Submitted:
30 December 2022
Posted:
05 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
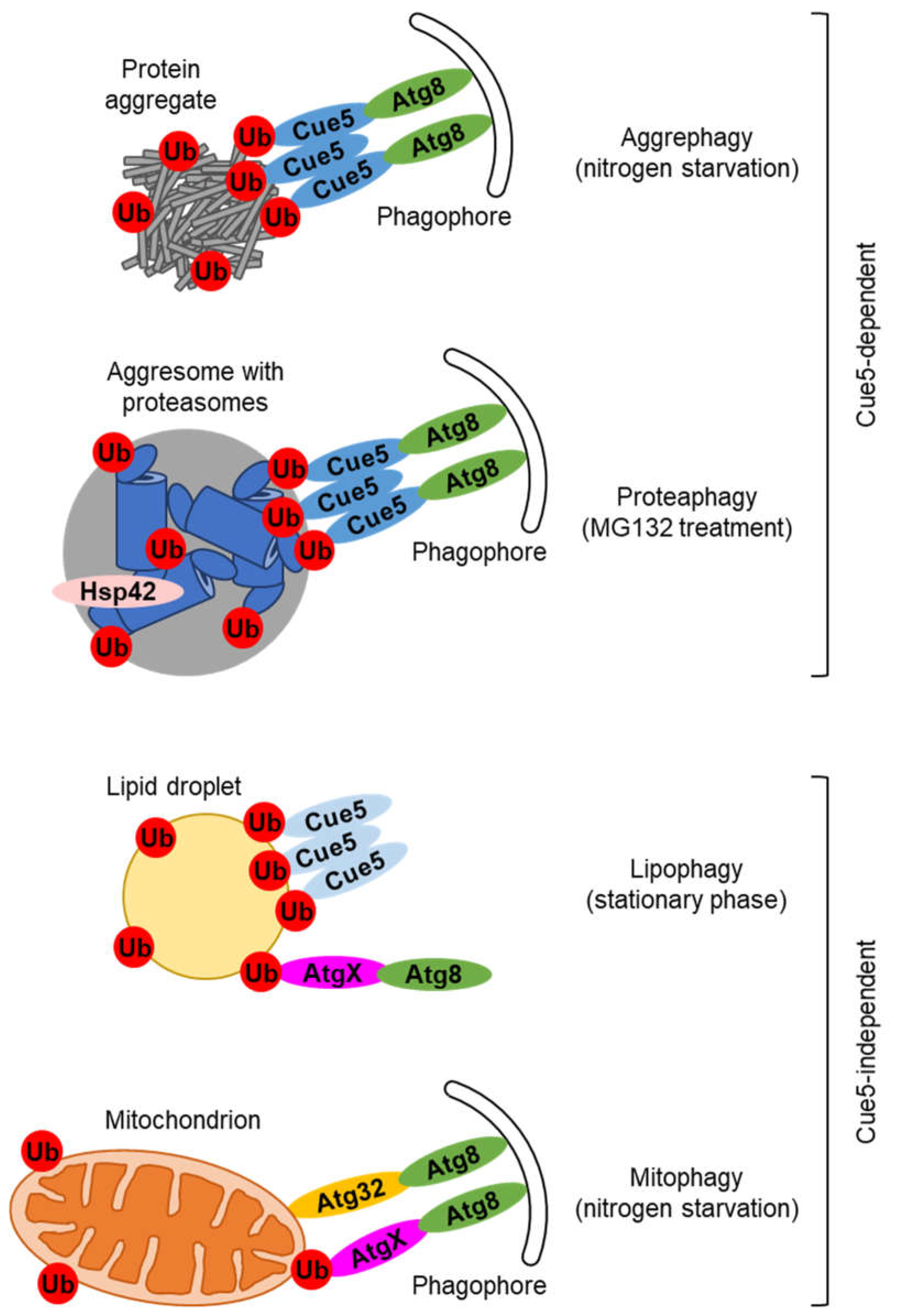
2. Cue5, the SQSTM1-like autophagic receptor in yeast
3. Ubiquitin-dependent but Cue5-independent autophagic pathways in yeast
4. Candidates for alternative ubiquitin-binding SARs in yeast
| Protein | Brief description | Ubiquitin-binding domain | Physical interactions | Autophagy regulator | ||
|---|---|---|---|---|---|---|
| Atg11 | Self | Total | ||||
| Arc40 | Subunit of the Arp2/3 complex | - | - | - | 51 | - |
| Ccr4 | Component of the Ccr4-Not complex | - | Yes | Yes | 3,035 | Yes [23] |
| Cdc48 | AAA ATPase with protein-unfoldase activity | - | - | Yes | 200 | Yes [29-31] |
| Cue5 | Ubiquitin-binding protein | CUE | - | Yes | 29 | Yes [9,10] |
| Dhh1 | DEAD-box helicase/mRNA decapping activator | - | Yes | Yes | 3,608 | Yes [24,25] |
| Dsk2 | Ubiquitin-like polyubiquitin-binding protein | UBA | - | Yes | 43 | Yes [26] |
| Ede1 | Endocytic adaptor | UBA | Yes [28] | Yes | 62 | Yes [28] |
| Ent1 | Epsin-like protein | UIM (x2) | - | - | 20 | - |
| Ent2 | Epsin-like protein | UIM (x2) | - | - | 48 | - |
| Nab2 | Nuclear polyadenylated RNA-binding protein | - | - | Yes | 2,641 | - |
| Nup84 | Subunit of the Nup84 subcomplex of NPC | - | - | Yes | 74 | - |
| Rsp5 | NEDD4 family E3 ubiquitin ligase | HECT | - | Yes | 354 | Yes [9,19,32] |
| Shp1 | UBX domain-containing protein, binds Cdc48 | UBA | - | - | 65 | Yes [30] |
| Sla1 | Cytoskeletal protein binding protein | - | - | Yes | 189 | - |
| Ubx5 | UBX domain-containing protein, binds Cdc48 | UBA, UIM [22] | - | - | 26 | Yes [22] |
| Ydj1 | Type I Hsp40 co-chaperone | - | - | Yes | 133 | Yes [27] |
5. Conclusions
Funding
Disclosure Statement
References
- Leber, R.; Silles, E.; Sandoval, I.V.; Mazón, M.J. Yol082p, a Novel CVT Protein Involved in the Selective Targeting of Aminopeptidase I to the Yeast Vacuole. J. Biol. Chem. 2001, 276, 29210–29217. [Google Scholar] [CrossRef]
- Scott, S.V.; Guan, J.; Hutchins, M.U.; Kim, J.; Klionsky, D.J. Cvt19 Is a Receptor for the Cytoplasm-to-Vacuole Targeting Pathway. Mol. Cell 2001, 7, 1131–1141. [Google Scholar] [CrossRef]
- Bjorkoy, G.; Lamark, T.; Brech, A.; et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–14. [Google Scholar] [CrossRef]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–45. [Google Scholar] [CrossRef] [PubMed]
- Kirkin, V.; Rogov, V.V. A Diversity of Selective Autophagy Receptors Determines the Specificity of the Autophagy Pathway. Mol. Cell 2019, 76, 268–285. [Google Scholar] [CrossRef]
- Shroff, A.; Nazarko, T.Y. SQSTM1, lipid droplets and current state of their lipophagy affairs. Autophagy 2022, 19, 720–723. [Google Scholar] [CrossRef]
