1. Introduction
The effect of anthropogenic chemical pollution on ecosystems has attracted growing attention from the scientific community in recent years. Antarctica is regarded as an "irreplaceable natural laboratory" due to its geographic and climatic isolation, as it is the only non-anthropized continent subject to scarce local sources of pollution but relatively high loads of contaminants from other areas [
1,
2].
Because of their potential toxicity, per - and poly-fluoroalkyl substances (PFAS), a heterogeneous group of synthetic chemicals, are of particular concern as emerging pollutants due to their worldwide presence, propensity for transport, bioaccumulation, environmental persistence, and potential toxicity. These factors are caused by their unique chemical and physical properties, which give them thermal and chemical stability as well as resistance to biodegradation. [
3]. PFOA is part of the emerging, most prevalent, and widespread PFAS (included in 2019 in the list of POPs regulated by the Stockholm Convention) [
4]. It is an ionizable substance, with greater solubility than chlorinated POPs and, consequently, it can reach even the most remote areas of the planet, like Antarctica, mostly through oceanic currents (OCT) [
5]. For this reason, the aquatic ecosystem is one of the major sinks of environmental PFAS, because of their high-water solubility in comparison to traditional POPs [
6]. However, to our knowledge, the data on the occurrence of PFAS in Antarctic biota are very limited, so more studies are required to investigate their occurrence and trophodynamic behaviours in Antarctic ecosystems [
7].
This research is part of the "AntaGPS" project, funded by the PNRA (National Research Program in Antarctica), which uses Antarctica as a global pollution sensor and its endemic organisms as bioindicators. One of the objectives of the project is the study of the antioxidant defence components of Antarctic fish and how this system can be influenced, at the biomolecular level, by exposure to some chemical stress factors [
8].
Species belonging to the suborder
Notothenioidei were chosen as experimental models given that this group of teleosts is the most widespread among the Antarctic fish fauna [
9]. These endemic organisms have great ecological importance since they developed physiological adaptations to extreme and stable environmental conditions [
10]. They may consequently exhibit less phenotypic flexibility and hence be more susceptible to environmental perturbations [
9]. Furthermore, the antioxidant system of these fish is considered very efficient, thanks to their adaptation to the low temperatures and high oxygen concentration of the Antarctic waters [
11].
It is known that all aerobic organisms have evolved metabolic strategies aimed at reducing the toxicity of reactive oxygen species (ROS), natural by-products of aerobic metabolism and continuously produced by cells, both at the cytoplasmic and mitochondrial levels [
12]. This complex defence system includes both low molecular weight scavengers and antioxidant enzymes (e.g. superoxide dismutase, SOD; glutathione peroxidase, GPx) for the detoxification and removal of ROS [
13].
Several studies confirm the correlation between toxicity induced by xenobiotics (such as PFAS) and an increase in ROS formation, with a consequent greater risk of oxidative stress [
14]. For these reasons, variations in the content and the activity of antioxidant enzymes can be used as biomarkers for contaminant-mediated oxidative stress in several marine organisms [
15]. PFAS can bioaccumulate and biomagnify along food webs and act at the molecular level, causing an alteration in ROS production, followed by a cellular redox imbalance [
7]. This can cause DNA damage, modifications of enzymatic activity and a subsequent cascade of oxidative damage to the whole organism, even up to cell apoptosis [
16,
17].
In this work, the analysis of mRNA accumulation (by RT-PCR) allowed us to evaluate the gene expression at the transcriptional level of various isoforms of SOD (1 and 2) and GPx (1 and 4), in Trematomus newnesi, the first time in an Antarctic fish species.
Two experimental groups were set up: one of the treated organisms, exposed to 1 PFOA, and one control group of unexposed individuals. The considered organs were the liver and kidney, due to their physiological role respectively in the accumulation and excretion of xenobiotics [
3].
The obtained results represent a starting point for using the expression of antioxidant enzymes as biomarkers, both of oxidative stress and exposure to PFAS, using notothenioids as bioindicators in future biomonitoring campaigns on the Antarctic marine environment, in line with the aim of the AntaGPS project.
2. Materials and Methods
2.1. Ethical Procedures
The sample collection and animal research conducted in this study comply with the Italian Ministry of Education, University, and Research regulations concerning activities and environmental protection in Antarctica and with the Protocol on Environmental Protection to the Antarctic Treaty, Annex II, Art. 3. All experiments were performed under the U.K. Animals (Scientific Procedures) Act, 1986 and associated guidelines; EU Directive 2010/63/EU; and Italian DL 2014/26 for animal experiments.
2.2. Sampling activity
The current study is based on a fieldwork activity performed in the proximity of Mario Zucchelli Station in Terra Nova Bay, Antarctica (74°42’S, 167°7’E) during the XXXVII Italian Expedition to Antarctica. Adult specimens of T. newnesi were caught at Tethys Bay (74°42,001'S, 164°02,640'E) at a depth of approximately 62 m.
After the sampling, the specimens were held in thermostated aquariums filled with oxygenated seawater at a temperature of about -1°C, immediately. After a distressing period of four days, two experimental groups were established: a control group, consisting of ten untreated organisms, and an experimental group of ten organisms exposed to 1.5 μg/L of PFOA for ten days. Their specimens were 20-23.5 cm in length and 83.8-149.7 g in weight. All the sampled organisms were females.
After this exposure time, the organisms were sacrificed with an overdose of ethyl 3-aminobenzoate methanesulfonate salt (0.065 g/L; Sigma; St. Louis, MO, United States) and then dissected for the collection of the liver and kidney, which were then frozen in liquid nitrogen and stored at -80°C.
