1. Introduction
Human T-lymphotropic virus type 1 (HTLV–I) is an oncogenic human retrovirus initially discovered in an adult T-cell leukemia/lymphoma (ATLL) patient [1]. It has been estimated that 5–10 million people around the world have HTLV–I, but it is important to note that the number is still mostly unknown in places such as India, China, Russia, Australia, and several African countries [2]. HTLV–I infection can cause a spectrum of clinical symptoms, from asymptomatic infection to malignant ATLL and HTLV–I-associated myelopathy/tropical spastic paraparesis (HAM/TSP) [3]. Most HTLV–I patients remain asymptomatic for life, but others advance to a preleukemic phase characterized by small numbers of circulating leukemic cells in the peripheral blood and skin lesions but no involvement of other organ systems [4]. Only 2.5-5% of virus carriers acquire ATLL after a long period of asymptomatic infection [5]. Molecular studies have shown that viral proteins disrupt biological activities, such as immortalization and IL-2-independent proliferation of T cells triggered by TAX protein [6]. ATLL leukemogenesis may be influenced by genetic and epigenetic changes, including DNA methylation, as well as the host's immune system [7]. Despite T-cell immortalization, the long incubation period (>30 years) before ATLL shows that genetic modifications other than viral infection contribute to pathogenesis [8].
Certain findings support the transcription of nonprotein coding portions of the mammalian genome [9], forming a network of transcripts that are intricately entangled with one another, including noncoding RNAs (ncRNAs). These molecules play significant roles in normal biological processes and human illnesses such as diabetes [10], aging heart [11], and cancer [12]. In eukaryotes and prokaryotes, small RNAs (sRNAs) regulate posttranscriptional gene expression [13]. The structural and functional complexity of sRNAs allows for the division of these molecules into regulatory and structural ncRNAs [14]. Transfer RNA (tRNA), ribosomal RNA, small nucleolar RNAs (snoRNAs), small cytoplasmic RNAs (scRNA), ribonuclease P (RNase P), mitochondrial RNA processing RNA, signal recognition particle RNA, and telomerase RNA are examples of structural noncoding RNAs [15]. MicroRNAs (miRNAs/miRs), P-element-induced wimpy testis-interacting RNAs (piRNAs), and long ncRNAs are examples of regulatory ncRNAs [16]. MiRNAs are the most thoroughly studied small ncRNAs. miRNAs are single-stranded RNA molecules with 18 to 25 nucleotides. They have become the main posttranscriptional regulators of gene expression and are important in many cellular processes, such as cell growth, differentiation, and apoptosis [17]. HTLV–I may alter ATLL by dysregulating host cell miRNAs. Some studies have found that cellular miRNAs play a role in HTLV-I-infected T-cell proliferation and survival. To date, HTLV–I-infected cell lines have been studied using microarray and PCR-based approaches to identify and characterize cellular miRNAs [18]. Our group recently revealed distinct sRNA signatures for ATLL and proposed that these signatures could be employed as biomarkers to detect ATLL at an early stage [19]. Furthermore, in patients with active ATLL, we observed that miR-451-3p was the miRNA that was most significantly downregulated, suggesting that this miR could be a promising therapeutic target in ATLL via the AMPK/Notch signaling pathway.
