1. Introduction
In a stable and unrestricted melieu, the minimum genetic requirement for a vital cell capable of autonomous growth counts no less than 300 genes to ensure at least cytosolic metabolism and genomic expression and preservation (Hutchison III et al., 2016; Thornburg et al., 2022). The huge structural and functional demands of extant living cells are difficult to reconcile with the enigmatic steps from inorganic to biological chemistry. In particular, the origin of the translation system is a chicken-and-egg conundrum: which came first, genes or proteins? (Wolf and Koonin 2007). An influential hypothesis put forward by Gilbert (1986) suggests that RNA was the first macromolecule in the prebiotic world, long before functional proteins and hence DNA (Kirschning, 2021). The RNA-world hypothesis assumes that life on Earth started with self-recognizing and self-replicating RNA molecules, supposedly RNA rybozimes or prototypical ribonucleotide reductases (Fusz et al., 2005; Lundin et al., 2015; Schneider et al., 2018; Becker et al., 2019). Only later proteins became the catalysts of life, contributing to the genetic takeover of DNA as the information-storage molecule (Müller et al., 2022). Variant theories suggest the occurrence of pristine pre-RNA systems such as, e.g., a heterogeneous chimeric nucleic acid structure where purine deoxyribonucleosides, pyrimidine ribonucleosides and non-canonical bases may have coexisted in the same geochemical scenario (Schneider et al., 2018; Bhowmik and Krishnamurthy, 2019; Xu et al., 2020). However, objections have been raised to the RNA world hypothesis, being RNA polymers’ catalytic repertoire small, chemically homogeneous and constrained by low yield/slow turnover (Kirschning, 2021).
Otherwise, the extant cooperation among peptides, small molecules, coenzymes and metal-based cofactors points towards primeval coevolution of different proto-metabolic biomolecules at a system level (Kroiss et al., 2020; Kirschning, 2021). Also, it has been suggested a primordial peptide synthesis without ribosomes, evoking a primeval RNA-peptide world. Alternatively, there existed a stage prior to the RNA world termed the “polypeptide” or the “protein world” (Milner-White, 2019), in which amino acids pairing was mandatory to allow peptide-to-peptide information transfer and ensuing synthesis of one protein sequence from another (Root-Bernstein, 1982). The hypothesis is attractive, since amino acids and even peptides are easier to chemically synthesize from simple chemicals than nucleotides (Milner-White, 2019). Further, as we will see in the sequel, simple amino acidic polymers display better binding and catalytic properties compared with RNA chains (Milner-White, 2019).
Here we will tackle the chicken-egg problem regarding the duality between nucleic acids and proteins from a new peptidic perspective. Given the almost infinite possibilities of combinations among amino acids and nucleic acids in long timescales, it is likely that complex molecules equipped with biological activity spontaneously arose in ancient abiotic environments. The real problem is not so much the spontaneous formation of complex biomolecules, but their preservation through duplication mechanisms and subsequent transmission to the next generation. The central dogma of molecular biology states that information is transferred from DNA to protein through an RNA intermediate, apart from a few reverse transcriptases (Wolf and Koonin 2007; Chandramouly et al., 2021). Going against this dogma, we will describe the possibility that the first peptides were “retro-translated” to nucleic acid polymers. We will suggest the primeval occurrence of a short, amino acidic polymer termed “selective amino acid- and nucleotide-matching oligopeptide” (henceforward SANMAO). SANMAO was able to non-covalently bind the amino acids endowed in a peptidic chain and to selectively pair them with the single nucleosides scattered in the abiotic medium. We will describe the chemical features of this hypothetical oligopeptide, its biological plausibility and evolutionary virtues.
2. Materials and Methods
The RNA world hypothesis suggests that the peptidic synthesis under prebiotic conditions was initiated by RNA molecules (Wolf and Koonin 2007). It has been proposed that non-canonical RNA bases established a rudimental and inaccurate peptide synthesis directly on RNA (Bhowmik and Krishnamurthy, 2019), starting from covalently connected chimeric molecules made of peptide and RNA (Müller et al., 2022). Since the formation of peptide bonds via direct binding of amino acids to RNA templates (Yarus 1998) is stereochemically implausible, it is more likely that the primeval RNA acted as a template to synthetize aminoacylating ribozymes (Wochner et al., 2011; Torres de Farias and José, 2020; Müller et al., 2022). Initially, RNA must have gained the ability to catalyze the synthesis of just small peptides (Müller et al., 2022). Translation might have started with ribozyme-catalyzed reactions involving abiogenic amino acids, followed by aspecific, non-templated synthesis of increasingly versatile peptides (Wolf and Koonin 2007). The specific amino acids recognition by proto-tRNAs might have depended just on the physical affinity between amino acids and their cognate codons/anticodons (Wolf and Koonin 2007). In the sequel, we will suggest another theory for the onset of peptide translation.
A scenario for the evolution of translation: towards a proteic beginning. We propose a peptide-first model in which oligopeptides capable of molecular recognition spontaneously appeared. Terminologically, we will consider oligopeptides as chains of aminoacids < 20 residues, while polypeptides >20 (Apostolopoulos et al., 2021). Here follows a list of the virtues of a peptide-first model:
Peptides’ and proteins’ polymers are easier to abiotically synthetize than nucleic acids’ (Fusz et al., 2005).
Peptide bonds are more chemically stable than RNA phosphodiester bonds, the latter being more prone to hydrolysis (Schiller, 2016).
The extant peptides and proteins have a wider breadth of chemical moieties for molecular recognition, displaying strong affinities for various types of small molecules.
Peptides and proteins have a much broader repertoire and efficiency as catalysts than RNAs and rybozimes. For example, the Ser-His and Gly-Gly oligopeptide can catalyze peptide bond formation (Schiller, 2016), while the Pro-Pro dipeptide and the Pro-x-x-Phe tetrapeptide (x=any amino acid) display aldol condensation activity, i.e., the most significant carbon-carbon bond-forming reaction (Schiller, 2016).
Proteins have a greater potential for the evolution of a variety of binding and catalytic capacities than RNA (Wolf and Koonin 2007).
Action of peptides as templates has been already demonstrated for self-replicating α-helix peptides (Schiller, 2016).
It has been suggested that amino acids and peptides drove the evolution of translation through the intermediate stage of peptidic proto-ligases forming covalent bonds (Frenkel-Pinter et al, 2019).
Cationic side chains incorporated into proto-peptides possibly interacted with negatively charged RNA, catalyzing RNA oligomerization (Frenkel-Pinter et al, 2019).
