpha in essential thrombocythemia during pregnancy. Ann. Hematol. 2017, 96, 877–878. [Google Scholar] [CrossRef] [PubMed]
- Yazdani Brojeni, P.; Matok, I.; Garcia Bournissen, F.; Koren, G. A systematic review of the fetal safety of interferon alpha. Reprod. Toxicol. 2012, 33, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Pons, J.C.; Lebon, P.; Frydman, R.; Delfraissy, J.F. Pharmacokinetics of interferon-alpha in pregnant women and fetoplacental passage. Fetal Diagn. Ther. 1995, 10, 7–10. [Google Scholar] [CrossRef] [PubMed]
- BAI, H.; KAWAHARA, M.; TAKAHASHI, M.; IMAKAWA, K. Recent progress of Interferon-Tau Research and potential direction beyond pregnancy recognition. J. Reprod. Dev. 2022, 68, 299–306. [Google Scholar] [CrossRef] [PubMed]
- Yockey, L.J.; Jurado, K.A.; Arora, N.; Millet, A.; Rakib, T.; Milano, K.M.; Hastings, A.K.; Fikrig, E.; Kong, Y.; Horvath, T.L.; et al. Type I interferons instigate fetal demise after Zika virus infection. Science immunology 2018, 3, eaao1680. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Aldo, P.; Roberts, C.M.; Stabach, P.; Liu, H.; You, Y.; Qiu, X.; Jeong, J.; Maxwell, A.; Lindenbach, B.; et al. Placenta-derived interferon-stimulated gene 20 controls ZIKA virus infection. EMBO Rep. 2021, 22, e52450. [Google Scholar] [CrossRef] [PubMed]
- Azamor, T.; Cunha, D.P.; da Silva, A.; Bezerra, O.; Ribeiro-Alves, M.; Calvo, T.L.; Kehdy, F.; Manta, F.; Pinto, T.; Ferreira, L.P.; et al. Congenital Zika Syndrome Is Associated With Interferon Alfa Receptor 1. Front. Immunol. 2021, 12, 764746. [Google Scholar] [CrossRef]
- Hijano, D.R.; Vu, L.D.; Kauvar, L.M.; Tripp, R.A.; Polack, F.P.; Cormier, S.A. Role of Type I Interferon (IFN) in the Respiratory Syncytial Virus (RSV) Immune Response and Disease Severity. Front. Immunol. 2019, 10, 566. [Google Scholar] [CrossRef] [PubMed]
- Claudio, G. Gallo, Sirio Fiorino, Giovanni Posabella, Donato Antonacci, Antonio Tropeano, Emanuele Pausini, Carlotta Pausini, Tomasso Guarniero, Marco Zancanaro (2020), COVID-19: Role of the Interferons.
- Borden, E.C. Interferons α and β in cancer: Therapeutic opportunities from new insights. Nat Rev Drug Discov 2019, 18, 219–234. [Google Scholar] [CrossRef] [PubMed]
- Koech, D.K.; Obel, A.O.; Minowada, J.; Hutchinson, V.A.; Cummins, J.M. Low dose oral alpha-interferon therapy for patients seropositive for human immunodeficiency virus type-1 (HIV-1). Mol. Biother. 1990, 2, 91–95. [Google Scholar]
- Babiuch, L.; Mian, M.; Kamińska, E.; Szymańska, B.; Georgiades, J.A. An interim report on the effect of natural human interferon alpha (IFN-alpha) lozenges in patients seropositive for the human immunodeficiency virus type 1 (HIV-1). Arch. Immunol. Et Ther. Exp. 1993, 41, 213–219. [Google Scholar]
- Emilie, D.; Burgard, M.; Lascoux-Combe, C.; Laughlin, M.; Krzysiek, R.; Pignon, C.; Rudent, A.; Molina, J.M.; Livrozet, J.M.; Souala, F.; Chene, G.; Grangeot-Keros, L.; Galanaud, P.; Sereni, D.; Rouzioux, C. Primoferon A Study Group Early control of HIV replication in primary HIV-1 infection treated with antiretroviral drugs and pegylated IFN alpha: Results from the Primoferon A (ANRS 086) Study. AIDS 2001, 15, 1435–1437. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.; Quereda, C.; Moreno, A.; Perez-Elías, M.J.; Antela, A.; Casado, J.L.; Dronda, F.; Mateos, M.L.; Bárcena, R.; Moreno, S. Pegylated interferon alpha2b plus ribavirin for the treatment of chronic hepatitis C in HIV-infected patients. AIDS 2004, 18, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Gargan, S.; Ahmed, S.; Mahony, R.; Bannan, C.; Napoletano, S.; O’Farrelly, C.; Borrow, P.; Bergin, C.; Stevenson, N.J. HIV-1 Promotes the Degradation of Components of the Type 1 IFN JAK/STAT Pathway and Blocks Anti-viral ISG Induction. EBioMedicine 2018, 30, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Poli, G.; Biswas, P.; Fauci, A.S. Interferons in the pathogenesis and treatment of human immunodeficiency virus infection. Antivir. Res. 1994, 24, 221–233. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Tran, J.T.; Sanchez, D.J. Manipulation of Type I Interferon Signaling by HIV and AIDS-Associated Viruses. J. Immunol. Res. 2019, 2019, 8685312. [Google Scholar] [CrossRef] [PubMed]
- Gargan, S.; Ahmed, S.; Mahony, R.; Bannan, C.; Napoletano, S.; O’Farrelly, C.; Borrow, P.; Bergin, C.; Stevenson, N.J. HIV-1 Promotes the Degradation of Components of the Type 1 IFN JAK/STAT Pathway and Blocks Anti-viral ISG Induction. EBioMedicine 2018, 30, 203–216. [Google Scholar] [CrossRef]
- Francis, M.L.; Meltzer, M.S.; Gendelman, H.E. Interferons in the persistence, pathogenesis, and treatment of HIV infection. AIDS Res. Hum. Retroviruses 1992, 8, 199–207. [Google Scholar] [CrossRef]
- Akiyama, H.; Ramirez, N.P.; Gibson, G.; Kline, C.; Watkins, S.; Ambrose, Z.; Gummuluru, S. Interferon-Inducible CD169/Siglec1 Attenuates Anti-HIV-1 Effects of Alpha Interferon. J. Virol. 2017, 91, e00972-17. [Google Scholar] [CrossRef] [PubMed]
- Fenton-May, A.E.; Dibben, O.; Emmerich, T.; Ding, H.; Pfafferott, K.; Aasa-Chapman, M.M.; Pellegrino, P.; Williams, I.; Cohen, M.S.; Gao, F.; et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology 2013, 10, 146. [Google Scholar] [CrossRef] [PubMed]