2. Case Description
In August 2013, a 30-year-old Brazilian woman was admitted to a public hospital in Bahia, northeast Brazil, because of fever, arthralgia, vomiting and abdominal pain for 2 weeks prior to admission. On physical examination, the patient had cervical, submental and inguinal lymphadenopathy. She reported no history of this type of swelling in the past and no history of intravenous drug use. Her family medical history was significant for leukemia. During hospitalization at that time, the laboratory findings revealed normal complete blood cells except for leukocyte counts of 51 × 10
3/mm
3, with 2.5 × 10
3/mm
3 banded neutrophils, 6.8 × 10
3/mm
3 segmented neutrophils, 4.2 × 10
3/mm
3 monocytes, and 33 × 10
3/mm
3 lymphocytes (
Table 1). The laboratory evaluation also reported marked hypercalcemia (ionized calcium 8.02 mg/dL). The patient was initially diagnosed with chronic lymphoproliferative disease and was referred to our hospital in September 2013 for further work up. On admission to our institute, the patient was found to have leukocyte counts of 67.41 × 10
3/mm
3 with 60.67 × 10
3/mm
3 lymphocytes, Hb 10.1 g/dL, and Hct 32.1%. The morphological and immunophenotypic features of the neoplastic cells in peripheral blood revealed that 75% of lymphocytes had convoluted nuclei and condensed chromatin. Flow cytometric analyses of surface markers of peripheral blood lymphocytes showed that 66% of the pathological T cells expressed the CD45 antigen, which expresses the T lymphoid antigens CD3 at low intensity, CD2+, CD5+, and CD4+ with coexpression of CD25+ and loss of CD7 expression. This population was negative for CD8 T lymphoid antigen, CD13, CD14, CD33, CD64 myelomonocytic antigens, CD19, CD20, CD22 B lymphoid antigens and TdT precursor cells CD34. Other peripheral blood findings were as follows: aspartate aminotransferase, 62 U/L; alanine aminotransferase, 