In sum, several reasons plead for a versatile polypeptide world at the very origin of life. We will see in the sequel that the main problem is to understand how primeval peptides were able produce their own copies. We suggest that, in a proteic world at the dawn of life, the issue of information transmission could be tackled through the production of nucleic acids’ polymers from spontaneously generated peptidic templates. We require a sort of “reverse translation”, a primitive “protein-dependent RNA polymerase” which might provide some kind of chemical correspondence between amino acids and their extant cognate triplets (Wolf and Koonin 2007; (Biro 2007).
Information transmission from amino acids to nucleotides? We explore the possibility that some peptides were “retro-translated” into nucleic acid polymers by an oligopeptide, namely SANMAO, able to selectively pair the polymeric amino acids and the single nucleotides scattered in the abiotic medium. Once a specific series of nucleotides was brought near, covalent links were established, speeding up non-stochastic formation of biologically relevant RNA chains.
Our aim is to look for the likely chemical and biological attributes of SANMAO. In has been suggested that the first peptides were short, glycine-rich peptidic chains supplied with randomly alternating L/D racemic amino acids. Since short polypeptides flicker between structureless conformations rather than staying fixed in one, enantiomeric ring/chain structures devoid of tertiary structure were favored at the expense of helical conformations (Milner-White and Russell, 2008). Like a few extant peptides that are metamorphic, i.e., quickly switch between different folds with different functions (Lella and Mahalakshmi, 2017; Dishman et al, 2021), it is feasible that the early oligopeptides explored many different conformations and moved so quickly between different states that no fixed conformation was detectable (Milner-White, 2019). The commonest primeval polypeptide conformation might have been the nest, i.e., a biologicaly active 3-6 residue peptide (Milner-White and Russell, 2008). For example, the synthetic hexapeptide Ser–Gly–Ala–Gly–Lys–Thr has been shown to bind inorganic phosphate, while other nests can either bind iron–sulfur centers for electrons storage/transfer, or form polymeric channels characterized by potassium-driven transmembrane potentials (2008; Milner-White, 2019).
SANMAO must be formed by the amino acids available on the Hadean Hearth. Since the primordial soup probably contained ~10 natural amino acids and peptides < 5 amino acids each, the possible SANMAO’s sequence combinations amount to ~110,000 (Schiller, 2016). To bring this number down, it should be noted that not all the possible amino acid pairs are available for bonding and higher affinities generate more persisting biomolecules (Root-Bernstein, 1982; Biro 2007; Lella and Mahalakshmi, 2017; Frenkel-Pinter et al, 2020). The most abundant amino acids are (and possibly were) Gln and the structurally versatile Gly and Ala (Trifonov 2004). Peptide flexibility is guaranteed by the small, rotating Gly, rigidity is guaranteed by Ala, Tyr, Trp, Phe and conformation changes are guaranteed by His. The hydrosulphiric bonds of Cys and Met confer resistance to denaturation, while Pro causes α- and β-chains disruption. The outer peptidic surfaces are rich of negative-charged Lys, Arg, His, while the inner surfaces are rich of hydrophobic amino acids. Occasionally, peptidic chains may exert opposite biological effects. To make an example, cationic poly(Leu-Lys) peptides favor mononucleotide diphosphates’ oligomerization and increase the abundance of long RNA oligomers (Frenkel-Pinter et al, 2020), while (Leu-Lys)n or (Leu-Lys-Lys-Leu)n peptides exhibit RNA hydrolysis activities (Frenkel-Pinter et al, 2020).
Amino acids with complex side chains such as the aromatic and the cationic ones are thought to be late recruitments to the genetic code (Wolf and Koonin 2007). Despite long-range electrostatics generated by cationic amino acids (i.e., Arg, His, and Lys) play at present a more dominant role than peptide conformation (Frenkel-Pinter et al, 2019), it must be stressed that the cationic amino acids were not abundant on the prebiotic Earth (Frenkel-Pinter et al, 2020). When coping with the primordial mediums, it must be also kept into account that different environments display different temperatures, pH and redox gradients (Holm et al, 2006; Baaske et al., 2007; Russell et al., 2010; Fryer, 2012; Postec et al., 2015; Lamadrid et al., 2017; Preiner et al., 2018; Cartwright and Russell, 2019; Boyd et al., 2020; Russell and Ponce, 2020). These variations deeply influence peptides formation, kinetics, folding and biological activity.
Our SANMAO’s scenario requires a mechanism for the elongation of the nuclosides’ growing chain similar to the extant mechanism for biological nucleic acids synthesis, where the 3′ end of the incoming nucleotide forms a covalent phosphodiester with the 5′ end of the previous nucleotide. It is worth mentioning that SANMAO does not require polymerase, hydrolase and ligase activity. Indeed, nucleotides’ polymers can spontaneously self-assembly even in abiotic environments, where the energy to form the phosphodiester bonds between the sugar base of one nucleotide and the phosphate group of the adjacent nucleotide is easily available. The biological task of SANMAO is not to create phosphodiester bonds, rather to guarantee and expedite the possibility that two given nucleotides get closer to each other, increasing the chances of their spontaneous polymerization.
Now, we are finally ready, in the next section of the Results, to build a SANMAO and describe its interactions.