2.3. Primers Design, Total RNA Extraction, sod1, sod2, gpx1 and gpx4 cDNA synthesis
The GenBank database, on the NCBI website, was used to get the nucleotide sequences for
sod1,
sod2,
gpx1 and
gpx4 as well as
gapdh for
T. newnesi. Primers were designed in the coding regions using the Primer3 software (
https://primer3.ut.ee/ (accessed on 1 June 2022)). The primer pairs were then examined using the IDT Oligo Analyzer tool (
https://eu.idtdna.com/pages/tools/oligoanalyzer (accessed on 1 June 2022)). Starting from the tissues mentioned in the previous paragraph, the total RNA was isolated using PRImeZOL™ reagent (Canvax, Córdoba, Spain) following the manufacturer’s instructions. When working with Antarctic fish samples, a further purification step with 8M LiCl is required to remove glucidic contaminants. RNA concentration was quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA); RNA integrity was evaluated by running an aliquot of RNA (1000ng/μL) on a denaturing gel stained. The cDNA synthesis was performed using a Biotechrabbit™ cDNA Synthesis Kit (Berlin, Germany) at 50°C for 1 h + 99°C for 5 minutes, from 1 μg of total RNA in a 20 μL reaction mixture, containing 2 μL of dNTP Mix (10 mM each), 0,5 μL of RNase Inhibitor, 40 U/ μL, 0,5 μL of OligodT anchor primer, 4 μL of 5x Reverse Transcriptase Buffer, 1 μL of RNA Template, 1 μL of RevertUPTM II Reverse Transcriptase and PCR Grade Water up to 20 μL. PCR reactions were performed with 50 ng of cDNA using various forward and reverse primers, listed in
Table S1, and GRS Taq DNA polymerase (Grisp, Porto, Portugal).
The PCR program was the following: 95°C for 5 minutes, 38 cycles of 95° for 30 s, 60° for 30 s and 72°C for 30 s. The final elongation step was performed at 72°C for 5 minutes.
2.4. qRT-PCR analysis
For each species and sample, Real-time PCR analysis was performed to evaluate the expression of cytoplasmic sod1 and gpx1 mRNAs as well as mitochondrial sod2 and gpx4 mRNAs. For this investigation, the housekeeping gene used was gapdh (GenBank accession number: KF915299.1). Each cDNA sample was analyzed with the exact primers listed in Table x1 with the following program: 95°C for 2 minutes, 40 x (95°C for 20 seconds and 60°C for 60 seconds) and then the dissociation stage 95°C for 15 s, 60°C for 1 minute, 95° for 15 s and 60°C for 15 s.