40 U/L; alkaline phosphatase, 555 U/L; iron saturation, 72%; lactate dehydrogenase, 2510 U/L; and calcium, 7.2 mg/dL. The symptoms were described as being consistent with peripheral T-cell lymphoma, unless otherwise specified. Contrast-enhanced computed tomography scans revealed bilateral pleural effusion and ascites with lymphadenopathy. Serological analysis showed a positive reaction against HTLV-1 antibodies, and molecular analysis demonstrated a high proviral load for the HTLV-1
tax DNA (4.5 × 10
3 copies per 1000 cells). The HTLV-1 clonal expansion assay revealed strong evidence of monoclonal T-cell expansion on DNA-based polymerase chain reaction (PCR) of the rearranged γ T-cell receptor (γTCR) gene. Therefore, the patient was diagnosed with acute ATLL with lymphadenopathy according to the Shimoyama classification criteria [20]. She was immediately treated with eight cycles of cyclophosphamide, adriamycin, vincristine, and prednisone (CHOP; September 20, 2013- March 10, 2014) followed by interferon-α and zidovudine maintenance therapy until June 2019. Five schedules of G-CSF administration starting 24 h after the end of the 7
th cycle were used. Another three schedules of G-CSF administration starting 24 h after the end of the 8
th cycle were used because the patient had neutropenia. The selected induction chemotherapy had effective and well-tolerated results. Bone marrow transplant was the greatest challenge in our patient, as no matching donor was available. The patient was last seen in September 2022, and she was still in remission with a good prognosis and performance status.
Several studies have suggested that small RNAs (sRNAs), particularly microRNAs (miRNAs), may have a significant impact on the cellular response to chemotherapy drugs and that a profile of these entities in peripheral blood may serve as potential biomarkers for the therapeutic response in certain cancers [21]. Therefore, we decided to profile the sRNA from peripheral blood mononuclear cells (PBMCs) at diagnosis (designated J1) and soon after maintenance therapy (designated J2) to determine if they were related to therapy response.