3. Results
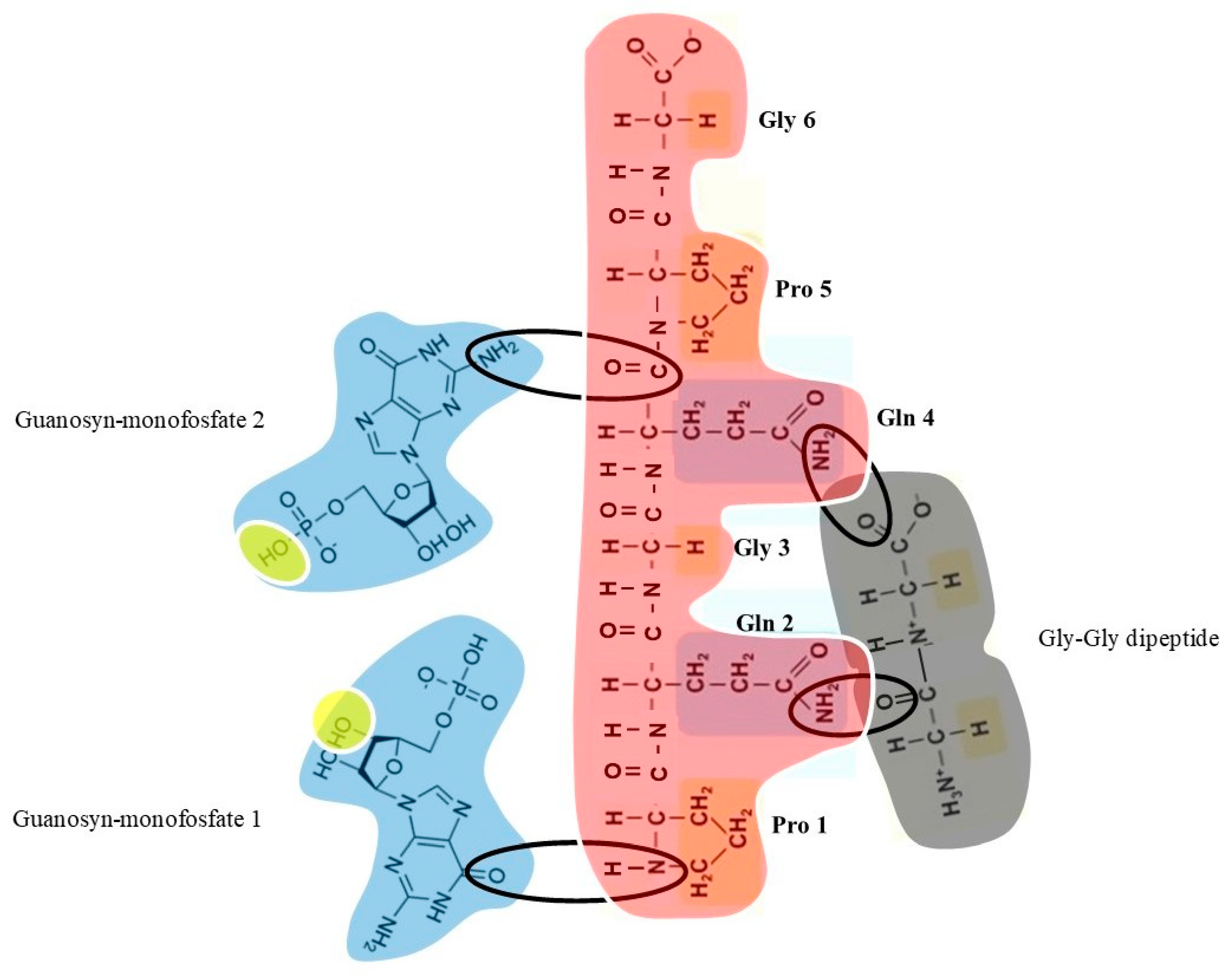
Here we simulate an oligopeptide supplied with the SANMAOs’ required biochemical features. SANMAO must display two active zones: the first active zone selectively links a specific amino acid located in a template polypeptide, while the second active zone selectively links isolated purines or pyrimidines.
To give a proof-of-concept example, we substantiate the chemical steps from Gly to its extant corresponding codon, i.e., the triplet codon GGG. Our scenario meets specific requirements:
We need a peptide acting as a template for the synthesis of nucleic acids chains. For sake of simplicity, we will use a gly-gly dipeptide, the simplest to generate in every plausible primeval scenario.
Also, we need the spontaneous occurrence of a pool of different nucleotides dispersed in the primordial milieu, including at least a few guanosine monofosfate molecules.
Then, we need an oligopepeptide (namely, SANMAO) able to link selectively via non-covalent interactions both the Gly and the Guanosyn monofosfate.
Among the countless feasible SANMAO conformations, we suggest a six-amino acids long oligopeptide capable of binding both Gly and Guanosyn monofosfate. Starting from the N-terminus, our SANMAO displays the following PQGQPG sequence:
Pro1-Gln2-Gly3-Gln4-Pro5-Gly6.
The hydrophilic Glu2/Gln4 and the hydrophobic Gly3/Gln6 are small, devoid of steric hindrance and provide SANMAO’s conformation flexibility. Our SANMAO allows the following non-covalent interactions (
Figure 1):
- a)
The acidic Glu2 and Glu 4 bind the two carbonyl groups of the gly-gly dipeptide.
- b)
The hydrophobic Pro1 and Gln2 bind the first molecule of guanosine monofosfate.
- c)
The hydrophobic Pro5 and Gly6 bind the second molecule of guanosine monofosfate.
SANMAO attains proximity between the two molecules of guanosine monofosfate, thus expediting the formation of a covalent phosphodiester bond between the 5′ end of the first guanosine monofosfate and the 3′ end of the second guanosine monofosfate.
Checking for the biological plausibility of the PQGQPG sequence, we ran UniProtKB for peptide search. The PQGQPG sequence can be found in bacteria, fungi and eukaryotes such as, e.g., Acinetobacter baumannii 99063, Streptomyces sp. PRh5, Oceanicoccus sagamiensis, the fungal plant pathogen Colletotrichum fioriniae PJ7, the roundworm Ancylostoma ceylanicum, the African malaria mosquito Anopheles gambiae, the common eastern firefly Photinus pyralis, the Atlantic salmon Salmo salar, the European red deer Cervus elaphus hippelaphus and also Homo sapiens. In humans and other mammals, the PQGQPG sequence can be detected in various collagen proteins. Also, the RCSB Protein Data Bank observed the PQGQPG sequence in the Pseudomonas aeruginosa’s minor pseudopilin XcpW, a type II secretion machinery carrying virulence factors into the extracellular space (Franz et al., 2011). The PQGQPG sequence, which deletion does not impair protein function, is located in a pseudolipin’s unstructured proteic extremity that is relatively hydrophobic and intrinsically disordered.
Therefore, our theoretical SANMAO sequence is not just biologically plausible, but can also be currently found in different taxonomical domains and kingdoms.
Thanks to SANMAO, the extremities of the two nucleotides can be arranged in close proximity, thus facilitating spontaneous covalent bond formation. The transient interactions provided by SANMAO allow peptidic chains to act as templates to build increasingly longer chains of nucleic acids that roughly correspond to the amino acids of the template polypeptide. At this stage, it does not matter whether the nucleic acids chains are made either of RNA, DNA, uncanonical nucleosides or unconventional mixtures.
Once achieved nucleic acids from a prototype peptidic template, SANMAO allows the reverse operation too. The novel nucleic acid polymer may serve as template for building a copy of the prototype polypeptide.