Transcript levels were expressed (in arbitrary units, a.u.) as the ratio between each gene and gapdh expression in the same sample.
2.5. Statistical analysis
All data were expressed as the average of five specimens ± standard deviation (SD). Statistical analyses were performed with the PRIMER statistical program. One-way ANOVA was followed by the Student-Newman-Keuls test to assess significant differences (p < 0.05).
3. Results
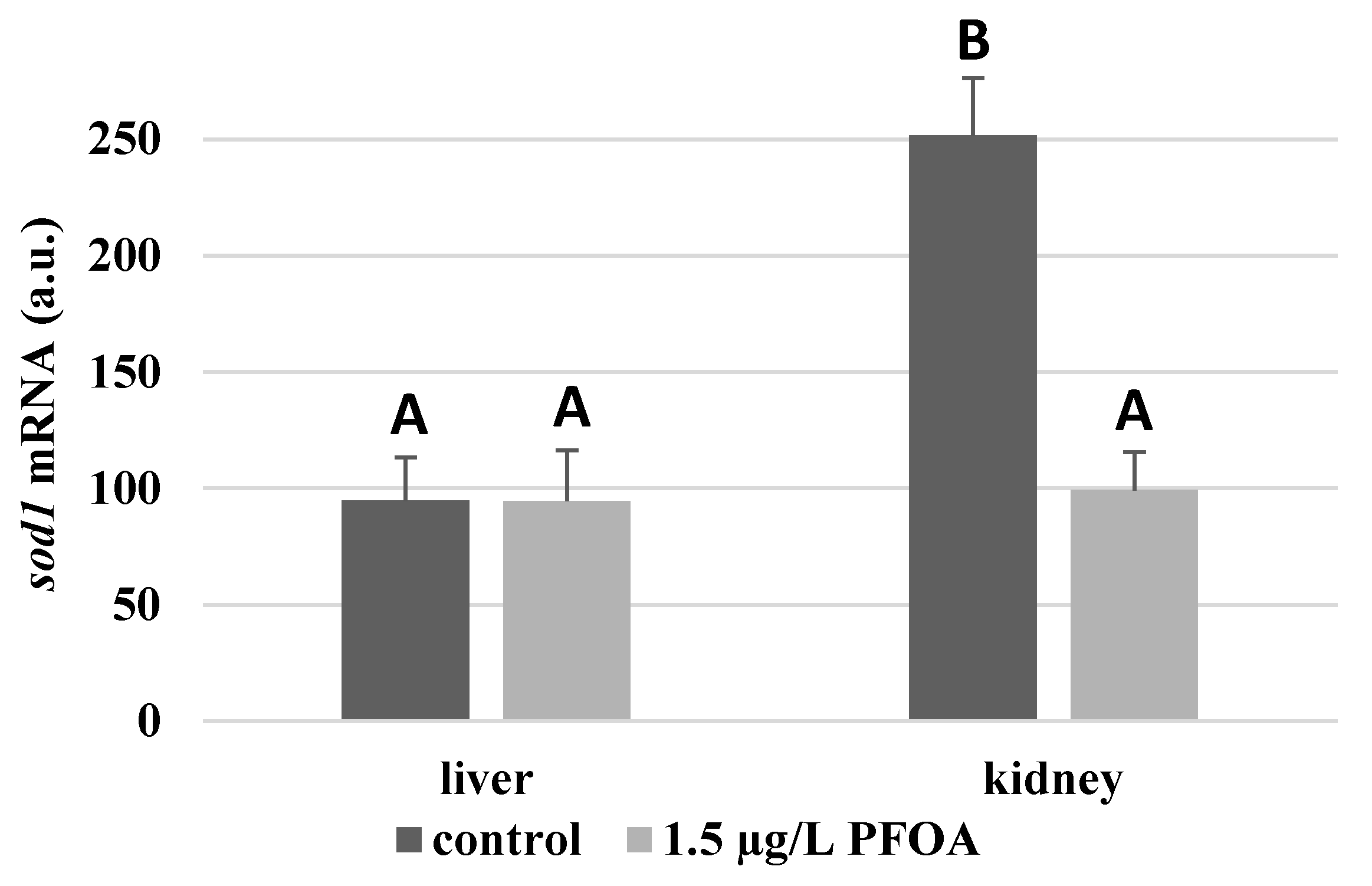
For each antioxidant enzyme, the kidney of control specimens always shows a greater level of expression (p < 0.05) than the respective liver. In particular, the renal mRNA levels for the
sod1 enzyme are about three times higher than the liver (
Figure 1), while those for the
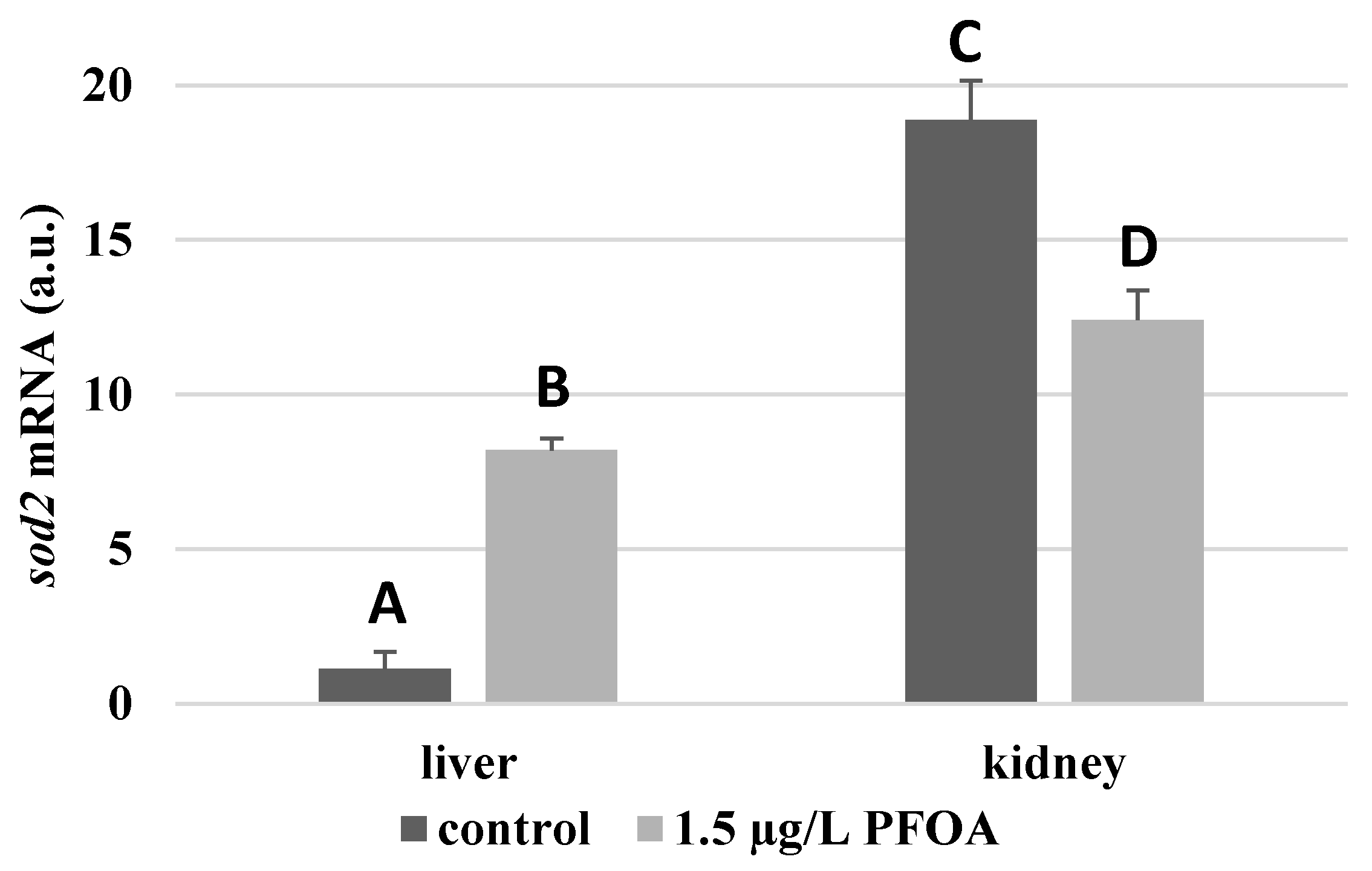
sod2 enzyme are about seventeen times higher (
Figure 2), the
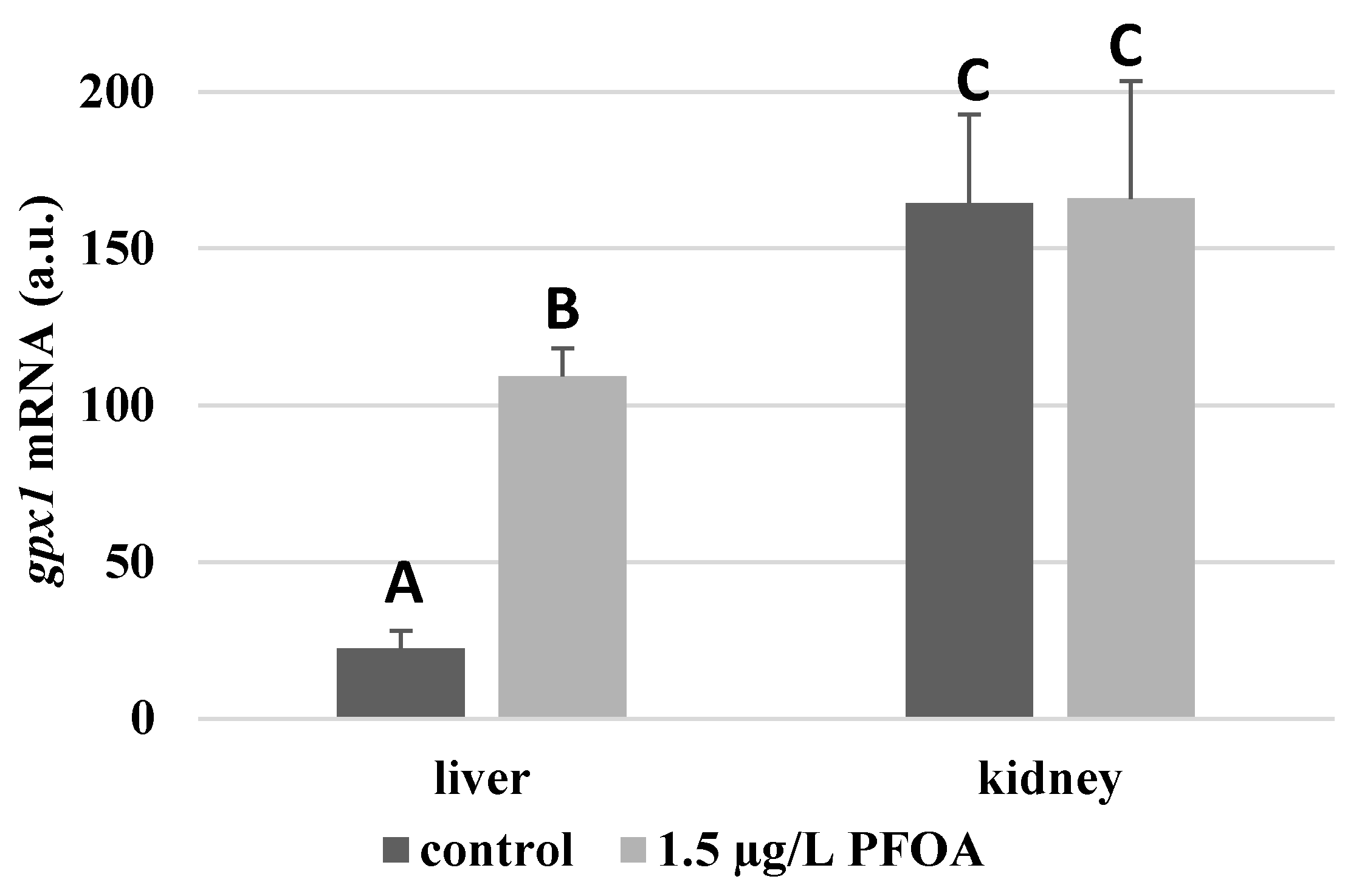
gpx1 enzyme is about seven times higher (
Figure 3), and the
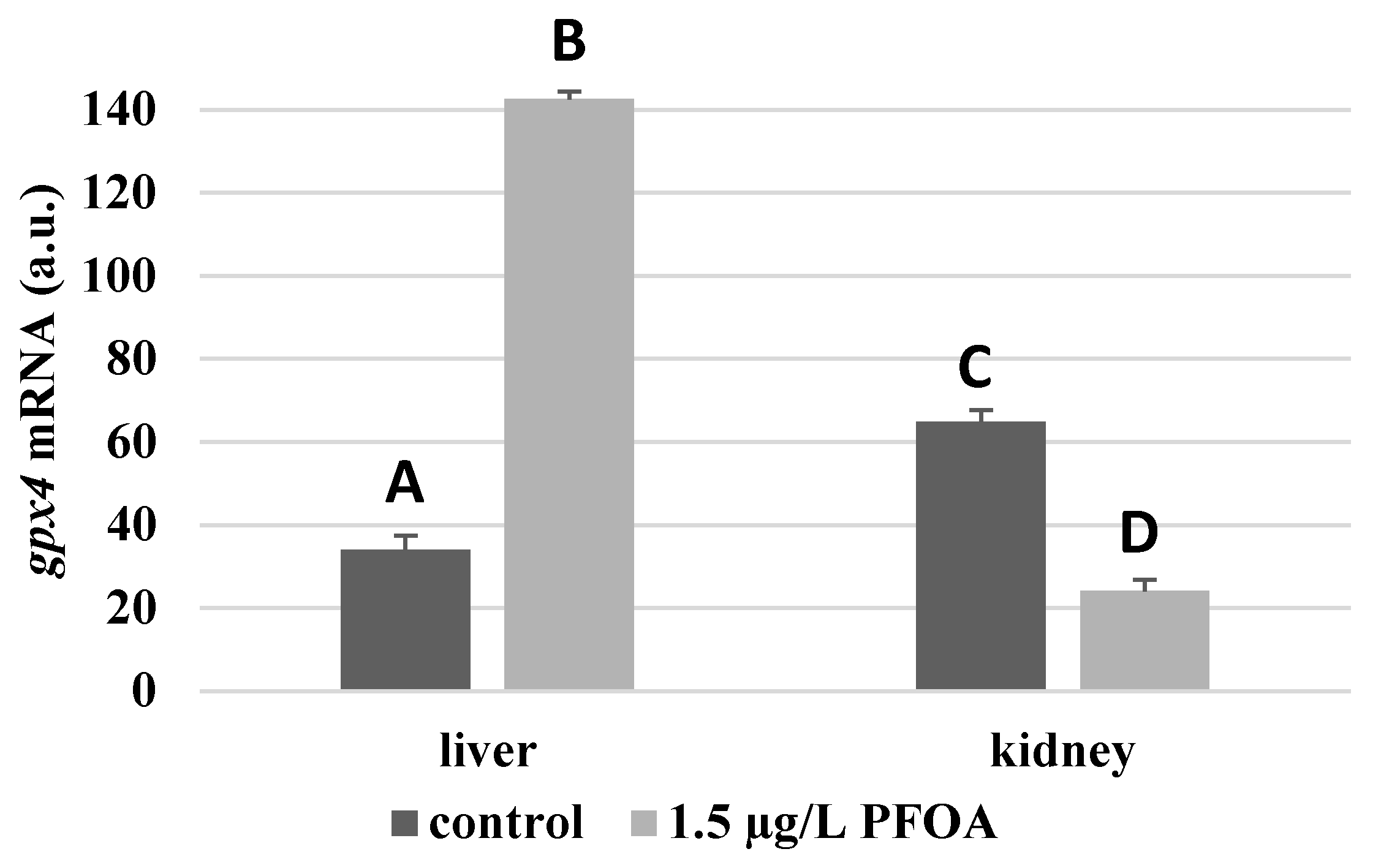
gpx4 enzyme is about two times higher (
Figure 4).