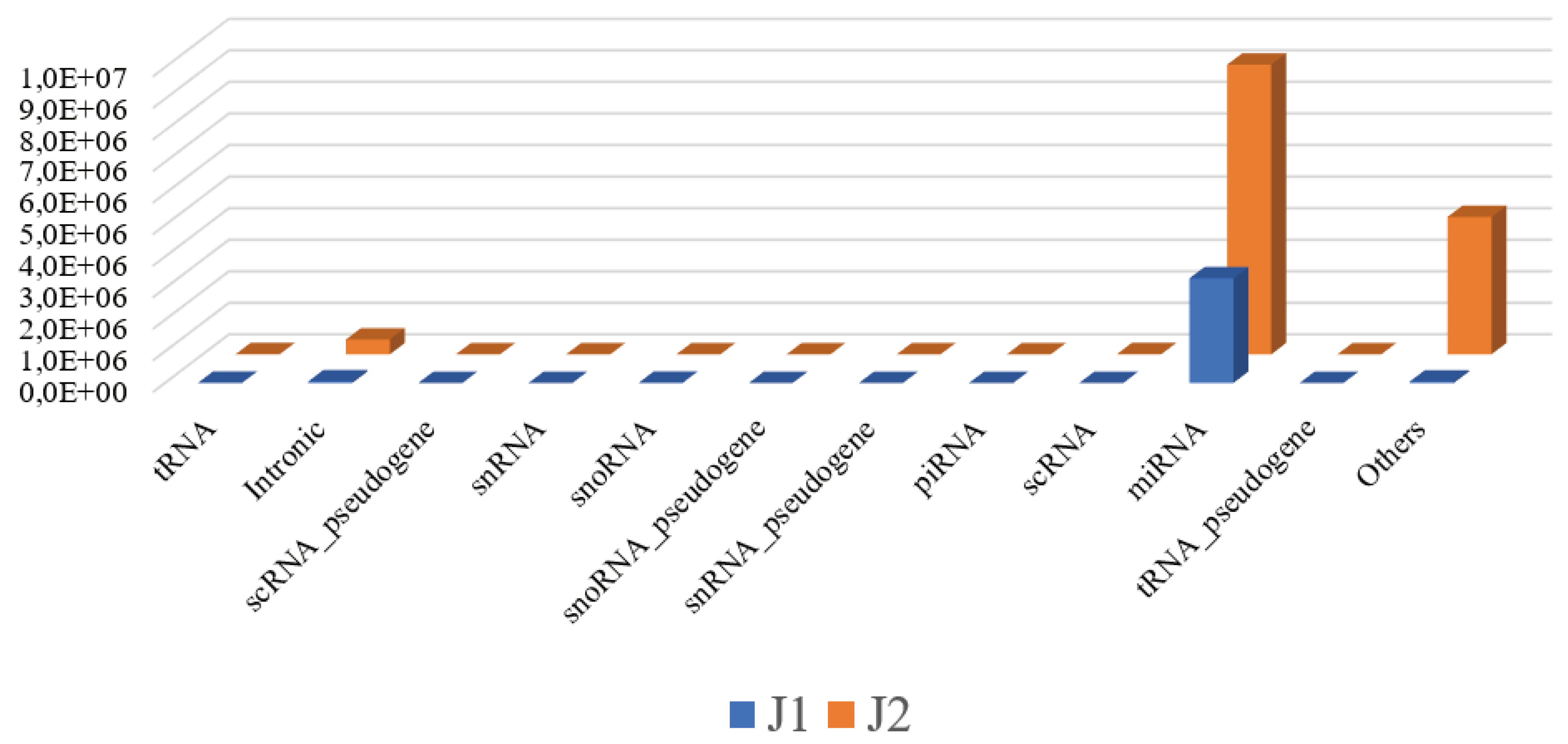
Extraction of sRNA, library preparation with the TruSeq Small RNA Sample Preparation Kit (Illumina, San Diego, CA), and sequencing on the MiSeq platform were performed according to the manufacturer’s instructions (Illumina, San Diego, CA) and a previous protocol [22; 23]. Only high-quality sequencing reads with a Sanger score of 30 or higher were considered for further analysis. The reads were aligned against the whole genome build hg19 using Strand NGS version 3.1 software (Strand Life Science) according to the small RNA alignment and small RNA analysis pipeline using standard parameters. The initial analysis showed that the number of reads mapping to different genic regions for J1 and J2 were 3,416,083 and 14,018,875, respectively, and that most of them were assigned to miRNAs (
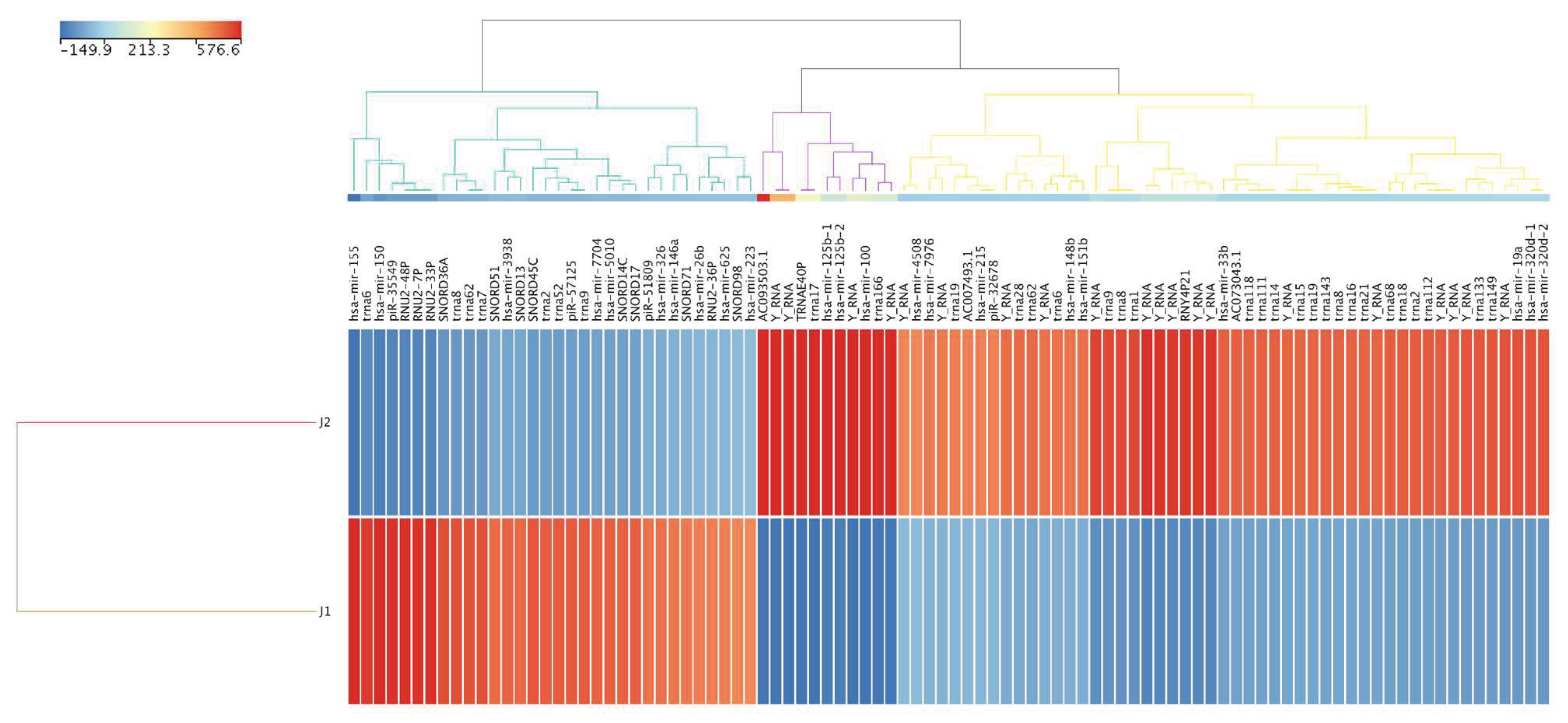
Figure 1). The numbers of reads that passed quality control for J1 and J2 were 2,469,842 and 12,429,708, respectively. After normalization, the differential expression of known sRNAs, novel entities and active miRNAs between J1 and J2 were compared. This strategy identified 94 known sRNAs that were substantially dysregulated, with 62 and 32 up- and downregulated following therapy, respectively, with fold change (FC) values ≥10 (
Figure 2, and
Supplementary Table S1). Of the 363 predicted novel sRNAs, 348 were upregulated and 15 were downregulated by at least FC values of ≥ 10 (
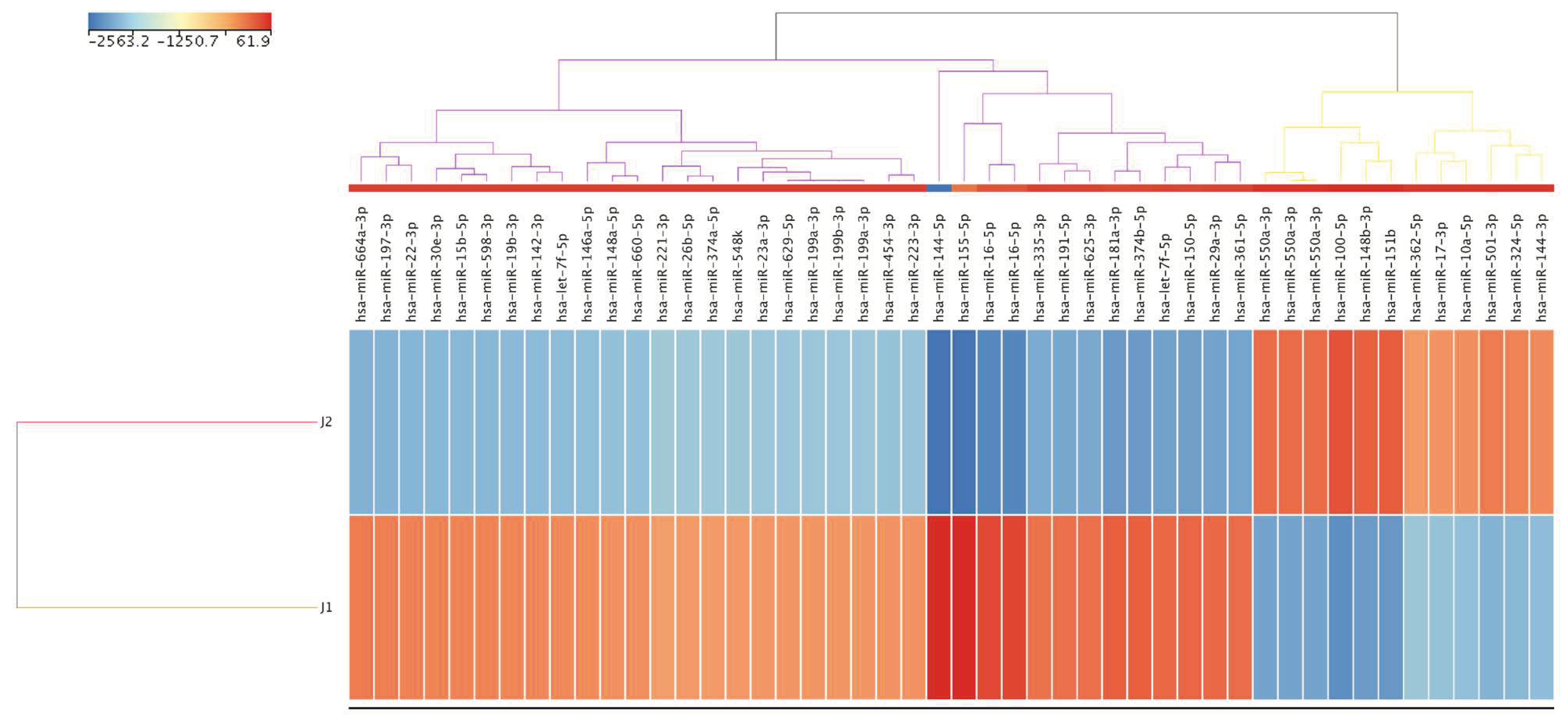
Supplementary Table S2). Notably, 48 mature miRNAs were differentially regulated 5 years after therapy. Of these, 12 were upregulated, while 36 were downregulated by at least FC values ≥ 9.8 (