- Meylan, P.R.; Guatelli, J.C.; Munis, J.R.; Richman, D.D.; Kornbluth, R.S. Mechanisms for the inhibition of HIV replication by interferons-alpha, -beta, and -gamma in primary human macrophages. Virology 1993, 193, 138–148. [Google Scholar] [CrossRef]
- Shirazi, Y.; Pitha, P.M. Interferon alpha-mediated inhibition of human immunodeficiency virus type 1 provirus synthesis in T-cells. Virology 1993, 193, 303–312. [Google Scholar] [CrossRef]
- Korth, M.J.; Taylor, M.D.; Katze, M.G. Interferon inhibits the replication of HIV-1, SIV, and SHIV chimeric viruses by distinct mechanisms. Virology 1998, 247, 265–273. [Google Scholar] [CrossRef]
- Agy, M.B.; Acker, R.L.; Sherbert, C.H.; Katze, M.G. Interferon treatment inhibits virus replication in HIV-1- and SIV-infected CD4+ T-cell lines by distinct mechanisms: Evidence for decreased stability and aberrant processing of HIV-1 proteins. Virology 1995, 214, 379–386. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.D.; Korth, M.J.; Katze, M.G. Interferon treatment inhibits the replication of simian immunodeficiency virus at an early stage: Evidence for a block between attachment and reverse transcription. Virology 1998, 241, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Oliva, A.; Kinter, A.L.; Vaccarezza, M.; Rubbert, A.; Catanzaro, A.; Moir, S.; Monaco, J.; Ehler, L.; Mizell, S.; Jackson, R.; et al. Natural killer cells from human immunodeficiency virus (HIV)-infected individuals are an important source of CC-chemokines and suppress HIV-1 entry and replication in vitro. J. Clin. Investig. 1998, 102, 223–231. [Google Scholar] [CrossRef]
- Fehniger, T.A.; Herbein, G.; Yu, H.; Para, M.I.; Bernstein, Z.P.; O’Brien, W.A.; Caligiuri, M.A. Natural killer cells from HIV-1+ patients produce C-C chemokines and inhibit HIV-1 infection. J. Immunol. 1998, 161, 6433–6438. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, Y.; Fang, S.; Su, J.; Jiang, J.; Liang, B.; Huang, J.; Zhou, B.; Zang, N.; Ho, W.; et al. Downregulation of autophagy-related gene ATG5 and GABARAP expression by IFN-λ1 contributes to its anti-HCV activity in human hepatoma cells. Antivir. Res. 2017, 140, 83–94. [Google Scholar] [CrossRef]
- Aboulnasr, F.; Hazari, S.; Nayak, S.; Chandra, P.K.; Panigrahi, R.; Ferraris, P.; Chava, S.; Kurt, R.; Song, K.; Dash, A.; et al. IFN-λ Inhibits MiR-122 Transcription through a Stat3-HNF4α Inflammatory Feedback Loop in an IFN-α Resistant HCV Cell Culture System. PLoS ONE 2015, 10, e0141655. [Google Scholar] [CrossRef] [PubMed]
- Castro-Jiménez, T.K.; Gómez-Legorreta, L.C.; López-Campa, L.A.; Martínez-Torres, V.; Alvarado-Silva, M.; Posadas-Mondragón, A.; Díaz-Lima, N.; Angulo-Mendez, H.A.; Mejía-Domínguez, N.R.; Vaca-Paniagua, F.; et al. Variability in Susceptibility to Type I Interferon Response and Subgenomic RNA Accumulation Between Clinical Isolates of Dengue and Zika Virus From Oaxaca Mexico Correlate With Replication Efficiency in Human Cells and Disease Severity. Front. Cell. Infect. Microbiol. 2022, 12, 890750. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Jiang, J.D.; Peng, Z. Recent advances in the anti-HCV mechanisms of interferon. Acta Pharm. Sinica. B 2014, 4, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.; Brodin, H.; Cave, C.; Waugh, N.; Price, A.; Gabbay, J. Pegylated interferon alpha-2a and -2b in combination with ribavirin in the treatment of chronic hepatitis C: A systematic review and economic evaluation. Health technology assessment 2004, 8, iii–125. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Roby, J.A.; Dobyns, W.B.; Rajagopal, L.; Gale, M., Jr.; Adams Waldorf, K.M. Immune Evasion Strategies Used by Zika Virus to Infect the Fetal Eye and Brain. Viral Immunol. 2020, 33, 22–37. [Google Scholar] [CrossRef] [PubMed]
- Grant, A.; Ponia, S.S.; Tripathi, S.; Balasubramaniam, V.; Miorin, L.; Sourisseau, M.; Schwarz, M.C.; Sánchez-Seco, M.P.; Evans, M.J.; Best, S.M.; García-Sastre, A. Zika Virus Targets Human STAT2 to Inhibit Type I Interferon Signaling. Cell Host Microbe 2016, 19, 882–890. [Google Scholar] [CrossRef] [PubMed]
- Iaconis, G.; Jackson, B.; Childs, K.; Boyce, M.; Goodbourn, S.; Blake, N.; Iturriza-Gomara, M.; Seago, J. Rotavirus NSP1 Inhibits Type I and Type III Interferon Induction. Viruses 2021, 13, 589. [Google Scholar] [CrossRef]
- de Witte, L.; Abt, M.; Schneider-Schaulies, S.; van Kooyk, Y.; Geijtenbeek, T.B. Measles virus targets DC-SIGN to enhance dendritic cell infection. J. Virol. 2006, 80, 3477–3486. [Google Scholar] [CrossRef] [PubMed]
- Koga, R.; Ohno, S.; Ikegame, S.; Yanagi, Y. Measles virus-induced immunosuppression in SLAM knock-in mice. J. Virol. 2010, 84, 5360–5367. [Google Scholar] [CrossRef] [PubMed]
- Avota, E.; Gassert, E.; Schneider-Schaulies, S. Measles virus-induced immunosuppression: From effectors to mechanisms. Med. Microbiol. Immunol. 2010, 199, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Avota, E.; Koethe, S.; Schneider-Schaulies, S. Membrane dynamics and interactions in measles virus dendritic cell infections. Cell. Microbiol. 2013, 15, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhu, F.C.; Shu, Y.L.; Zhou, R.; Liu, L.Q.; Zhang, L.L.; Shi, Z.Y.; Tang, Z.; Lin, L.Z.; Yu, Z.A.; et al. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi Zhonghua Shiyan He Linchuang Bingduxue Zazhi Chin. J. Exp. Clin. Virol. 2005, 19, 220–222.