Except for
sod1 (
Figure 1), whose mRNA levels seem unaffected by the exposure to PFOA, all the enzymes considered experience a statistically significant increase (p < 0.05) in gene expression in the liver after the treatment. In particular, PFOA-exposed specimens show
sod2 mRNA levels that are about 12-times greater in the liver than in the kidney (
Figure 2),
gpx1 levels that are about 5-times higher (
Figure 3) and gpx4 levels that are about 4-times higher (
Figure 4).
In the kidney, except for
gpx1 (
Figure 3), whose mRNA levels stay unaltered following the treatment, exposure to PFOA causes a general statistically significant reduction (p<0.05) in mRNA accumulation. In particular, PFOA-exposed specimens have
sod1 enzyme levels that are about 60% lower compared to the liver (
Figure 1),
sod2 enzyme mRNA levels that are about 34% lower (
Figure 2), and
gpx4 enzyme mRNA levels that are about 63% lower (
Figure 4).
4. Discussion
The Antarctic environment has unique features related to its isolation and distance from other continents of the Earth. It is considered pristine even if contaminants can reach Antarctica by long-range atmospheric transport [
1].
Antarctic animals evolved in this environment, exposed to peculiar chemical and physical conditions, such as low temperatures and high oxygen concentration in the marine waters [
18]. These conditions probably affected the adaptive strategies of these organisms during the last 10–12 million years [
19] and, in particular, led to the evolution of very efficient cellular defence systems against oxidative stress [
20,
21].
One of the main scientific questions about the ecophysiology of Antarctic organisms is whether they have evolved acclimatization capacities towards a variation of the environmental concentrations of contaminants of emerging interest, given that they evolved without a significant selective pressure represented by the presence of these chemical elements. In particular, in this paper, we aimed to verify whether and how short-time exposure to high PFOA concentrations can be reflected in the implementation of the gene expression of antioxidant enzymes.
PFAS are suspected to be carcinogenic, and one possible mechanism of action is that they cause oxidative stress [
16]. In fact, according to earlier studies, the observed changes in the activities of antioxidant enzymes suggested that PFAS exposure may upset the antioxidant system's delicate balance, increasing the production of ROS, which affects the mitochondria and triggers a cascade of events that amplifies cell apoptosis [
3,
22,
23].
According to these studies, our findings demonstrate that the genes of almost all considered antioxidant enzymes (except
SOD1, which shows no statistically significant difference) were transcriptionally activated in the liver following 10-day exposure to 1.5 µg/l of PFOA. The detoxifying role of the liver is enhanced if organisms are exposed to a contaminant like PFOA. This was expected since it is known that it is an organ that exhibits high metabolic activity as it is crucial to the preservation of cellular homeostasis [
24]. The liver's rapid metabolism results in the generation of ROS, whose abundance is monitored by its extraordinary capacity to express antioxidants [
25]. However, it should be highlighted that the prominent ROS production that distinguishes the liver of Antarctic teleosts is probably more related to the detoxifying function of this organ than to its digestive activity, which is relatively limited and occasional due to the sporadic feeding of these fish. In fact, the liver is known to bioaccumulate xenobiotics, particularly metal ions like Cd and Cu, which are present in high concentrations in the Antarctic seabed as a natural condition [
26,
27]. According to recent research, the liver is the organ that accumulates PFOA in high concentrations in fish living in PFAS-polluted environments [
17].
In the liver, GPx4 is the enzyme that exhibits the greatest increase in gene expression after exposure to PFOA, followed by GPx1 and SOD2, whereas SOD1 is unaffected. These results are compatible with an increase in ROS formation at the mitochondrial level, since the very specific cellular localization of GPx4 and SOD2 [
28], and confirm that mitochondria are one of the main targets of PFAS toxicity [
16,
22,
23], also in fish [
17]. SOD1 is the most abundant expressed antioxidant enzyme in untreated fish, and this result may be related to a high level of
•O
2‒ production at the cytoplasmic level under normal conditions [
29]. The decrease in the expression of this gene during PFOA exposure, in favour of an increase in the expression of GPx1, could instead indicate that in such circumstances there is a reduction in the rate of
•O
2‒ formation and an increase in that of the H
2O
2.