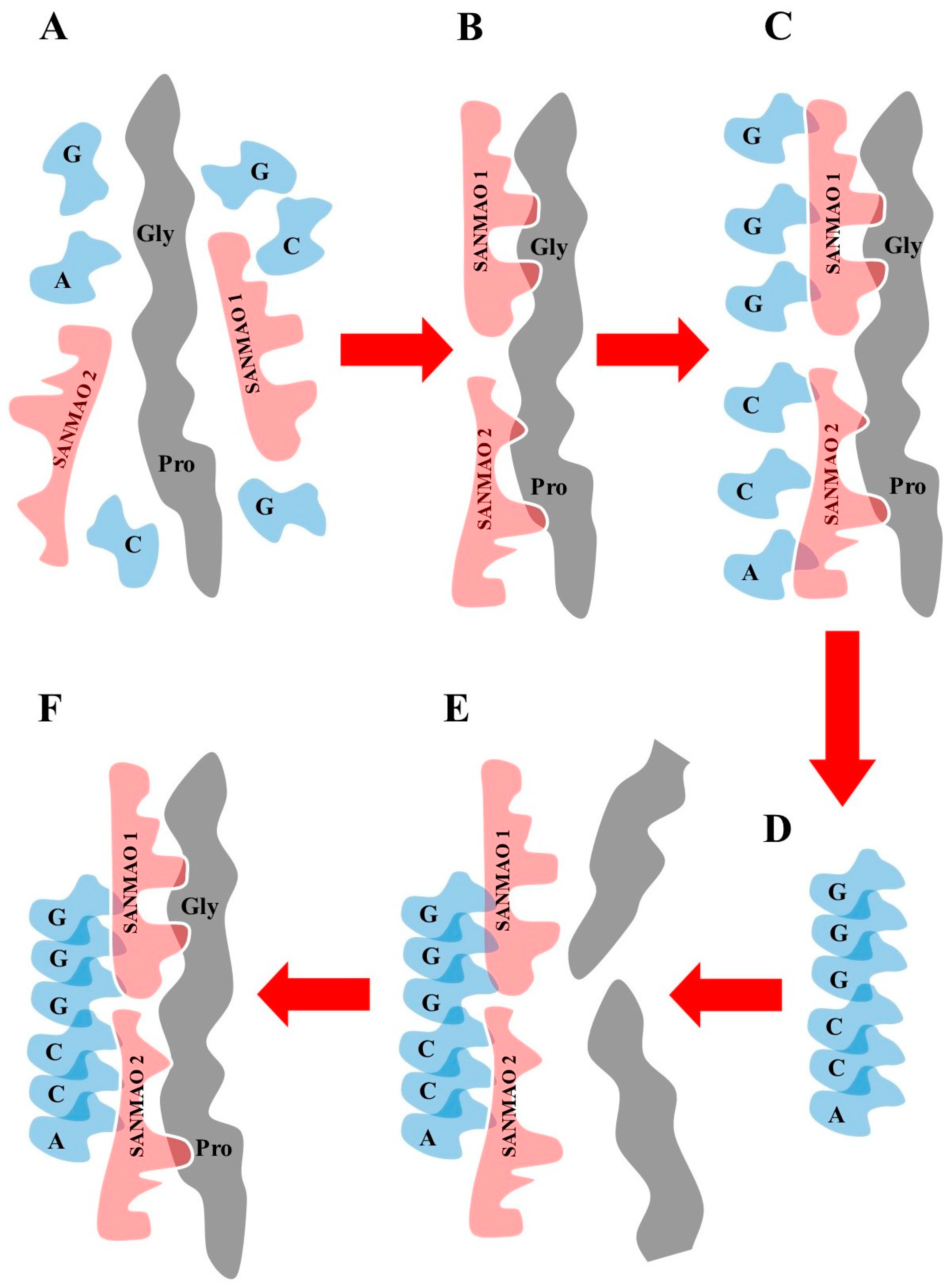
Figure 2 summarizes the complete theoretical sequence. In the primordial milieu containing biomolecules and polymers (
Figure 2A), different SANMAOs link specific amino acids placed inside a template peptide (
Figure 2B). Then, every SANMAO links the corresponding nucleotidic triplet, giving rise to a progressively longer nucleic acid polymer (
Figure 2C). Next, the nucleic acid polymer (
Figure 2D), through the same SANMAOs (
Figure 2E), uses the scattered amino acids available in the primordial milieu to produce a copy of the template peptide (
Figure 2F). Note that every SANMAO (say SANMAO 1) may also act as template peptide for another SANMAO (say SANMAO 2), the whole procedure leading to the duplication of the same SANMAO 1.
4. Conclusions
We suggest the random appearance on the Hadean Earth (about 4 billion years ago) of oligopeptides (SANMAOs) able to link both the amino acids and the purines/pyrimidines scattered in the primeval abiotic medium. SANMAOs acted as a buffer between amino acids and nucleic acids, promoting a sort of “reverse traslation” from peptidic templates to nucleotidic polymers.
Three fundamental premises underlie our scientific effort to address the issue of the first cellular replication and to tackle the proteins/nucleic acids’ chicken-egg problem via the SANMAO hypothesis:
The occurrence of a primeval environment with a continuous source of freely available energy is required for the spontaneous synthesis of biomolecules’ polymers. Many energetically plausible paths from inorganic settings to the first living cells have been outlined (Sousa et al., 2013). For example, the energetic sources for early organic synthesis and systems reactions could have been derived from the proton gradients at the hydrothermal vents-ocean alkaline interface (Sousa et al., 2013). The nanopores formed by the Fe-rich saponite sheets might have acted as confined microenvironments capable of preserving biopolymers precursors from the detrimental effects of the aqueous medium (Ménez et al., 2018). Then, manifold isolated chemical compartments generated mingled agglomerates able to last for long until the SANMAO’s replication processes began.
Physical constraints must be kept into account. Physical rules allow biomolecular mixtures, in particular peptides, to self-assemble and cooperate, generating self-organizing hierarchies of polymeric materials (Jacobs, 2021; Trivedi et al., 2022; McMullen et al., 2022).
The previous prebiotic formation of complex biomolecules is mandatory. The availability of amino acids, polypeptides, proteins, purines, pyrimidines, nucleotides and nucleotides is required under prebiotically plausible conditions on the Hadean Earth. The challenge is to trace plausible physical-chemical processes that permit biomolecules’ abiotic formation and self-regenerating catalytic cycles from a handful of simple compounds such as cyanide, water ammonia, and so on (Becker et al., 2019; Wołos et al. 2020).