- How, J.; Hobbs, G. Use of Interferon Alfa in the Treatment of Myeloproliferative Neoplasms: Perspectives and Review of the Literature. Cancers 2020, 12, 1954. [Google Scholar] [CrossRef]
- Illés, Á.; Pinczés, L.I.; Egyed, M. A pharmacokinetic evaluation of ropeginterferon alfa-2b in the treatment of polycythemia vera. Expert Opin. Drug Metab. Toxicol. 2021, 17, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Sebők, C.; Walmsley, S.; Tráj, P.; Mackei, M.; Vörösházi, J.; et al. Immunomodulatory effects of chicken cathelicidin-2 on a primary hepatic cell co-culture model. PLoS ONE 2022, 17, e0275847. [Google Scholar] [CrossRef] [PubMed]
- Scott, L.J.; Perry, C.M. Interferon-alpha-2b plus ribavirin: A review of its use in the management of chronic hepatitis C. Drugs 2002, 62, 507–556. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, J.; Jones, J.; Hartwell, D.; Davidson, P.; Price, A.; Waugh, N. Interferon alpha (pegylated and non-pegylated) and ribavirin for the treatment of mild chronic hepatitis C: A systematic review and economic evaluation. Health technology assessment 2007, 11, 1–iii. [Google Scholar] [CrossRef]
- Fatemi Naeini, F.; Yazdanpanah, M.; Mohaghegh, F.; Rajabi, P.; Tabatabaei, E.T. Interferon α-induced lupus-like reaction in a mycosis fungoides patient: A case report. Int. J. Clin. Pharmacol. Ther. 2022, 60, 306–310. [Google Scholar] [CrossRef] [PubMed]
- Albrecht, J.D.; Ninosu, N.; Barry, D.; Albrecht, T.; Schaarschmidt, M.L.; Goerdt, S.; Nicolay, J.P. Non-pegylated and Pegylated Interferon Alpha-2a in Cutaneous T-cell Lymphoma and the Risk of Severe Ocular Side-effects. Acta dermato-venereologica 2022, 102, adv00722. [Google Scholar] [CrossRef] [PubMed]
- Edahiro, Y.; Ohishi, K.; Gotoh, A.; Takenaka, K.; Shibayama, H.; Shimizu, T.; Usuki, K.; Shimoda, K.; Ito, M.; VanWart, S.A.; et al. Efficacy and safety of ropeginterferon alfa-2b in Japanese patients with polycythemia vera: An open-label, single-arm, phase 2 study. Int. J. Hematol. 2022, 116, 215–227. [Google Scholar] [CrossRef]
- Shabunin, S.; Gritsyuk, V.; Vostroilova, G.; Shabanov, D.; Khokhlova, N.; Korchagina, A. Study of Mutagenic Antitoxic Properties of, G.e.n.t.a.b.i.f.e.r.o.n.-B. Macedonian Veterinary Review 2022, 45 79-87. [CrossRef]
- Acevedo Ospina, H.; Guay-Vincent, M.-M.; Descoteaux, A. Macrophage mitochondrial biogenesis and metabolic reprogramming induced by leishmania donovani require lipophosphoglycan and type I interferon signaling. MBio 2022. [CrossRef]
- Lodi, L.; Mastrolia, M.V.; Bello, F.; Rossi, G.M.; Angelotti, M.L.; Crow, Y.J.; Romagnani, P.; Vaglio, A. Type I interferon-related kidney disorders. Kidney Int. 2022, 101, 1142–1159. [Google Scholar] [CrossRef]
- Ma, Z.; Qin, Y.; Jia, Y.; Xie, Y.; Qi, X.; Guo, Y.; He, J.; Zhang, Y.; Li, F.; Yu, J.; et al. Thyroid dysfunction incidence and risk factors in Chinese chronic hepatitis B patients treated with pegylated interferon alpha: A long-term follow-up study. J. Viral Hepat. 2022, 29, 412–419. [Google Scholar] [CrossRef]
- Carella, C.; Mazziotti, G.; Morisco, F.; Manganella, G.; Rotondi, M.; Tuccillo, C.; Sorvillo, F.; Caporaso, N.; Amato, G. Long-term outcome of interferon-alpha-induced thyroid autoimmunity and prognostic influence of thyroid autoantibody pattern at the end of treatment. J. Clin. Endocrinol. Metab. 2001, 86, 1925–1929. [Google Scholar] [CrossRef]
- Vital, E.M.; Merrill, J.T.; Morand, E.F.; Furie, R.A.; Bruce, I.N.; Tanaka, Y.; Manzi, S.; Kalunian, K.C.; Kalyani, R.N.; Streicher, K.; et al. Anifrolumab efficacy and safety by type I interferon gene signature and clinical subgroups in patients with SLE: Post hoc analysis of pooled data from two phase III trials. Ann. Rheum. Dis. 2022, 81, 951–961. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.; Tsay, E.; Federman, N.; Lechner, M.G.; Su, M.A. Polyendocrine Autoimmunity and Diabetic Ketoacidosis Following Anti-PD-1 and Interferon α. Pediatrics 2022, 149, e2021053363. [Google Scholar] [CrossRef] [PubMed]
- Martins, D.; Dipasquale, O.; Davies, K.; Cooper, E.; Tibble, J.; Veronese, M.; Frigo, M.; Williams, S.; Turkheimer, F.; Cercignani, M.; Harrison, N.A. Transcriptomic and cellular decoding of functional brain connectivity changes reveal regional brain vulnerability to pro- and anti-inflammatory therapies. Brain, behavior, and immunity 2022, 102, 312–323. [Google Scholar] [CrossRef]
- Dipasquale, O.; Cooper, E.A.; Tibble, J.; Voon, V.; Baglio, F.; Baselli, G.; Cercignani, M.; Harrison, N.A. Interferon-α acutely impairs whole-brain functional connectivity network architecture—A preliminary study. Brain, behavior, and immunity 2016, 58, 31–39. [Google Scholar] [CrossRef]
- Vignau, J.; Karila, L.; Costisella, O.; Canva, V. Hépatite C, Interféron alpha et dépression: Principales hypothèses physiopathologiques [Hepatitis C, interferon a and depression: Main physiopathologic hypothesis]. L’Encephale 2005, 31, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.A.; Cooper, E.; Voon, V.; Tibble, J.; Cercignani, M.; Harrison, N.A. Interferon and anti-TNF therapies differentially modulate amygdala reactivity which predicts associated bidirectional changes in depressive symptoms. Mol. Psychiatry 2021, 26, 5150–5160. [Google Scholar] [CrossRef] [PubMed]
- Xue, B.; Li, H.; Liu, S.; Feng, Q.; Xu, Y.; Deng, R.; Chen, S.; Wang, J.; Li, X.; Wan, M.; Tang, S.; Zhu, H. The redox cycling of STAT2 maintains innate immune homeostasis. Cell Rep. 2022, 40, 111215. [Google Scholar] [CrossRef] [PubMed]