Contrarily, the kidney is not affected by PFOA exposure, and treated samples even show a tendency to have lower levels of mRNA accumulation coding for all antioxidant enzymes (except for GPx1, which expression did not change). Although the kidney plays a detoxifying role too, this result is most likely due to a non-involvement of this organ during the short (10-day) exposure period.
That said, a non-variation in kidney mRNA levels could be expected, yet our results show a decrease. SOD1 is the enzyme in the kidney that exhibits the greatest reduction in gene expression after exposure to PFOA, followed by GPx4 and SOD2, whereas GPx1 is unaffected. There are primarily two reasons why the cell reduces the messenger production for a certain protein. The first happens when the organism does not require the protein. The second happens when the cell needs to save energy that may be used at other locations in the body, the liver in this case, where it is necessary to mitigate a high oxidative risk, through the biosynthesis of SOD and GPx. Both factors probably influence the outcome. Given its role in xenobiotic excretion and the possible PFAS-induced toxicity, the kidney is indeed a target organ for chemical substances. Although the exposure to PFOA in our experiment is acute rather than chronic, therefore we can hypothesize a renal response in more than 10 days.
5. Conclusions
The antioxidant enzymes considered in this study had already been characterized in various Antarctic fish species, such as
T. bernacchii [
30,
31] and
T. eulepidotus from a structural and functional point of view [
32]. Knowledge about
T. newnesi physiology was very scarce and almost exclusively limited to the anti-freezing properties of its body fluids, a feature shared with other species of Antarctic fish [
33].
To our knowledge, the gene expression results provided here are the first on T. newnesi and among the first considering the mRNA accumulation in Antarctic fish, even though numerous research have already focused on the evaluation of antioxidant enzyme activity in Antarctic fish.
The collected data on gene expression demonstrates how the examined species can express molecular defences against excessive ROS production, which is more pronounced under natural conditions in the Antarctic marine environment.
The findings also give insight into the physiological responses in Antarctic fish to PFAS exposure, specifically PFOA, and on the tissues that are the most crucial for the detoxification of these chemicals. SOD and GPx within the antioxidant defence system play a crucial role in the scavenging of superoxide radical (
•O
2‒) and hydrogen peroxide (H
2O
2), respectively, in normal conditions as well as under stress, but it is just one part of the comprehensive system of defence against ROS. Therefore, in the future, it will be required to assess the function that additional antioxidant components, both enzymatic and non-enzymatic, especially those acting at the mitochondrial level [
34], played to have a complete picture.
The information provided in this work can contribute significantly to the prediction of the physiological responses of these organisms to environmental changes, which can improve the general understanding of the molecular and functional evolution of Antarctic fishes. Furthermore, our analysis of gene expression may provide the basis for using antioxidant enzymes as indicators for both oxidative stress and PFOA exposure.
Regarding the prospects for the future, it is clear that the cellular analysis carried out in this work must be combined with biochemical assays to correlate the physiological response of the cell with measurements of cellular processes, such as enzymatic activity or the amount of ROS present in the tissue. Since it is known from research on other species that organisms surviving in naturally unfavourable conditions, but not under acute stress, have evolved post-transcriptional control of the gene expression of antioxidant enzymes, quantifying enzymatic activity would be certainly informative [
35]. In fact, it can be inferred that a portion of the transcript is not immediately translated when significant levels of mRNA are present without a comparable presence of active protein. According to this theory, the transcript is kept in intracellular compartments like P-bodies or stress granules, where the messengers may go through degradation or future translation, respectively [
35,
36]. This circumstance enables the tissues to react to acute stress extraordinarily quickly, in this case by producing more of a particular antioxidant enzyme in response to an abrupt rise in the rate of the associated ROS generation. This theory has been strengthened by recent research on the stress granule nucleation proteins also in marine animals [
37], including Antarctic fish [
38], which shows that this situation occurs particularly in organs like the liver and muscle, which can experience stress even in a short amount of time and as a result need a quick defensive action made explicit by antioxidants.
In the future, it will also be appropriate to increase research on the cellular production of ROS and the detoxifying function of antioxidant enzymes in physiological and non-physiological settings, such as exposure to environmental pollutants of anthropogenic origin capable of carrying out a pro-oxidant action, like PFAS. These kinds of studies continue to be conducted in our facilities and are the focus of the scientific community.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Table S1: Primer pairs used for qRT-PCR. Amplicon sizes and annealing temperatures (Ta) are also indicated.
Author Contributions
Conceptualization, G.S.; methodology, G.S.; validation, S.P., E.P. and S.S.; formal analysis, S.P., E.P.; investigation, S.P., E.P., S.S., P.I., D.P. and G.S.; resources, P.I., D.P. and G.S.; data curation, S.P. and E.P.; writing—original draft preparation, S.P. and E.P.; writing—review and editing, S.S, P.I., D.P. and G.S.; visualization, D.P. and G.S.; supervision, D.P. and G.S.; project administration, P.I., D.P. and G.S.; fund-ing acquisition, D.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Italian National Program for Antarctic Research (PNRA). Project identification code: 2018/B2Z1.01.
Institutional Review Board Statement
All the activities on animals performed during the Italian Antarctic Expedition are under the control of a PNRA Ethics Referent, which acts on behalf of the Italian Ministry of Foreign Affairs. In particular, the required data are the following: Project identification code: 2018/B2Z1.01. Name of the ethics committee or institutional review board: Italian Ministry of Foreign Affairs. Name of PNRA Ethics Referent: Dr. Carla Ubaldi, ENEA Antarctica, Technical Unit (UTA). Date of approval: 13 October 2021.