Hence, the availability of the amino acids and peptides in prebiotic settings must be investigated. Primordial amino acids and Gly-gly and cyclo(gly-gly) oligopeptides were exogenously delivered to the Hadean Earth (and possibly to the Noachian Mars) by comets and asteroids through hypervelocity impact shocks (Shimoyama et al., 2002; Holm et al, 2015; Ménez et al., 2018; Eigenbrode, et al., 2018). In the interstellar medium, carbon atoms’ condensation on the surface of cold cosmic dust leads to the spontaneous formation of isomeric polyglycine peptides (Krasnokutski et al., 2022; Broadley et al., 2022). Simulations of impacts of Fe-bearing meteorites/asteroids on CO2- and N2-rich oceans suggest that glycine and alanine can be spontaneously synthetized from inorganic mixtures of Fe, Ni, Mg2SiO4, H2O, CO2 and N2 (Takeuchi et al., 2020). Also, amino acids and peptides may have appeared on the Hadean earth under different geochemical scenario, including Fischer-Tropsch-type synthesis (Russell et al., 2010; Rouméjon and Cannat, 2014), Miller and Urey’s spark discharge, Sutherland’s cyanosulfdic protometabolism (Ferris, 1992; Holm et al, 2015; Takeuchi et al., 2020; Kirschning, 2021), Strecker synthesis (Ménez et al., 2018), alkylation of indole with pyruvate followed by amination under hydrothermal conditions (Ménez et al., 2018), etc. The starting blocks for building amino acids are usually pyruvate, carbohydrate-containing molecules, carboxylic acids, ammonium, glutamate, glutamine, aspartate, H2S and thiosulfate (Kirschning, 2021). Complex amino acid such as tryptophan can be abiotically formed beneath the Atlantis Massif during a serpentinites’ late alteration stage, via Friedel-Crafts reactions catalyzed by iron-rich saponite clays (Ménez et al., 2018). It is noteworthy that amino acids synthesis may occur under various conditions of water supply, Ph and temperature, leading, e.g., to the production of phenols that are a moiety of tyrosine (Russell et al., 2010), of phenylalanyl-phenylalanine and tyrosyl-tyrosine oligomers (Milner-White and Russell, 2008), of high yields of alanine, glutamate, phenylalanine and tyrosine (Milner-White and Russell, 2008). Polypeptides can be produced abiotically also in extreme conditions. For example, dry glycine polymers are formed at pressures up to ~250 atm during thermal cycling between 0-250° C (Milner-White and Russell, 2008). Further, possible amino acids ancestors like depsipeptides produce polypeptides during dry-down cycles under mild conditions (Frenkel-Pinter et al, 2019).
In a SANMAO scenario, the first peptides, utilizing the nucleotides spontaneously generated in the primeval medium, acted as a template for nucleic acid polymerization. Therefore, the next step is to investigate the availability of nucleic acids in prebiotic settings.
The building blocks of RNA and DNA, i.e., the purine/pyrimidine nucleobases and their structural isomers, have been detected in carbonaceous meteorites, possibly generated by photochemical reactions in the interstellar medium and subsequently incorporated into asteroids (Oba et al., 2022). When incubated with impact and volcanic rock glasses at room temperature, ribonucleoside triphosphates are converted to a steady polyribonucleic acid of 90–150 nucleotides in length (Jerome et al., 2022). Still, purines and pyrimidines, in particular adenine, may be abiotically formed both in seafloor hydrothermal environments and shallow ponds. Purines synthesis is feasible along a pathway based on the reaction of formamidopyrimidine precursors with ribose, whereas pyrimidines require a further step involving aminooxazoles (Becker et al., 2019). Fischer-Tropsch type reactions on carbonaceous chondrites in the presence of NH3 and short heating up to 600° C generate the familiar nucleobases, although at very low conversions (Preiner et al., 2018). An abiotic one-pot scenario has been suggested to generate purines, pyrimidines and also 5′-mono- and diphosphates in the same environment, driven solely by wet-dry cycles occurring at low temperatures in a few separated shallow ponds (Becker et al., 2019). Wet–dry cycles allow mononucleotide mixtures’ polymerization up to >100 nucleotides in length (Kirschning, 2021). While isocyanates in combination with sodium nitrite generate methylated nucleosides (Schneider et al., 2018), the uranosyl-selective prebiotic synthesis of purine deoxyribonucleosides uses intermediates from the prebiotic synthesis of pyrimidines, leading to a mixture of deoxyadenosine, deoxyinosine, cytidine and uridine (Xu et al., 2020). Dry heating combinations of glycine, alanine, valine, lysine, asparagine and glutamine produce pterin riboside at 160-200° C for 4-6 h (Preiner et al., 2018). When ultramafic rocks are exposed to fluids, the alkalinity may promote abiotic formation of pentoses, particularly ribose (Holm et al, 2006). Also, water-soluble polyphosphates like cyclotriphosphate can be generated in the vicinity of volcanoes (Kirschning, 2021).
In sum, experimental and theoretical approaches suggest that key biomolecules such as amino acids, polypeptides, purines, pyrimidines, nucleotides and nucleotides were available under various prebiotically plausible conditions, including meteorites delivery, shallow ponds and hydrothermal vents scenarios. Nevertheless, biomolecules’ prebiotic formation is not an easy process, being bonds formation kinetically frustrated by high energy barriers (Frenkel-Pinter et al, 2019). This would mean that spontaneously generated biomolecular polymers of reasonable length were present on the Hadean Hearth, but not abundant (Preiner et al., 2018; Takeuchi et al., 2020).
Another problem is the scarce fidelity of the first SANMAO mechanism of protein translation. Indeed, structural organization and cell order distribution of granular systems (Wanjura et al., 2019) suggest that every hierarchical level adds complexity during assembly at the expense of increases in random errors (Michel and Yunker, 2019). This would mean that the first efforts to transmit the genetic material to the offspring, i.e., the first random couplings between amino acids and nucleosides, were unavoidably prone to errors. Being the primeval translational mechanisms not as accurate and efficient as the modern triplet-based version (Wolf and Koonin 2007; Schiller, 2016), the ancient peptides gave rise to polymerized strains of low-fidelity RNA. Nevertheless, the many mistakes of the primeval “reverse-transational” mechanism were possibly effective to enlarge the number of codon combinations. Then, the natural selection of strong affinity intermolecular interactions and the decreased entropy of the best couplings led to increased stabilization over long timescales (Schiller, 2016). Due to decoding systems’ evolutionary pressures (Otten et al., 2020; Torres de Farias and José, 2020), the subsequent emergence and selection of RNA drove the separation of catalytic and genetic functions (Schiller, 2016). Then, repetition and duplication of simple peptidic and nucleic acids polymers took progressively place (Milner-White, 2019). Synergy and co-evolution among diverse classes of molecules including polyesters and depsipeptides contributed to attenuate hydrolysis rates, promote folding/solubility and accelerate ligand binding/catalysis (Frenkel-Pinter et al, 2020). Actually, cationic proto-peptides significantly increase the thermal stability of folded RNA structures, while RNA increases by >30-fold the depsipeptides’ lifetime (Frenkel-Pinter et al, 2020).