- Baruch, K.; Deczkowska, A.; David, E.; Castellano, J.M.; Miller, O.; Kertser, A.; Berkutzki, T.; Barnett-Itzhaki, Z.; Bezalel, D.; Wyss-Coray, T.; et al. Aging. Aging-induced type I interferon response at the choroid plexus negatively affects brain function. Science 2014, 346, 89–93. [Google Scholar] [CrossRef]
- Mesquita, S.D.; Ferreira, A.C.; Gao, F.; Coppola, G.; Geschwind, D.H.; Sousa, J.C.; Correia-Neves, M.; Sousa, N.; Palha, J.A.; Marques, F. The choroid plexus transcriptome reveals changes in type I and II interferon responses in a mouse model of Alzheimer’s disease. Brain Behav. Immun. 2015, 49, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Pestka, S.; Krause, C.D.; Walter, M.R. Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 2004, 202, 8–32. [Google Scholar] [CrossRef] [PubMed]
- Deczkowska, A.; Baruch, K.; Schwartz, M. Type I/II Interferon Balance in the Regulation of Brain Physiology and Pathology. Trends Immunol. 2016, 37, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Sakai, S.; Shichita, T. Role of alarmins in poststroke inflammation and neuronal repair. Seminars in immunopathology 2022. Advance online publication. [CrossRef]
- Andersson, U.; Yang, H.; Harris, H. High-mobility group box 1 protein (HMGB1) operates as an alarmin outside as well as inside cells. Semin. Immunol. 2018, 38, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Goh, J.; Behringer, M. Exercise alarms the immune system: A HMGB1 perspective. Cytokine 2018, 110, 222–225. [Google Scholar] [CrossRef]
- Nie, Y.; Yang, D.; Oppenheim, J.J. Alarmins and Antitumor Immunity. Clin. Ther. 2016, 38, 1042–1053. [Google Scholar] [CrossRef] [PubMed]
- Garg, A.D.; Agostinis, P. Cell death and immunity in cancer: From danger signals to mimicry of pathogen defense responses. Immunol. Rev. 2017, 280, 126–148. [Google Scholar] [CrossRef] [PubMed]
- Blank, T.; Prinz, M. Type I interferon pathway in CNS homeostasis and neurological disorders. Glia 2017, 65, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Tresse, E.; Riera-Ponsati, L.; Jaberi, E.; Sew, W.; Ruscher, K.; Issazadeh-Navikas, S. IFN-β rescues neurodegeneration by regulating mitochondrial fission via STAT5, PGAM5, and Drp1. EMBO J. 2021, 40, e106868. [Google Scholar] [CrossRef] [PubMed]
- McDonough, A.; Lee, R.V.; Weinstein, J.R. Microglial Interferon Signaling and White Matter. Neurochem. Res. 2017, 42, 2625–2638. [Google Scholar] [CrossRef]
- Ejlerskov, P.; Hultberg, J.G.; Wang, J.; Carlsson, R.; Ambjørn, M.; Kuss, M.; Liu, Y.; Porcu, G.; Kolkova, K.; Friis Rundsten, C.; et al. Lack of Neuronal IFN-β-IFNAR Causes Lewy Body- and Parkinson’s Disease-like Dementia. Cell 2015, 163, 324–339. [Google Scholar] [CrossRef] [PubMed]
- Magalhaes, J.; Tresse, E.; Ejlerskov, P.; Hu, E.; Liu, Y.; Marin, A.; Montalant, A.; Satriano, L.; Rundsten, C.F.; Carlsen, E.; et al. PIAS2-mediated blockade of IFN-β signaling: A basis for sporadic Parkinson disease dementia. Mol. Psychiatry 2021, 26, 6083–6099. [Google Scholar] [CrossRef] [PubMed]
- Csépány, T.; Bereczki, D. Immunmoduláns kezelés sclerosis multiplexben [Immunomodulatory therapy in multiple sclerosis]. Ideggyogy. Szle. 2004, 57, 401–416. [Google Scholar]
- Kieseier, B.C.; Hartung, H.P. Current disease-modifying therapies in multiple sclerosis. Semin. Neurol. 2003, 23, 133–146. [Google Scholar] [CrossRef] [PubMed]
- Flachenecker, P. Disease-modifying drugs for the early treatment of multiple sclerosis. Expert Rev. Neurother. 2004, 4, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Tintoré, M. New options for early treatment of multiple sclerosis. Journal of the neurological sciences 2009, 277 Suppl 1, S9–S11. [Google Scholar] [CrossRef]
- Brochet, B. Activité à long terme de l’acétate de glatiramère dans le traitement de la sclérose en plaques: État des connaissances [Long-term effects of glatiramer acetate in multiple sclerosis]. Rev. Neurol. 2008, 164, 917–926. [Google Scholar] [CrossRef] [PubMed]
- Gorlé, N.; Vandenbroucke, R.E. Interferons: A molecular switch between damage and repair in ageing and Alzheimer’s disease. Mech. Ageing Dev. 2019, 183, 111148. [Google Scholar] [CrossRef] [PubMed]
- Feng, E.; Balint, E.; Poznanski, S.M.; Ashkar, A.A.; Loeb, M. Aging and Interferons: Impacts on Inflammation and Viral Disease Outcomes. Cells 2021, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Frisch, S.M.; MacFawn, I.P. Type I interferons and related pathways in cell senescence. Aging Cell 2020, 19, e13234. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef] [PubMed]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; et al. Author Correction: L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 572, E5. [Google Scholar] [CrossRef]
- Childs, B.; van Deursen, J. Inhibition of ‘jumping genes’ promotes healthy ageing. Nature 2019, 566, 46–48. [Google Scholar] [CrossRef]
- Gilbert, C.; Lefeuvre, C.; Preisser, L.; Pivert, A.; Soleti, R.; Blanchard, S.; Delneste, Y.; Ducancelle, A.; Couez, D.; Jeannin, P. Age-Related Expression of IFN-λ1 Versus IFN-I and Beta-Defensins in the Nasopharynx of SARS-CoV-2-Infected Individuals. Front. Immunol. 2021, 12, 750279. [Google Scholar] [CrossRef] [PubMed]
- Wan, D.; Jiang, W.; Hao, J. Research Advances in How the cGAS-STING Pathway Controls the Cellular Inflammatory Response. Front. Immunol. 2020, 11, 615. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Colonna, M. Microglia in Alzheimer’s disease at single-cell level. Are there common patterns in humans and mice? J. Exp. Med. 2021, 218, e20202717. [Google Scholar] [CrossRef] [PubMed]