Figure 3 and
Supplementary Table S3). Six of the 12 upregulated mature miRNA families were categorized as highly abundant (miR-100-5p, miR-151b, miR-148b-3p, miR-550a-3p, miR-501-3p, and miR-324-5p). The top six downregulated mature miRNAs were miR-144-5p, miR-155, miR-16-5p, miR-181a-3p, miR-374b-5p, and miR-150-5p.
Gene set enrichment analysis implemented in the mirWALK v.3 tool was used to analyze the top six up- and downregulated active miRNAs in response to therapy to identify biological networks and functions enriched in the dataset. The analysis predicted 90 target genes for the 11 differentially expressed miRNAs (
Table S4). All target genes in the respective dataset were then used to compute the Reactome, KEGG pathways, and gene ontology (GO) annotations. The Reactome pathway identified nine significantly enriched pathways. Of the pathways, three were considered very relevant for the investigated phenotype, with the highest associated corrected
p values (
Table S5). They are briefly presented here. The “macroautophagy” pathway (Corr p value = 0.0177) was identified to be enriched via the association with ATLL therapy of four genes: UBE2V1, ATG9A, MTMR3, and PEDS1-UBE2V1. The “Hemostasis” pathway (Corr p value = 0.018) and a subpathway to “Hemostasis” called “Factors involved in megakaryocyte development and platelet production” (Corr p value = 0.003) were enriched via the genes TP53, VEGFA, PHACTR2, MYB, KIF1B, PRKCA, KIF3B, CAPZA2, SLC7A11, and CDC37L1. According to KEGG, the dysregulated miRNAs were mostly enriched in the p53 signaling pathway, human cytomegalovirus infection, and cancer microRNAs. As shown in
Table S5, GO annotation showed that 17 GO terms were significantly (FDR<0.05) associated with biological process (BP), four with cellular components (CC), and eight with molecular function (MF).