Catalytic and reproducing systems prone to Darwinian selection eventually developed, generating versatile and resilient metabolic “hypercycles” that circumvented the potential “error catastrophe” due to the formation of long strings of random sequences (Mossela, and Steel, 2005). Further, thousands of metabolic innovations on earlier phylogenetic branches were facilitated by horizontal gene transfers, breeding stepwise adaptation to successive environments and ensuing regulatory adaptations (Pang and Lercher, 2018).
It has been argued that life emerged many times via multiple different historical pathways (Kempes and Krakauer, 2021). If life spontaneously arose on the Hadean Hearth from rather trivial non-organic assemblies, why it has not been produced de novo after the appearance of the last universal common ancestor (LUCA)? We suggest a straightforward answer: any viable effort after LUCA to produce unprecedented life was doomed to fail because of natural selection. After LUCA, the likely newcomers generated from inorganic precursors would have been too fragile to compete with the well-adapted LUCA’s heirs equipped with consolidated selective advantage and plastic immune memory (Ferro et al., 2019; Bernheim et al., 2021; Wein and Sorek, 2022).
In sum, in touch with the literature, we assume that life is a spontaneous process consisting of self-assembly of complex inorganic molecules. Focusing on the crucial step of protein translation, we propose, this time against the literature and the central dogma of molecular biology, an unusual but plausible scenario in which peptides lead to nucleic acid polymers.
Author Contributions
The Authors equally contributed to: study concept and design, acquisition of data, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, obtained funding, administrative, technical, and material support, study supervision.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Ethics approval and consent to participate
This research does not contain any studies with human participants or animals performed by the Authors.
Consent for publication
The Authors transfer all copyright ownership, in the event the work is published. The undersigned authors warrant that the article is original, does not infringe on any copyright or other proprietary right of any third part, is not under consideration by another journal, and has not been previously published.
Availability of data and materials
All data and materials generated or analyzed during this study are included in the manuscript. The Authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Conflicts of Interest
The Authors do not have any known or potential conflict of interest including any financial, personal or other relationships with other people or organizations within three years of beginning the submitted work that could inappropriately influence, or be perceived to influence, their work.
References
- Apostolopoulos, Vasso; Joanna Bojarska, Tsun-Thai Chai, Sherif Elnagdy, Krzysztof Kaczmarek, et al. A Global Review on Short Peptides: Frontiers and Perspectives . Molecules. 2021 Jan 15;26(2):430. [CrossRef]
- Baaske P, Weinert F, Duhr S, Lemke K, RussellMJ, Braun D. (2007). Extreme accumulation of nucleotides in simulated hydrothermal pore systems. Proceedings of the National Academy of Sciences of the USA 104, 9346–9351.
- Becker, Sidney; Jonas Feldmann, Stefan Wiedemann, Hidenori Okamura, Christina Schneider, et al. Unified prebiotically plausible synthesis of pyrimidine and purine RNA ribonucleotides. Science. 2019 Oct 4;366(6461):76-82. [CrossRef]
- Bernheim, A., Millman, A., Ofir, G. et al. Prokaryotic viperins produce diverse antiviral molecules. Nature 589, 120–124 (2021). [CrossRef]
- Bhowmik, S., Krishnamurthy, R. The role of sugar-backbone heterogeneity and chimeras in the simultaneous emergence of RNA and DNA. Nat. Chem. 11, 1009–1018 (2019). [CrossRef]
- Biro, Jan C. The Proteomic Code: a molecular recognition code for proteins. Theor Biol Med Model. 2007 Nov 13;4:45. [CrossRef]
- Boyd, Eric S; Maximiliano J. Amenabar, Saroj Poudel, and Alexis S. Templeton. Bioenergetic constraints on the origin of autotrophic metabolism. Philos Trans A Math Phys Eng Sci. 2020 Feb 21; 378(2165): 20190151. Published online 2020 Jan 6. [CrossRef]
- Broadley, M.W., Bekaert, D.V., Piani, L. et al. Origin of life-forming volatile elements in the inner Solar System. Nature 611, 245–255 (2022). [CrossRef]
- Cartwright Julyan H. E. and Russell Michael J. 2019. The origin of life: the submarine alkaline vent theory at 30. Interface Focus.92019010420190104. http://doi.org/. [CrossRef]
- Chandramouly, Gurushankar; Jiemin Zhao, Shane McDevitt, Timur Rusanov, Trung Hoang, et al. Polθ reverse transcribes RNA and promotes RNA-templated DNA repair. Sci Adv. 2021 Jun; 7(24): eabf1771. Published online 2021 Jun 11. [CrossRef]
- Dishman, Acacia F; Robert C Tyler, Jamie C Fox, Andrew B Kleist, Kenneth E Prehoda, et al. Evolution of fold switching in a metamorphic protein. Science. 2021 Jan 1;371(6524):86-90. [CrossRef]
- Eigenbrode, Jennifer L; Roger E Summons, Andrew Steele, Caroline Freissinet, Maëva Millan, et al. Organic matter preserved in 3-billion-year-old mudstones at Gale crater, Mars. Science. 2018 Jun 8;360(6393):1096-1101. [CrossRef]
- Ferro, Kevin; Robert Peuß, Wentao Yang, Philip Rosenstiel, Hinrich Schulenburg, Joachim Kurtz. Experimental evolution of immunological specificity. Proc Natl Acad Sci U S A . 2019 Oct 8;116(41):20598-20604. [CrossRef]