- Levine, M.E.; Lu, A.T.; Quach, A.; Chen, B.H.; Assimes, T.L.; Bandinelli, S.; Hou, L.; Baccarelli, A.A.; Stewart, J.D.; Li, Y.; et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 2018, 10, 573–591. [Google Scholar] [CrossRef] [PubMed]
- Lim, U.; Song, M.A. DNA Methylation as a Biomarker of Aging in Epidemiologic Studies. Methods Mol. Biol. 2018, 1856, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Rauchhaus, J.; Robinson, J.; Monti, L.; Di Antonio, M. G-quadruplexes Mark Sites of Methylation Instability Associated with Ageing and Cancer. Genes 2022, 13, 1665. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.J.; Hwang, J.A.; Yang, E.J.; Kim, E.C.; Kim, J.R.; Kim, S.Y.; Kim, Y.Z.; Park, S.C.; Lee, Y.S. Nintedanib induces senolytic effect via STAT3 inhibition. Cell Death Dis. 2022, 13, 760. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.; Shin, S.; Cho, E.; Ryu, D.; Garandeau, D.; Chajra, H.; Fréchet, M.; Park, D.; Jung, E. Senotherapeutic-like effect of Silybum marianum flower extract revealed on human skin cells. PLoS ONE 2021, 16, e0260545. [Google Scholar] [CrossRef]
- Kim, J.C.; Park, T.J.; Kang, H.Y. Skin-Aging Pigmentation: Who Is the Real Enemy? Cells 2022, 11, 2541. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jang, J.; Song, M.J.; Kim, G.; Park, C.H.; Lee, D.H.; Lee, S.H.; Chung, J.H. Attenuation of intrinsic ageing of the skin via elimination of senescent dermal fibroblasts with senolytic drugs. J. Eur. Acad. Dermatol. Venereol. JEADV 2022, 36, 1125–1135. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Jang, J.; Song, M.J.; Park, C.H.; Lee, D.H.; Lee, S.H.; Chung, J.H. Inhibition of matrix metalloproteinase expression by selective clearing of senescent dermal fibroblasts attenuates ultraviolet-induced photoaging. Biomed. Pharmacother. = Biomed. Pharmacother. 2022, 150, 113034. [Google Scholar] [CrossRef]
- Choo, O.-S.; Lee, Y.Y.; Kim, Y.S.; Kim, Y.J.; Lee, D.H.; Kim, H.; Jang, J.H.; Choung, Y.-H. Effect of statin on age-related hearing loss via drug repurposing. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2022, 1869, 119331. [Google Scholar] [CrossRef] [PubMed]
- Dacia Plant (2021), Interferonate tablets. https://doi.org/2020). Association between anti-interferon-alpha autoantibodies and COVID-19 in systemic lupus erythematosus. medRxiv: The preprint server for health sciences, 2020.10.29.20222000. [CrossRef]
- Solórzano, A.; Webby, R.J.; Lager, K.M.; Janke, B.H.; García-Sastre, A.; Richt, J.A. Mutations in the NS1 protein of swine influenza virus impair anti-interferon activity and confer attenuation in pigs. J. Virol. 2005, 79, 7535–7543. [Google Scholar] [CrossRef] [PubMed]
- Ngunjiri, J.M.; Buchek, G.M.; Mohni, K.N.; Sekellick, M.J.; Marcus, P.I. Influenza virus subpopulations: Exchange of lethal H5N1 virus NS for H1N1 virus NS triggers de novo generation of defective-interfering particles and enhances interferon-inducing particle efficiency. J. Interferon Cytokine Res. Off. J. Int. Soc. Interferon Cytokine Res. 2013, 33, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Ives Charlie-Silva, Amanda, P.C. Araújo, Abraão, T.B. Guimarães, Flávio P Veras, Helyson, L.B. Braz, Letícia, G. de Pontes, Roberta J. B. Jorge, Marco A. A. Belo, Bianca H, V. Fernandes, Rafael H. Nóbrega, Giovane Galdino, Antônio Condino-Neto, Jorge Galindo-Villegas, Glaucia M. Machado-Santelli, Paulo R. S. Sanches, Rafael M. Rezende, Eduardo M. Cilli, Guilherme Malafaia, An insight into neurotoxic and toxicity of spike fragments SARS-CoV-2 by exposure environment: A threat to aquatic health?, available at: bioRxiv 2021.01.11.425914. [CrossRef]
- Raghavan, S.; Kenchappa, D.B.; Leo, M.D. (2021, May 19). SARS-COV-2 spike protein induces degradation of junctional proteins that maintain endothelial barrier integrity. Frontiers. Retrieved December 5, 2022, from https://www.frontiersin.org/articles/10.3389/fcvm.2021.687783/full. [CrossRef]
- Jafarzadeh, A.; Chauhan, P.; Saha, B.; Jafarzadeh, S.; Nemati, M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: Lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020, 257, 118102. [Google Scholar] [CrossRef]
- Brown, M.; Bhardwaj, N. Super(antigen) target for SARS-CoV-2. Nat. Rev. Immunol. 2021, 21, 72. [Google Scholar] [CrossRef]
- Huang, Y.; Xie, J.; Guo, Y.; Sun, W.; He, Y.; Liu, K.; Yan, J.; Tao, A.; Zhong, N. SARS-CoV-2: Origin, Intermediate Host and Allergenicity Features and Hypotheses. Healthcare 2021, 9, 1132. [Google Scholar] [CrossRef] [PubMed]
- Grifoni, A.; Weiskopf, D.; Ramirez, S.I.; Mateus, J.; Dan, J.M.; Moderbacher, C.R.; Rawlings, S.A.; Sutherland, A.; Premkumar, L.; Jadi, R.S.; et al. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell 2020, 181, 1489–1501.e15. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.H.; Zhang, S.; Porritt, R.A.; Rivas, M.N.; Paschold, L.; Willscher, E.; Binder, M.; Arditi, M.; Bahar, I. Superantigenic character of an insert unique to SARS-CoV-2 spike supported by skewed TCR repertoire in patients with hyperinflammation. Proc. Natl. Acad. Sci. 2020, 117, 25254–25262. [Google Scholar] [CrossRef] [PubMed]
- Bruttel, V.; Washburne, A.; VanDongen, A. Endonuclease fingerprint indicates a synthetic origin of SARS-COV-2. 2022. [CrossRef]
- Fleming, D.R.M. (2021). Is COVID-19 a Bioweapon?: A Scientific and Forensic investigation. Skyhorse Publishing (NY). ISBN 978-1-5107-7019-5.