3. Discussion
To the best of our knowledge, this is the first case study to reveal the miRNAs associated with ATLL patients who received CHOP chemotherapy followed by interferon and zidovudine as maintenance regimens. Patients with aggressive types ATLL (acute and lymphoma) have the poorest prognosis, with median survival of only 6–10 months even with intensive chemotherapy [24]. The worst prognosis is because of the inherent chemoresistance, a substantial tumor burden, hypercalcemia, and/or recurrent infection problems due to significant immune weakness [25]. Another possible explanation is that miRNAs control the expression of proteins that cause therapeutic failure. This enables cancer cells to acquire the desired trait [26]. In the present study, we screened for the expression of sRNA in PBMCs from HTLV-1-infected patient with acute ATLL who was treated with interferon-α and CHOP chemotherapy. A comparison of small RNA levels before chemotherapy and after the maintenance phase of the therapy revealed 94 significantly dysregulated known sRNAs, 363 predicted novel sRNAs, and 48 mature miRNAs. The majority of target genes predicted in this study were involved in various biological processes (BPs), the most significant of which was the response to hypoxia. There is overwhelming evidence that proof activation of the hypoxia-inducible factor 1 transcription factor is a common pathway affected by human oncogenic viruses, including HTLV-1 [27]. Positive regulation of cell cycle arrest was another significant BP in ATLL therapy. Previous experimental studies have shown that arsenic trioxide, in combination with IFN, induces cell cycle arrest and apoptosis in HTLV-I–infected and freshly isolated leukemia cells from ATLL patients [28] via a rapid shut-off of the NF-B pathway and a delayed shut-off of cell cycle–associated genes due to Tax degradation by the proteasome [29; 30]. Our findings lend support to these studies, implying that the combination of chemotherapy and IFN likely influenced miRNA expression, hence promoting the expression of target genes that trigger cell cycle arrest and apoptosis.
With regard to the pathways predicted in this study, it has recently been reported that activation of the macroautophagy pathway is an essential mechanism of the activated NF-B signaling pathway in ATLL [31]. Poiesz et al. [1] showed that the granzyme B-Tax transgene prevents p53 from working in the early stages of the development of a large granular lymphocytic tumor, while p53-inactivating mutations occur in later stages of the growth of the tumor. It has been postulated that progression to ATLL can likely occur only if a mutation develops that disables one of the essential cellular checkpoints before the activated virus is resuppressed. This notion is reinforced by evidence that most ATLL patients' leukemic cells include one or more mutations that disrupt the synthesis or function of biological components involved in T-cell replication, cell cycle arrest, or apoptosis [32; 33; 34].
After the maintenance phase of the therapy, the most significantly elevated miRNAs were miR-100-5p and miR-151b. Meanwhile, our findings revealed that miR-144-5p and miR155 were the most downregulated miRNAs following treatment. A recent study by Hassan et al. [35] investigated differences in miR-100-5p expression among three forms of ALL: pre-B-ALL, B-ALL, and T-ALL. The expression of miR-100-5p was determined in bone marrow aspirates from 85 pediatric ALL patients and 12 healthy donors. Their findings demonstrated that miR-100-5p was overexpressed in B-ALL but considerably downregulated in T-ALL. Based on these results, the authors suggested that reduced miR-100-5p expression was associated with high-risk prognostic factors. Similarly, Li et al [36] documented that patients with high-risk prognostic markers, T-ALL patients, and patients with MLL rearrangement and BCR-ABL fusion genes had reduced expression of miR-100-5p, implying that the expression of miR-100-5p is cell-type specific. In line with the existing evidence of miR-100-5p in ALL, MLL and other cancers, we assumed that miR-100-5p may carry out similar processes in ATLL and that high expression of this miR confers a favorable prognosis. MiR-100-5p is thought to have just a few mRNA targets, one of which is mTOR (mammalian target of rapamycin) [37]. A previous study showed that inhibiting PI3K/Akt/mTOR signaling causes growth arrest in ATLL cells [38]. There is a relationship between mTOR and tau, which is associated with the function of cyclin-dependent kinase 5 (CDK5) and autophagy. Reduced mTOR signaling may relieve pathologically elevated tau phosphorylation. While Caccamo et al [39] reported preclinical data suggesting that decreasing mTOR signaling may be a feasible therapeutic option for tauopathies, our findings imply that this salvage pathway may already be engaged in our ATLL patient due to miR-100-5p overexpression.