- Kim, J.; Huang, W.P.; Stromhaug, P.E.; et al. Convergence of multiple autophagy and cytoplasm to vacuole targeting components to a perivacuolar membrane compartment prior to de novo vesicle formation. J. Biol. Chem. 2002, 277, 763–73. [Google Scholar] [CrossRef]
- Shintani, T.; Huang, W.-P.; Stromhaug, P.E.; Klionsky, D.J. Mechanism of Cargo Selection in the Cytoplasm to Vacuole Targeting Pathway. Dev. Cell 2002, 3, 825–837. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Psakhye, I.; Jentsch, S. Autophagic Clearance of PolyQ Proteins Mediated by Ubiquitin-Atg8 Adaptors of the Conserved CUET Protein Family. Cell 2014, 158, 549–563. [Google Scholar] [CrossRef]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic Turnover of Inactive 26S Proteasomes in Yeast Is Directed by the Ubiquitin Receptor Cue5 and the Hsp42 Chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef]
- Lu, K.; den Brave, F.; Jentsch, K.L.F.D.B.S. Receptor oligomerization guides pathway choice between proteasomal and autophagic degradation. Nat. Cell Biol. 2017, 19, 732–739. [Google Scholar] [CrossRef]
- Robichaud, S.; Fairman, G.; Vijithakumar, V.; Mak, E.; Cook, D.P.; Pelletier, A.R.; Huard, S.; Vanderhyden, B.C.; Figeys, D.; Lavallée-Adam, M.; et al. Identification of novel lipid droplet factors that regulate lipophagy and cholesterol efflux in macrophage foam cells. Autophagy 2021, 17, 3671–3689. [Google Scholar] [CrossRef]
- Kumar, R.; Shroff, A.; Nazarko, T.Y. Komagataella phaffii Cue5 Piggybacks on Lipid Droplets for Its Vacuolar Degradation during Stationary Phase Lipophagy. Cells 2022, 11, 215. [Google Scholar] [CrossRef]
- van Zutphen, T.; Todde, V.; de Boer, R.; Kreim, M.; Hofbauer, H.F.; Wolinski, H.; Veenhuis, M.; van der Klei, I.J.; Kohlwein, S.D. Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 2014, 25, 290–301. [Google Scholar] [CrossRef]
- Wang, C.-W.; Miao, Y.-H.; Chang, Y.-S. A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J. Cell Biol. 2014, 206, 357–366. [Google Scholar] [CrossRef]
- Seo, A.Y.; Lau, P.W.; Feliciano, D.; et al. AMPK and vacuole-associated Atg14p orchestrate mu-lipophagy for energy production and long-term survival under glucose starvation. Elife 2017, 6. [Google Scholar] [CrossRef]
- Kumar, R.; Rahman, M.A.; Nazarko, T.Y. Nitrogen Starvation and Stationary Phase Lipophagy Have Distinct Molecular Mechanisms. Int. J. Mol. Sci. 2020, 21, 9094. [Google Scholar] [CrossRef]
- Müller, M.; Kötter, P.; Behrendt, C.; Walter, E.; Scheckhuber, C.Q.; Entian, K.-D.; Reichert, A.S. Synthetic Quantitative Array Technology Identifies the Ubp3-Bre5 Deubiquitinase Complex as a Negative Regulator of Mitophagy. Cell Rep. 2015, 10, 1215–1225. [Google Scholar] [CrossRef]