- Lyons-Weiler, J. Pathogenic priming likely contributes to serious and critical illness and mortality in COVID-19 via autoimmunity. J Transl Autoim 2020, 3, 100051. [Google Scholar] [CrossRef]
- Navaratnarajah, C.K.; Pease, D.R.; Halfmann, P.J.; Taye, B.; Barkhymer, A.; Howell, K.G.; Charlesworth, J.E.; Christensen, T.A.; Kawaoka, Y.; Cattaneo, R.; Schneider, J.W. Wanek Family Program for HLHS-Stem Cell Pipeline Highly Efficient SARS-CoV-2 Infection of Human Cardiomyocytes: Spike Protein-Mediated Cell Fusion and Its Inhibition. J. Virol. 2021, 95, e0136821. [Google Scholar] [CrossRef] [PubMed]
- Buzhdygan, T.P.; DeOre, B.J.; Baldwin-Leclair, A.; Bullock, T.A.; McGary, H.M.; Khan, J.A.; Razmpour, R.; Hale, J.F.; Galie, P.A.; Potula, R.; et al. The SARS-CoV-2 spike protein alters barrier function in 2D static and 3D microfluidic in-vitro models of the human blood-brain barrier. Neurobiol. Dis. 2020, 146, 105131. [Google Scholar] [CrossRef] [PubMed]
- Ken Shirato and Takako Kizaki (2021), SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages, Heliyon 7 e06187, pp 2-9. [CrossRef]
- Kim, E.S.; Jeon, M.T.; Kim, K.S.; Lee, S.; Kim, S.; Kim, D.G. Spike Proteins of SARS-CoV-2 Induce Pathological Changes in Molecular Delivery and Metabolic Function in the Brain Endothelial Cells. Viruses 2021, 13, 2021. [Google Scholar] [CrossRef] [PubMed]
- Kalashnyk, O.; Lykhmus, O.; Izmailov, M.; Koval, L.; Komisarenko, S.; Skok, M. SARS-Cov-2 spike protein fragment 674-685 protects mitochondria from releasing cytochrome c in response to apoptogenic influence. Biochem. Biophys. Res. Commun. 2021, 561, 14–18. [Google Scholar] [CrossRef] [PubMed]
- Segreto, R.; Deigin, Y. The genetic structure of SARS-CoV-2 does not rule out a laboratory origin: SARS-COV-2 chimeric structure and furin cleavage site might be the result of genetic manipulation. BioEssays: News Rev. Mol. Cell. Dev. Biol. 2021, 43, e2000240. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.K.; Kulsum, U.; Rufai, S.B.; Mudliar, S.R.; Singh, S. Mutations in SARS-CoV-2 Leading to Antigenic Variations in Spike Protein: A Challenge in Vaccine Development. J. Lab. Physicians 2020, 12, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Arvin, A.M.; Fink, K.; Schmid, M.A.; Cathcart, A.; Spreafico, R.; Havenar-Daughton, C.; Lanzavecchia, A.; Corti, D.; Virgin, H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature 2020, 584, 353–363. [Google Scholar] [CrossRef]
- Ricke, D.O. Two Different Antibody-Dependent Enhancement (ADE) Risks for SARS-CoV-2 Antibodies. Front. Immunol. 2021, 12, 640093. [Google Scholar] [CrossRef] [PubMed]
- Bournazos, S.; Gupta, A.; Ravetch, J.V. The role of IgG Fc receptors in antibody-dependent enhancement. Nat. Rev. Immunol. 2020, 20, 633–643. [Google Scholar] [CrossRef] [PubMed]
- Halstead, S.B. Vaccine-Associated Enhanced Viral Disease: Implications for Viral Vaccine Development. BioDrugs: Clin. Immunother. Biopharm. Gene Ther. 2021, 35, 505–515. [Google Scholar] [CrossRef] [PubMed]
- Bigay, J.; Le Grand, R.; Martinon, F.; Maisonnasse, P. Vaccine-associated enhanced disease in humans and animal models: Lessons and challenges for vaccine development. Front. Microbiol. 2022, 13, 932408. [Google Scholar] [CrossRef] [PubMed]
- Gartlan, C.; Tipton, T.; Salguero, F.J.; Sattentau, Q.; Gorringe, A.; Carroll, M.W. Vaccine-Associated Enhanced Disease and Pathogenic Human Coronaviruses. Front. Immunol. 2022, 13, 882972. [Google Scholar] [CrossRef] [PubMed]
- Costa Silva, R.; Bandeira-Melo, C.; Paula Neto, H.A.; Vale, A.M.; Travassos, L.H. COVID-19 diverse outcomes: Aggravated reinfection, type I interferons and antibodies. Med. Hypotheses 2022, 167, 110943. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.S.; Hughes, S.H. HIV-1 reverse transcription. Cold Spring Harb. Perspect. Med. 2012, 2, a006882. [Google Scholar] [CrossRef] [PubMed]
- Desfarges, S.; Ciuffi, A. (2012). Viral Integration and Consequences on Host Gene Expression. In: Witzany, G. (eds) Viruses: Essential Agents of Life. Springer, Dordrecht. [CrossRef]
- Sciamanna, I.; De Luca, C.; Spadafora, C. The Reverse Transcriptase Encoded by LINE-1 Retrotransposons in the Genesis, Progression, and Therapy of Cancer. Front. Chem. 2016, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Richards, A.; Khalil, A.; Wogram, E.; Ma, H.; Young, R.A.; Jaenisch, R. SARS-CoV-2 RNA reverse-transcribed and integrated into the human genome. bioRxiv [Preprint]. 2020 Dec 13:2020.12.12.422516. [CrossRef]
- Tarasova, O.; Ivanov, S.; Filimonov, D.A.; Poroikov, V. Data and Text Mining Help Identify Key Proteins Involved in the Molecular Mechanisms Shared by SARS-CoV-2 and HIV-1. Molecules 2020, 25, 2944. [Google Scholar] [CrossRef] [PubMed]
- Doerfler, W. Adenoviral Vector DNA- and SARS-CoV-2 mRNA-Based Covid-19 Vaccines: Possible Integration into the Human Genome—Are Adenoviral Genes Expressed in Vector-based Vaccines? Virus Res. 2021, 302, 198466. [Google Scholar] [CrossRef] [PubMed]
- Kowarz, E.; Krutzke, L.; Reis, J.; Bracharz, S.; Kochanek, S.; Marschalek, R. (2021). Vaccine-induced COVID-19 mimicry syndrome: Splice reactions within the SARS-CoV-2 spike open reading frame result in spike protein variants that may cause thromboembolic events in patients immunized with vector-based vaccines. Nature-portfolio [preprint]. [CrossRef]
- Krutzke, L.; Roesler, R.; Wiese, S.; Kochanek, S. (2021). Process-related impurities in the ChAdOx1 nCov-19 vaccine. Nature-portfolio [preprint]. [CrossRef]
- NatBiotech, Research not fit to print. Nat Biotechnol 2016, 34, 115. [CrossRef] [PubMed]
- Aldén, M.; Olofsson Falla, F.; Yang, D.; Barghouth, M.; Luan, C.; Rasmussen, M.; De Marinis, Y. Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line. Curr. Issues Mol. Biol. 2022, 44, 1115–1126, https://www.mdpi.com/1467-3045/44/3/73/htm. [Google Scholar] [CrossRef] [PubMed]
- Luisetto, M.; Almukthar, N.; Tarro, G. Intracellular Reverse Transcription of COVID-19 mRNA Vaccine; LAP LAMBERT Academic Publishing: 2022, 1, 3-67. ISBN: 978-620-0-31572-4.