- Franz, L.P., Douzi, B., Durand, E., Dyer, D.H., Voulhoux, R., Forest, K.T. Structure of the minor pseudopilin XcpW from the Pseudomonas aeruginosa type II secretion system. (2011) Acta Crystallogr D Biol Crystallogr 67: 124-130. [CrossRef]
- Frenkel-Pinter, Moran; Jay W Haynes, Martin C, Anton S Petrov, Bradley T Burcar, et al. Selective incorporation of proteinaceous over nonproteinaceous cationic amino acids in model prebiotic oligomerization reactions. Proc Natl Acad Sci U S A. 2019 Aug 13;116(33):16338-16346. [CrossRef]
- Frenkel-Pinter, M., Haynes, J.W., Mohyeldin, A.M. et al. Mutually stabilizing interactions between proto-peptides and RNA. Nat Commun 11, 3137 (2020). [CrossRef]
- Fryer, Patricia. Serpentinite mud volcanism: observations, processes, and implications. Ann Rev Mar Sci. 2012;4:345-73. [CrossRef]
- Fusz, Stefan, Alexander Eisenführ, Seergazhi G Srivatsan, Alexander Heckel, Michael Famulok. A ribozyme for the aldol reaction. Chem Biol. 2005 Aug;12(8):941-50. [CrossRef]
- Gilbert, W. Origin of life: The RNA world. Nature 319, 618 (1986). [CrossRef]
- Gjorevski, N; M Nikolae, T E Brown, O Mitrofanova, N Brandenberg et al. Tissue geometry drives deterministic organoid patterning. Science. 2022 Jan 7;375(6576):eaaw9021. Epub 2022 Jan 7. [CrossRef]
- Holm, Nils G; Marion Dumont, Magnus Ivarsson & Cécile Konn. Alkaline fluid circulation in ultramafic rocks and formation of nucleotide constituents: a hypothesis. Geochemical Transactions volume 7, Article number: 7 (2006). [CrossRef]
- Holm, N.G.; C. Oze, O. Mousis, J.H. Waite, and A. Guilbert-Lepoutre. Serpentinization and the Formation of H2 and CH4 on Celestial Bodies (Planets, Moons, Comets). Astrobiology. 2015 Jul 1; 15(7): 587–600. [CrossRef]
- Hutchison III, Clyde A; Ray-Yuan Chuang, Vladimir N Noskov, Nacyra Assad-Garcia, Thomas J Deerinc, et al. Design and synthesis of a minimal bacterial genome. Science, 25 Mar 2016, Vol 351, Issue 6280. [CrossRef]
- Jacobs, William M. Self-Assembly of Biomolecular Condensates with Shared Components. Phys. Rev. Lett. 126, 258101 – Published 24 June 2021. [CrossRef]
- Jerome, Craig A; Hyo-Joong Kim, Stephen J Mojzsis, Steven A Benner, Elisa Biondi. Catalytic Synthesis of Polyribonucleic Acid on Prebiotic Rock Glasses. Astrobiology. 2022 Jun;22(6):629-636. [CrossRef]
- Kempes, Christopher P. & David C. Krakauer. The Multiple Paths to Multiple Life. Journal of Molecular Evolution volume 89, pages415–426 (2021). [CrossRef]
- Kirschning, Andreas. The coenzyme/protein pair and the molecular evolution of life. Natural Product Reports, Volume 38, Issue 5, May 2021, Pages 993-1010. [CrossRef]
- Klatt, M.A., Lovrić, J., Chen, D. et al. Universal hidden order in amorphous cellular geometries. Nat Commun 10, 811 (2019). [CrossRef]
- Krasnokutski, S. A.; K.-J. Chuang, C. Jäger, N. Ueberschaar & Th. Henning. A pathway to peptides in space through the condensation of atomic carbon. Nature Astronomy volume 6, pages381–386 (2022)Cite this article. [CrossRef]
- Kroiss, D.; Ashkenasy, G.; Braunschweig, A. B.; Tuttle, T.; Ulijn R. V. Catalyst: Can systems chemistry unravel the mysteries of the chemical origins of life? Chem. 2019, 5, 1917−1920.
- Lamadrid, Hector M.; J. Donald Rimstidt, Esther M. Schwarzenbach, Frieder Klein, Sarah Ulrich, et al. Effect of water activity on rates of serpentinization of olivine. Nature Communications volume 8, Article number: 16107 (2017). [CrossRef]
- Lella, Muralikrishna; Mahalakshmi R. Metamorphic Proteins: Emergence of Dual Protein Folds from One Primary Sequence. Biochemistry 2017, 56, 24, 2971–2984, Publication Date:June 1, 2017. [CrossRef]
- Lundin, Daniel; Gustav Berggren, Derek T. Logan, Britt-Marie Sjöberg. The Origin and Evolution of Ribonucleotide Reduction. Life 2015, 5(1), 604-636. [CrossRef]
- Lyons, Nicholas A; Roberto Kolter. On the evolution of bacterial multicellularity. Curr Opin Microbiol . 2015 Apr;24:21-8. [CrossRef]
- Martinfilipa, William F.; L. Sousaand Nick Laneauthors. Analysis of the bioenergetics of primitive organisms suggests that life began at hydrothermal vents. Science 6 Jun 2014 Vol 344, Issue 6188 pp. 1092-1093. [CrossRef]
- McMullen, A., Muñoz Basagoiti, M., Zeravcic, Z. et al. Self-assembly of emulsion droplets through programmable folding. Nature 610, 502–506 (2022). [CrossRef]
- Ménez , Bénédicte ; Céline Pisapia, Muriel Andreani, Frédéric Jamme, Quentin P Vanbellingen, et al. Abiotic synthesis of amino acids in the recesses of the oceanic lithosphere. Nature. 2018 Dec;564(7734):59-63. Epub 2018 Nov 7. [CrossRef]
- Michel, Jonathan A; Peter J Yunker. Structural hierarchy confers error tolerance in biological materials. Proc Natl Acad Sci U S A . 2019 Feb 19;116(8):2875-2880. [CrossRef]
- Milner-White, E.J., Russell, M.J. Predicting the conformations of peptides and proteins in early evolution. Biol Direct 3, 3 (2008). [CrossRef]
- Milner-White, E. James. Protein three-dimensional structures at the origin of life. Interface Focus. 2019 Dec 6; 9(6): 20190057. Published online 2019 Oct 18. [CrossRef]
- Mossela, Elchanan; Mike Steel. Random biochemical networks: the probability of self-sustaining autocatalysis. Journal of Theoretical Biology, Volume 233, Issue 3, 7 April 2005, Pages 327-336. [CrossRef]
- Müller, Felix; Luis Escobar, Felix Xu, Ewa Węgrzyn, Milda Nainytė, et al. A prebiotically plausible scenario of an RNA–peptide world. Nature volume 605, pages279–284 (2022)Cite this article. [CrossRef]