There is still scarce information regarding miR-151b in the literature. Almeida et al [40] recently reported that the upregulation of miR-151bb together with other miRs in childhood T-ALL may be used for distinguishing childhood T- and B-ALL subtypes. Roth et al found that the expression signatures of a combination of several miRNAs, including miR-151b, in the blood are associated with the prognosis of primary CNS lymphoma patients and allow for good separation between short- and long-term survivors [41]. In cases with upper tract urinary cancer, the differential expression of miR-151b in serum may serve as a prognostic indicator [42]. It has also been suggested to serve as a blood-based biomarker for the detection of ischemic stroke patients [43]. It remains to be seen whether miR-151b together with the other miRNAs described here are mere fine-tuners or central players in ATLL disease, a question that can be addressed by future functional studies.
MiR-144 was shown to be downregulated in several types of leukemia, including acute lymphoblastic leukemia and chronic and acute myeloid leukemia, and it may potentially be linked to imatinib resistance in the chronic myeloid leukemia cell line K562R [44; 45]. Furthermore, it was shown that miR-144 was highly expressed in normal human T cells and that it inhibited the production of interferon-γ and tumor necrosis factor-α, which in turn decreased the proliferation of T cells [46]. In addition to its leukemogenic role, the downregulation of miR-144 leads to the overexpression of Myc in diffuse large B-cell lymphoma [47]. Notably, overexpression of Myc was linked to the pathogenesis of ATLL [48]. Thus, it is possible that miR-144 downregulation and HTLV1-encoded TAX oncogene may promote the expression of Myc.
According to the literature, miR-155 is an important miRNA that contributes greatly to the pathogenesis of diverse hematological malignancies and solid tumors [49]. Ishihara and colleagues [50] showed that overexpression of miR-155 was linked with poor prognosis in ATLL and suggested its utility as a new biomarker for disease stage. Thus, the downregulation of miR-155 after maintenance therapy in our patient presumably inhibits cancer growth since overexpression of this miR inhibits TGFβR2, which hinders TGFβ1's tumor suppressive effect in leukemic patients [50].
In conclusion, our results demonstrate that PBMC samples before and after therapy from acute ATLL patient show a differential sRNA expression pattern. We also suggest that the expression of miR-100-5p, miR-151b, miR-144-5p, and miR-155 in PBMC samples predicts the pathological response in ATLL patients and may be considered a novel potential minimally invasive biomarker for the prognosis of ATLL patients. Although independent validation of the data is necessary, the identification of miRNAs associated with good outcome in patients with acute ATLL may help in tailoring treatment and surveillance strategies in these patients.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
References
- B.J. Poiesz, F.W. Ruscetti, A.F. Gazdar, P.A. Bunn, J.D. Minna, and R.C. Gallo, Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 77 (1980) 7415-9. [CrossRef]
- Gessain, and O. Cassar, Epidemiological Aspects and World Distribution of HTLV-1 Infection. Frontiers in microbiology 3 (2012) 388. [CrossRef]