- Lurie, N.; Saville, M.; Hatchett, R.; Halton, J. Developing COVID-19 vaccines at pandemic speed. N Engl J Med 2020, 382, 1969–1973. [Google Scholar] [CrossRef]
- Kounis, N.G.; Koniari, I.; Mplani, V.; Plotas, P.; Velissaris, D. Hypersensitivity myocarditis and the pathogenetic conundrum of COVID 19 Vaccine Related Myocarditis. Cardiology 2022. Advance online publication. [CrossRef]
- Cadegiani, F.A. Catecholamines Are the Key Trigger of COVID-19 mRNA Vaccine-Induced Myocarditis: A Compelling Hypothesis Supported by Epidemiological, Anatomopathological, Molecular, and Physiological Findings. Cureus 2022, 14, e27883. [Google Scholar] [CrossRef] [PubMed]
- Hirose, S.; Hara, M.; Koda, K.; Natori, N.; Yokota, Y.; Ninomiya, S.; Nakajima, H. Acute autoimmune transverse myelitis following COVID-19 vaccination: A case report. Medicine 2021, 100, e28423. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.C.; Wang, C.H.; Chang, K.C.; Hung, M.J.; Chen, H.Y.; Liao, S.C. Guillain-Barré Syndrome Associated with COVID-19 Vaccination. Emerg Infect Dis 2021, 27, 3175–3178. [Google Scholar] [CrossRef] [PubMed]
- Farinholt, T.; Doddapaneni, H.; Qin, X.; Menon, V.; Meng, Q.; Metcalf, G.; Chao, H.; Gingras, M.C.; Avadhanula, V.; Farinholt, P.; et al. Transmission event of SARS-CoV-2 delta variant reveals multiple vaccine breakthrough infections. BMC Med 2021, 19, 255. [Google Scholar] [CrossRef] [PubMed]
- Goldman, S.; Bron, D.; Tousseyn, T.; Vierasu, I.; Dewispelaere, L.; Heimann, P.; Cogan, E.; Goldman, M. Rapid progression of angioimmunoblastic T Cell lymphoma following BNT162b2 mRNA vaccine booster shot: A case report. Front. Med. 2021, 8, 798095. [Google Scholar] [CrossRef] [PubMed]
- Jureidini, J.; McHenry, L.B. The illusion of evidence based medicine BMJ 2022, 376, o702. [CrossRef]
- Savulescu, J.; Giubilini, A.; Danchin, M. Global Ethical Considerations Regarding Mandatory Vaccination in Children. J. Pediatr. 2021, 231, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Schoenmaker, L.; Witzigmann, D.; Kulkarni, J.A.; Verbeke, R.; Kersten, G.; Jiskoot, W.; Crommelin, D. mRNA-lipid nanoparticle COVID-19 vaccines: Structure and stability. Int. J. Pharm. 2021, 601, 120586. [Google Scholar] [CrossRef] [PubMed]
- Pardi, N.; Weissman, D. (2017) Nucleoside Modified mRNA Vaccines for Infectious Diseases. In: Kramps, T.; Elbers, K. (eds) RNA Vaccines. Methods in Molecular Biology, vol 1499. Humana Press, New York, NY. [CrossRef]
- Kirchdoerfer, R.N.; Wang, N.; Pallesen, J.; Wrapp, D.; Turner, H.L.; Cottrell, C.A.; Corbett, K.S.; Graham, B.S.; McLellan, J.S.; Ward, A.B. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. Sci. Rep. 2018, 8, 15701. [Google Scholar] [CrossRef] [PubMed]
- Timothy, P. Riley, Hui-Ting Chou, Ruozhen Hu, Krzysztof P. Bzymek, Ana R. Correia, Alexander C. Partin, Danqing Li, Danyang Gong, Zhulun Wang, Xinchao Yu, Paolo Manzanillo and Fernando Garces (2021), Enhancing the Prefusion Conformational Stability of SARS-CoV-2 Spike Protein Through Structure-Guided Design, Frontiers in Immunology. [CrossRef]
- Li, C.; Chen, Y.; Zhao, Y.; Lung, D.C.; Ye, Z.; Song, W.; Liu, F.-F.; Cai, J.-P.; Wong, W.-M.; Yip, C.C.-Y.; et al. Intravenous Injection of Coronavirus Disease 2019 (COVID-19) mRNA Vaccine Can Induce Acute Myopericarditis in Mouse Model. Clin. Infect. Dis. 2021, 74, 1933–1950. [Google Scholar] [CrossRef] [PubMed]
- Gargano, J.W.; Wallace, M.; Hadler, S.C.; Langley, G.; Su, J.R.; Oster, M.E.; Broder, K.R.; Gee, J.; Weintraub, E.; Shimabukuro, T.; et al. Use of mRNA COVID-19 Vaccine After Reports of Myocarditis Among Vaccine Recipients: Update from the Advisory Committee on Immunization Practices—United States, MMWR Morb Mortal Wkly Rep 2021, 70, 977–982. [CrossRef]
- Menni, C.; Klaser, K.; May, A.; Polidori, L.; Nguzen, L.H.; Drew, D.A.; Merino, J.; Hu, C.; Selvachandran, S.; Antonelli, M.; et al. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: A prospective observational study. Lancet Infect Dis 2021, 21, 939–949. [Google Scholar] [CrossRef]
- Block, J. (2021). Vaccinating people who have had covid-19: Why doesn’t natural immunity count in the US? BMJ, n2101. [CrossRef]
- Jiang, S. Don’t rush to deploy COVID-19 vaccines and drugs without sufficient safety guarantees. Nature 2020, 579, 321. [Google Scholar] [CrossRef] [PubMed]
- Kostoff, R.N.; Calina, D.; Kanduc, D.; Briggs, M.B.; Vlachoyiannopoulos, P.; Svistunov, A.A.; Tsatsakis, A. Why are we vaccinating children against COVID-19? Toxicol Rep. 2021, 8, 1665–1684. [Google Scholar] [CrossRef]
- Cabanillas, B.; Akdis, C.; Novak, N. (2021). Allergic reactions to the first COVID-19 vaccine: A potential role of polyethylene glycol? Allergy, 76, 1617-1618. [CrossRef]