- Xu, J., Chmela, V., Green, N. et al. Selective prebiotic formation of RNA pyrimidine and DNA purine nucleosides. Nature 582, 60–66 (2020). [CrossRef]
- Xu, C., Martin, N., Li, M. et al. Living material assembly of bacteriogenic protocells. Nature 609, 1029–1037 (2022). [CrossRef]
- Oba, Yasuhiro; Yoshinori Takano, Yoshihiro Furukawa, Toshiki Koga, Daniel P. Glavin, et al. Identifying the wide diversity of extraterrestrial purine and pyrimidine nucleobases in carbonaceous meteorites. Nature Communications volume 13, Article number: 2008 (2022). [CrossRef]
- Otten, Renee; Ricardo A P Pádua, H Adrian Bunzel, Vy Nguyen, Warintra Pitsawong, et al. How directed evolution reshapes the energy landscape in an enzyme to boost catalysis. Science. 2020 Dec 18;370(6523):1442-1446. Epub 2020 Nov 19. [CrossRef]
- Pang, Tin Yau; Martin J. Lercher. Each of 3,323 metabolic innovations in the evolution of E. coli arose through the horizontal transfer of a single DNA segment. PNAS, December 18, 2018, 116 (1) 187-192. [CrossRef]
- Postec, Anne; Marianne Quéméneur, Méline Bes, Nan Mei, Fatma Benaïssa, et al. Microbial diversity in a submarine carbonate edifice from the serpentinizing hydrothermal system of the Prony Bay (New Caledonia) over a 6-year period. Front. Microbiol., 27 August 2015. [CrossRef]
- Preiner, Martina; Joana C. Xavier, Filipa L. Sousa, Verena Zimorski, Anna Neubeck, et al. Serpentinization: Connecting Geochemistry, Ancient Metabolism and Industrial Hydrogenation. Life 2018, 8(4), 41. [CrossRef]
- Root-Bernstein, Robert Scott. Amino acid pairing. Journal of Theoretical Biology. Volume 94, Issue 4, 21 February 1982, Pages 885-894. [CrossRef]
- Rouméjon, Stéphane; Mathilde Cannat. Serpentinization of mantle-derived peridotites at mid-ocean ridges: Mesh texture development in the context of tectonic exhumation. Geochemistry, Geophysiscs, Geosystems. Volume15, Issue6, June 2014, Pages 2354-2379. First published: 23 May 2014 Citations: 55. [CrossRef]
- Russell, M J; A J Hall, W Martin. Serpentinization as a source of energy at the origin of life. Geobiology. 2010 Dec;8(5):355-71. [CrossRef]
- Russell Michael J.; Adrian Ponce. Six ‘Must-Have’ Minerals for Life’s Emergence: Olivine, Pyrrhotite, Bridgmanite, Serpentine, Fougerite and Mackinawite. Life 2020, 10(11), 291. [CrossRef]
- Schiller, Martin R. The minimotif synthesis hypothesis for the origin of life. J Transl Sci. 2016; 2(5): 289–296. Published online 2016 Jul 19. [CrossRef]
- Schneider Christina; Sidney Becker, Hidenori Okamura, Antony Crisp, Tynchtyk Amatov, et al. Noncanonical RNA Nucleosides as Molecular Fossils of an Early Earth-Generation by Prebiotic Methylations and Carbamoylations. Angew Chem Int Ed Engl. 2018 May 14;57(20):5943-5946. Epub 2018 Apr 17. [CrossRef]
- Shimoyama, Akira; Ryo Ogasawara. Dipeptides and diketopiperazines in the Yamato-791198 and Murchison carbonaceous chondrites. Orig Life Evol Biosph. 2002 Apr;32(2):165-79. [CrossRef]
- Sousa, Filipa L, Thorsten Thiergart, Giddy Landan, Shijulal Nelson-Sathi, Inês A C Pereira, et al. Early bioenergetic evolution. Philos Trans R Soc Lond B Biol Sci. 2013 Jun 10;368(1622):20130088. Print 2013 Jul 19. [CrossRef]
- Takeuchi, Yuto; Yoshihiro Furukawa, Takamichi Kobayashi, Toshimori Sekine, Naoki Terada & Takeshi Kakegawa Impact-induced amino acid formation on Hadean Earth and Noachian Mars. Scientific Reports volume 10, Article number: 9220 (2020). [CrossRef]
- Thornburg, Zane R.; David M. Bianchi, Troy A. Brier, Hamilton O. Smith, John I. Glass, et al. Fundamental behaviors emerge from simulations of a living minimal cell. Cell Volume 185, Issue 2, P345-360.E28, January 20, 2022. [CrossRef]
- Torres de Farias, Sávio; Marco V José. Transfer RNA: The molecular demiurge in the origin of biological systems. Prog Biophys Mol Biol. 2020 Jul;153:28-34. Epub 2020 Feb 24. [CrossRef]
- Trifonov, E. N. The triplet code from first principles. J. Biomol. Struct. Dyn. 2004, 22, 1−11. [CrossRef]
- Trivedi, M., Saxena, D., Ng, W.K. et al. Self-organized lasers from reconfigurable colloidal assemblies. Nat. Phys. 18, 939–944 (2022). [CrossRef]
- Yarus M. Amino acids as RNA ligands: a direct-RNA-template theory for the code’s origin. J Mol Evol 1998, 47(1):109-117. [CrossRef]
- Wang, R., Fang, F., Cui, J. et al. Learning self-driven collective dynamics with graph networks. Sci Rep 12, 500 (2022). [CrossRef]
- Wanjura, Clara C.; Paula Gago, Takashi Matsushima, Raphael Blumenfeld. Structural Evolution of Granular Systems: Theory. arXiv:1904.06549 [Submitted on 13 Apr 2019 (v1), last revised 25 Sep 2019 (this version, v2)].
- Wein, T., Sorek, R. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat Rev Immunol 22, 629–638 (2022). [CrossRef]
- Wochner, Aniela; James Attwater, Alan Coulson, Philipp Holliger. Ribozyme-catalyzed transcription of an active ribozyme. Science. 2011 Apr 8;332(6026):209-12. [CrossRef]
- Wolf, Y.I., Koonin, E.V. On the origin of the translation system and the genetic code in the RNA world by means of natural selection, exaptation, and subfunctionalization. Biol Direct 2, 14 (2007). [CrossRef]
- Wołos Agnieszka; Rafał Roszak, Anna Żądło-Dobrowolska, Wiktor Beker, Barbara Mikulak-Klucznik. Synthetic connectivity, emergence, and self-regeneration in the network of prebiotic chemistry. Science. 2020 Sep 25;369(6511):eaaw1955. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).