Informed Consent Statement
Not applicable.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Corsolini, S.; Ademollo, N. POPs in Antarctic ecosystems: is climate change affecting their temporal trends? Environ. Sci. Process Impacts 2022, 24, 1631–1642. [Google Scholar] [CrossRef] [PubMed]
- Marrone, A.; La Russa, D.; Brunelli, E.; Santovito, G.; La Russa, M. F.; Barca, D.; Pellegrino, D. Antarctic Fish as a Global Pollution Sensor: Metals Biomonitoring in a Twelve-Year Period. Front. Mol. Biosci. 2021, 8, 794946. [Google Scholar] [CrossRef] [PubMed]
- Bonato, M.; Corrà, F.; Bellio, M.; Guidolin, L.; Tallandini, L.; Irato, P.; Santovito, G. PFAS Environmental Pollution and Antioxidant Responses: An Overview of the Impact on Human Field. Int. J. Environ. Res. Public Health 2020, 17, 8020. [Google Scholar] [CrossRef]
- Stockholm Convention: available online http://chm.pops.int/TheConvention/ThePOPs/AllPOPs/tabid/2509/Default.aspx (accessed on 04 November 2022).
- Casal, P.; González-Gaya, B.; Zhang, Y.; Reardon, A. J. F.; Martin, J. W.; Jiménez, B.; Dachs, J. Accumulation of Perfluoroalkylated Substances in Oceanic Plankton. Environ. Sci. Technol. 2017, 51, 2766–2775. [Google Scholar] [CrossRef]
- Kurwadkar, S.; Dane, J.; Kanel, S.R.; Nadagouda, M.N.; Cawdrey, R.W.; Ambade, B.; Struckhoff, G.C.; Wilkin, R. Per- and polyfluoroalkyl substances in water and wastewater: A critical review of their global occurrence and distribution. Sci. Total Environ. 2022, 809, 151003. [Google Scholar] [CrossRef] [PubMed]
- Gao, K.; Miao, X.; Fu, J.; Chen, Y.; Li, H.; Pan, W.; Fu, J.; Zhang, Q.; Zhang, A.; Jiang, G. Occurrence and Trophic Transfer of Per- and Polyfluoroalkyl Substances in an Antarctic Ecosystem. Environ. Pollut. 2020, 257, 113383. [Google Scholar] [CrossRef]
- Marrone, A. Antarctica as a Global Pollution Sensor: The AntaGPS Project. Detritus 2022, 19, 1. [Google Scholar] [CrossRef]
- O’Brien, K. M.; Oldham, C. A.; Sarrimanolis, J.; Fish, A.; Castellini, L.; Vance, J.; Lekanof, H.; Crockett, E. L. Warm Acclimation Alters Antioxidant Defences but Not Metabolic Capacities in the Antarctic Fish, Notothenia coriiceps. Conserv. Physiol. 2022, 10(1), coac054. [Google Scholar] [CrossRef]
- Daane, J.M.; Detrich, H.W. 3rd. Adaptations and Diversity of Antarctic Fishes: A Genomic Perspective. Annu. Rev. Anim. Biosci. 2022, 10, 39–62. [Google Scholar] [CrossRef]
- Bakiu, R.; Boldrin, F.; Pacchini, S.; Schumann, S.; Piva, E.; Tolomeo, A. M.; Ferro, D.; Grapputo, A.; Santovito, G.; Irato, P. Molecular Evolution of Metallothioneins of Antarctic Fish: A Physiological Adaptation to Peculiar Seawater Chemical Characteristics. Journal of Marine Science and Engineering 2022, 10(11), 1592. [Google Scholar] [CrossRef]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed]
- Irato, P.; Santovito, G. Enzymatic and non-enzymatic molecules with antioxidant function. Antioxidants 2022, 10, 579. [Google Scholar] [CrossRef]
- Di Giulio, R.D. Indices of Oxidative Stress as Biomarkers for Environmental Contamination. In Aquatic Toxicology and Risk Assessment: Fourteenth Volume; Mayes, M.A., Barron, M.G., Eds.; ASTM International: West Conshohocken, PA, USA, 1991; pp. 15–31. [Google Scholar] [CrossRef]
- Benedetti, M.; Giuliani, M.E.; Regoli, F. Oxidative metabolism of chemical pollutants in marine organisms: molecular and biochemical biomarkers in environmental toxicology. Ann. N. Y. Acad. Sci. 2015, 1340, 8–19. [Google Scholar] [CrossRef]
- Hu, X.-Z.; Hu, D.-C. Effects of Perfluorooctanoate and Perfluorooctane Sulfonate Exposure on Hepatoma Hep G2 Cells. Arch. Toxicol. 2009, 83(9), 851–861. [Google Scholar] [CrossRef]
- Piva, E.; Schumann, S.; Dotteschini, S.; Brocca, G.; Radaelli, G.; Marion, A.; Irato, P.; Bertotto, D.; Santovito, G. Antioxidant Responses Induced by PFAS Exposure in Freshwater Fish in the Veneto Region. Antioxidants 2022, 11(6), 1115. [Google Scholar] [CrossRef]
- Sidell, B.D. Life at body temperatures below 0 degrees C: The physiology and biochemistry of Antarctic fishes. Gravit. Space Biol. Bull. 2000, 13, 25–34. [Google Scholar]
- Peck, L.S. Prospects for surviving climate change in Antarctic aquatic species. Front. Zool. 2005, 2, 9. [Google Scholar] [CrossRef]
- Ricci, F.; Lauro, F.M.; Grzymski, J.J.; Read, R.; Bakiu, R.; Santovito, G.; Luporini, P.; Vallesi, A. The antioxidant defense system of the marine polar ciliate Euplotes nobilii: Characterization of the msrB gene family. Biology 2017, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Chatzidimitriou, E.; Bisaccia, P.; Corrà, F.; Bonato, M.; Irato, P.; Manuto, L.; Toppo, S.; Bakiu, R.; Santovito, G. Copper/zinc superoxide dismutase from the crocodile icefish Chionodraco hamatus: Antioxidant defense at constant sub-zero temperature. Antioxidants 2020, 9, 325. [Google Scholar] [CrossRef] [PubMed]