- Belgareh-Touzé, N.; Cavellini, L.; Cohen, M.M. Ubiquitination of ERMES components by the E3 ligase Rsp5 is involved in mitophagy. Autophagy 2017, 13, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, K.; Kondo-Okamoto, N.; Ohsumi, Y. Mitochondria-Anchored Receptor Atg32 Mediates Degradation of Mitochondria via Selective Autophagy. Dev. Cell 2009, 17, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Kanki, T.; Wang, K.; Cao, Y.; Baba, M.; Klionsky, D.J. Atg32 Is a Mitochondrial Protein that Confers Selectivity during Mitophagy. Dev. Cell 2009, 17, 98–109. [Google Scholar] [CrossRef]
- Marshall, R.S.; Hua, Z.; Mali, S.; et al. ATG8-Binding UIM Proteins Define a New Class of Autophagy Adaptors and Receptors. Cell 2019, 177, 766–781. [Google Scholar] [CrossRef]
- Yin, Z.; Zhang, Z.; Lei, Y.; Klionsky, D.J. Bidirectional roles of the Ccr4-Not complex in regulating autophagy before and after nitrogen starvation. Autophagy 2022, 19, 415–425. [Google Scholar] [CrossRef]
- Hu, G.; McQuiston, T.; Bernard, A.; Park, Y.-D.; Qiu, J.; Vural, A.; Zhang, N.; Waterman, S.R.; Blewett, N.H.; Myers, T.G.; et al. A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy. Nature 2015, 17, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yao, Z.; Jin, M.; Namkoong, S.; Yin, Z.; Lee, J.H.; Klionsky, D.J. Dhh1 promotes autophagy-related protein translation during nitrogen starvation. PLOS Biol. 2019, 17, e3000219. [Google Scholar] [CrossRef]
- Chuang, K.-H.; Liang, F.; Higgins, R.; Wang, Y. Ubiquilin/Dsk2 promotes inclusion body formation and vacuole (lysosome)-mediated disposal of mutated huntingtin. Mol. Biol. Cell 2016, 27, 2025–2036. [Google Scholar] [CrossRef]
- Higgins, R.; Kabbaj, M.-H.; Hatcher, A.; Wang, Y. The absence of specific yeast heat-shock proteins leads to abnormal aggregation and compromised autophagic clearance of mutant Huntingtin proteins. PLOS ONE 2018, 13, e0191490. [Google Scholar] [CrossRef]
- Wilfling, F.; Lee, C.-W.; Erdmann, P.S.; Zheng, Y.; Sherpa, D.; Jentsch, S.; Pfander, B.; Schulman, B.A.; Baumeister, W. A Selective Autophagy Pathway for Phase-Separated Endocytic Protein Deposits. Mol. Cell 2020, 80, 764–778. [Google Scholar] [CrossRef]
- Ossareh-Nazari, B.; Bonizec, M.; Cohen, M.; Dokudovskaya, S.; Delalande, F.; Schaeffer, C.; Van Dorsselaer, A.; Dargemont, C. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. Embo Rep. 2010, 11, 548–554. [Google Scholar] [CrossRef]
- Krick, R.; Bremer, S.; Welter, E.; et al. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 2010, 190, 965–73. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Kolaitis, R.M.; Taylor, J.P.; et al. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell 2013, 153, 1461–74. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. A trio of ubiquitin ligases sequentially drives ubiquitylation and autophagic degradation of dysfunctional yeast proteasomes. Cell Rep. 2022, 38, 110535. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