- Kim, M.A.; Lee, Y.W.; Kim, S.R.; Kim, J.H.; Min, T.K.; Park, H.S.; Shin, M.; Ye, Y.M.; Lee, S.; Lee, J.; et al. COVID-19 vaccine-associated anaphylaxis and allergic reactions: Consensus statements of the KAAACI Urticaria/Angioedema/Anaphylaxis Working Group. Allergy Asthma Immunol Res 2021, 13, 526–544. [Google Scholar] [CrossRef] [PubMed]
- Cerpa-Cruz, S.; Paredes-Casillas, P.; Landeros Navarro, E.; et al. Adverse events following immunization with vaccines containing adjuvants. Immunol Res 2013, 56, 299–303. [Google Scholar] [CrossRef] [PubMed]
- Sangaletti, P.; Doe, J.; Gatti, A.; Arvay, C.; Giuliani, L.; Lettner, H. SARS-CoV-2 and the Vaccination Hype. Int. J. Vaccine Theory Pract. Res. 2022, 2, 173–207. [Google Scholar] [CrossRef]
- Sumi, T.; Nagahisa, Y.; Matsuura, K.; Sekikawa, M.; Yamada, Y.; Nakata, H.; Chiba, H. Lung squamous cell carcinoma with hemoptysis after vaccination with tozinameran (BNT162b2, Pfizer-BioNTech). Thorac Cancer 2021, 12, 3072–3075, https://onlinelibrary.wiley.co. [Google Scholar] [CrossRef] [PubMed]
- Shimoyama, S.; Kanisawa, Y.; Ono, K.; Souri, M.; Ichinose, A. First and fatal case of autoimmune acquired factor XIII/13 deficiency after COVID-19/SARS-CoV-2 vaccination. Am J Hematol 2021, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Seneff, S.; Nigh, G. Worse than the disease? Reviewing some possible unintended consequences of the mRNA vaccines against COVID-19. International Journal of Vaccine Theory Practice and Research 2021, 2, 38–79, Retrieved from https://ijvtpr.com/index.php/IJVTPR/article/view/23. [Google Scholar] [CrossRef]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.A.; Kaneda, A.; The Genotype to Phenotype Japan (G2P-Japan) Consortium, Nakagawa, S.; Sato, K. Ineffective neutralization of the SARS-CoV-2 Mu variant by convalescent and vaccine sera. bioRxiv 2021, 2021.09.06.459005. [CrossRef]
- Inserra, F.; Tajer, C.; Antonietti, L.; Mariani, J.; Ferder, L.; Manucha, W. Vitamin D supplementation: An alternative to enhance the effectiveness of vaccines against SARS-CoV-2? Vaccine 2021, 39, 4930–4931. [Google Scholar] [CrossRef] [PubMed]
- Grenfell, B.T.; Pybus, O.G.; Gog, J.R.; Wood, J.L.; Daly, J.M.; Mumford, J.A.; Holmes, E.C. Unifying the epidemiological and evolutionary dynamics of pathogens. Science 2004, 303, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Villarreal, L.P. (2015) Virolution Can Help Us Understand the Origin of Life. In: Kolb V (ed). Astrobiology—An Evolutionary Approach. CRC Press, Boca Raton (FL); ISBN 13-978-1-4665-8462-4.
- Villarreal, L.P.; Witzany, G. (2021) Social networking of quasi-species consortia drive virolution via persistence. AIMS Microbiology, 7, 138-162. [CrossRef]
- Ho, M.-W. The new genetics and natural versus artificial genetic modification. Entropy 2013, 15, 4748–4781. [Google Scholar] [CrossRef]
- Ho, M.-W. (1999). Genetic Engineering, Dream or Nightmare? The Brave New World of Bad Science and Big Business, 2nd ed. Gateway Books, Dublin (IRL). ISBN 0-8264-1257-2.
- Mathieu, E.; Ritchie, H.; Ortiz-Ospina, E.; Roser, M.; Hasell, J.; Appel, C.; Giattino, C.; Ortiz-Ospina, E. (2021). A global database of COVID-19 vaccinations. Nat Hum Behav 5: 947-953. Data on COVID-19 (coronavirus) vaccinations by Our World in Data, accessible: https://github.com/owid/COVID-19-data/tree/master/public/data/vaccinations. [CrossRef]
- Cerezo, L.; Macià I Garau, M. Acute radiation syndrome and Fukushima: A watershed moment? Rep. Pract. Oncol. Radiother. J. Greatpoland Cancer Cent. Pozn. Pol. Soc. Radiat. Oncol. 2012, 17, 1–3. [Google Scholar] [CrossRef]
- Rios, C.I.; Cassatt, D.R.; Hollingsworth, B.A.; Satyamitra, M.M.; Tadesse, Y.S.; Taliaferro, L.P.; Winters, T.A.; DiCarlo, A.L. Commonalities Between COVID-19 and Radiation Injury. Radiat. Res. 2021, 195, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Moozhipurath, R.K.; Kraft, L.; Skiera, B. Evidence of protective role of Ultraviolet-B (UVB) radiation in reducing COVID-19 deaths. Sci Rep 2020, 10, 17705. [Google Scholar] [CrossRef] [PubMed]
- Moozhipurath, R.K.; Kraft, L. Association of lockdowns with the protective role of ultraviolet-B (UVB) radiation in reducing COVID-19 deaths. Sci Rep 2021, 11, 22851. [Google Scholar] [CrossRef] [PubMed]
- Rezaie, A.; Melmed, G.Y.; Leite, G.; et al. Endotracheal Application of Ultraviolet A Light in Critically Ill Patients with Severe Acute Respiratory Syndrome Coronavirus 2: A First-in-Human Study. Adv Ther 2021, 38, 4556–4568. [Google Scholar] [CrossRef] [PubMed]
- Stawicki, S.P. Could tracheo-bronchial ultraviolet C irradiation be a valuable adjunct in the management of severe COVID-19 pulmonary infections? Int J Acad Med [serial online] 2020 [cited 2022 Feb 28];6:156-8. Available from: https://www.ijam-web.org/text.asp?2020/6/2/156/287946.
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