- Choi, E. M.; Suh, K. S.; Rhee, S. Y.; Oh, S.; Woo, J.-T.; Kim, S. W.; Kim, Y. S.; Pak, Y. K.; Chon, S. Perfluorooctanoic Acid Induces Mitochondrial Dysfunction in MC3T3-E1 Osteoblast Cells. J. Environ. Sci. Health A 2017, 52, 281–289. [Google Scholar] [CrossRef]
- Suh, K. S.; Choi, E. M.; Kim, Y. J.; Hong, S. M.; Park, S. Y.; Rhee, S. Y.; Oh, S.; Kim, S. W.; Pak, Y. K.; Choe, W.; Chon, S. Perfluorooctanoic Acid Induces Oxidative Damage and Mitochondrial Dysfunction in Pancreatic β-Cells. Mol. Med. Rep. 2017, 15, 3871–3878. [Google Scholar] [CrossRef]
- Ku, T.; Zhou, M.; Hou, Y.; Xie, Y.; Li, G.; Sang, N. Tebuconazole Induces Liver Injury Coupled with ROS-Mediated Hepatic Metabolism Disorder. Ecotoxicol. Environ. Saf. 2021, 220, 112309. [Google Scholar] [CrossRef] [PubMed]
- Ploch, S. A.; Lee, Y.-P.; MacLean, E.; Di Giulio, R. T. Oxidative Stress in Liver of Brown Bullhead and Channel Catfish Following Exposure to Tert-Butyl Hydroperoxide. Aquat. Toxicol. 1999, 46, 231–240. [Google Scholar] [CrossRef]
- Motta, C.M.; Simoniello, P.; Di Lorenzo, M.; Migliaccio, V.; Panzuto, R.; Califano, E.; Santovito, G. Endocrine disrupting effects of copper and cadmium in the oocytes of the Antarctic Emerald rockcod Trematomus bernacchii. Chemosphere 2021, 268, 129282. [Google Scholar] [CrossRef]
- Bakiu, R.; Pacchini, S.; Piva, E.; Schumann, S.; Tolomeo, A.M.; Ferro, D.; Irato, P.; Santovito, G. Metallothionein Expression as a Physiological Response against Metal Toxicity in the Striped Rockcod Trematomus hansoni. Int. J. Mol. Sci. 2022, 23, 12799. [Google Scholar] [PubMed]
- Du, J.; Cai, J.; Wang, S.; You, H. Oxidative Stress and Apotosis to Zebrafish (Danio rerio) Embryos Exposed to Perfluorooctane Sulfonate (PFOS) and ZnO Nanoparticles. Int J Occup Med Environ Health 2017, 30(2), 213–229. [Google Scholar] [CrossRef]
- Lushchak, V. I.; Bagnyukova, T. V. Effects of Different Environmental Oxygen Levels on Free Radical Processes in Fish. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 2006, 144(3), 283–289. [Google Scholar] [CrossRef]
- Santovito, G.; Piccinni, E.; Boldrin, F.; Irato, P. Comparative study on metal homeostasis and detoxification in two Antarctic teleosts. Comp. Biochem. Physiol. C 2012, 155, 580–586. [Google Scholar] [CrossRef]
- Sattin, G.; Bakiu, R.; Tolomeo, A. M.; Carraro, A.; Coppola, D.; Ferro, D.; Patarnello, T.; Santovito, G. Characterization and Expression of a New Cytoplasmic Glutathione Peroxidase 1 Gene in the Antarctic Fish Trematomus bernacchii. Hydrobiologia 2015, 761(1), 363–372. [Google Scholar] [CrossRef]
- Sattin, G.; Santovito, G.; Cassini, A. Physiological Antioxidant Responses against High Environmental Oxygen Concentration: Glutathione Peroxidase from the Antarctic Teleost Trematomus eulepidotus. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 2008, 151 (1, Supplement), S27. [Google Scholar] [CrossRef]
- Fields, L.G.; DeVries, A.L. Variation in blood serum antifreeze activity of Antarctic Trematomus fishes across habitat temperature and depth. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 2015, 185, 43–50. [Google Scholar] [CrossRef]
- Tolomeo, A.M.; Carraro, A.; Bakiu, R.; Toppo, S.; Garofalo, F.; Pellegrino, D.; Gerdol, M.; Ferro, D.; Place, S.P.; Santovito, G. Molecular characterization of novel mitochondrial peroxiredoxins from the Antarctic emerald rockcod and their gene expression in response to environmental warming. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2019, 255, 108580. [Google Scholar] [CrossRef]
- Lavut, A.; Raveh, D. Sequestration of Highly Expressed MRNAs in Cytoplasmic Granules, P-Bodies, and Stress Granules Enhances Cell Viability. PLoS Genet 2012, 8(2), e1002527. [Google Scholar] [CrossRef]
- Olszewska, M.; Bujarski, J. J.; Kurpisz, M. P-Bodies and Their Functions during MRNA Cell Cycle: Mini-Review. Cell Biochem Funct 2012, 30(3), 177–182. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Peronato, A.; Franchi, N.; Ballarin, L.; Bakiu, R.; Santovito, G. Stress granules in Ciona robusta: First evidences of TIA-1-related nucleolysin and tristetraprolin gene expression under metal exposure. Comp. Biochem. Physiol. C 2021, 243, 108977. [Google Scholar] [CrossRef] [PubMed]
- Nicorelli, E.; Gerdol, M.; Buonocore, F.; Pallavicini, A.; Scapigliati, G.; Guidolin, L.; Irato, P.; Corrà, F.; Santovito, G. First Evidence of T Cell Restricted Intracellular Antigen (TIA) Protein Gene Expression in Antarctic Fish. 2018. http://hdl.handle.net/2067/33483.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).