- K. Verdonck, E. Gonzalez, S. Van Dooren, A.M. Vandamme, G. Vanham, and E. Gotuzzo, Human T-lymphotropic virus 1: recent knowledge about an ancient infection. The Lancet. Infectious diseases 7 (2007) 266-81. [CrossRef]
- G. Franchini, R.F. Ambinder, and M. Barry, Viral Disease in Hematology. Hematology. American Society of Hematology. Education Program (2000) 409-423. [CrossRef]
- E.L. Murphy, B. Hanchard, J.P. Figueroa, W.N. Gibbs, W.S. Lofters, M. Campbell, J.J. Goedert, and W.A. Blattner, Modelling the risk of adult T-cell leukemia/lymphoma in persons infected with human T-lymphotropic virus type I. International journal of cancer 43 (1989) 250-3. [CrossRef]
- M. Higuchi, and M. Fujii, Distinct functions of HTLV-1 Tax1 from HTLV-2 Tax2 contribute key roles to viral pathogenesis. Retrovirology 6 (2009) 117. [CrossRef]
- M. Yoshida, M. Seiki, K. Yamaguchi, and K. Takatsuki, Monoclonal integration of human T-cell leukemia provirus in all primary tumors of adult T-cell leukemia suggests causative role of human T-cell leukemia virus in the disease. Proceedings of the National Academy of Sciences of the United States of America 81 (1984) 2534-7. [CrossRef]
- Y. Nagata, K. Kontani, T. Enami, K. Kataoka, R. Ishii, Y. Totoki, T.R. Kataoka, M. Hirata, K. Aoki, K. Nakano, A. Kitanaka, M. Sakata-Yanagimoto, S. Egami, Y. Shiraishi, K. Chiba, H. Tanaka, Y. Shiozawa, T. Yoshizato, H. Suzuki, A. Kon, K. Yoshida, Y. Sato, A. Sato-Otsubo, M. Sanada, W. Munakata, H. Nakamura, N. Hama, S. Miyano, O. Nureki, T. Shibata, H. Haga, K. Shimoda, T. Katada, S. Chiba, T. Watanabe, and S. Ogawa, Variegated RHOA mutations in adult T-cell leukemia/lymphoma. Blood 127 (2016) 596-604. [CrossRef]
- S. Djebali, C.A. Davis, A. Merkel, A. Dobin, T. Lassmann, A. Mortazavi, A. Tanzer, J. Lagarde, W. Lin, F. Schlesinger, C. Xue, G.K. Marinov, J. Khatun, B.A. Williams, C. Zaleski, J. Rozowsky, M. Roder, F. Kokocinski, R.F. Abdelhamid, T. Alioto, I. Antoshechkin, M.T. Baer, N.S. Bar, P. Batut, K. Bell, I. Bell, S. Chakrabortty, X. Chen, J. Chrast, J. Curado, T. Derrien, J. Drenkow, E. Dumais, J. Dumais, R. Duttagupta, E. Falconnet, M. Fastuca, K. Fejes-Toth, P. Ferreira, S. Foissac, M.J. Fullwood, H. Gao, D. Gonzalez, A. Gordon, H. Gunawardena, C. Howald, S. Jha, R. Johnson, P. Kapranov, B. King, C. Kingswood, O.J. Luo, E. Park, K. Persaud, J.B. Preall, P. Ribeca, B. Risk, D. Robyr, M. Sammeth, L. Schaffer, L.H. See, A. Shahab, J. Skancke, A.M. Suzuki, H. Takahashi, H. Tilgner, D. Trout, N. Walters, H. Wang, J. Wrobel, Y. Yu, X. Ruan, Y. Hayashizaki, J. Harrow, M. Gerstein, T. Hubbard, A. Reymond, S.E. Antonarakis, G. Hannon, M.C. Giddings, Y. Ruan, B. Wold, P. Carninci, R. Guigo, and T.R. Gingeras, Landscape of transcription in human cells. Nature 489 (2012) 101-8. [CrossRef]
- Higuchi, A. Nakatsuka, J. Eguchi, S. Teshigawara, M. Kanzaki, A. Katayama, S. Yamaguchi, N. Takahashi, K. Murakami, D. Ogawa, S. Sasaki, H. Makino, and J. Wada, Identification of circulating miR-101, miR-375 and miR-802 as biomarkers for type 2 diabetes. Metabolism: clinical and experimental 64 (2015) 489-97. [CrossRef]
- E. van Rooij, L.B. Sutherland, X. Qi, J.A. Richardson, J. Hill, and E.N. Olson, Control of stress-dependent cardiac growth and gene expression by a microRNA. Science 316 (2007) 575-9. [CrossRef]
- W.C. Zhang, T.M. Chin, H. Yang, M.E. Nga, D.P. Lunny, E.K. Lim, L.L. Sun, Y.H. Pang, Y.N. Leow, S.R. Malusay, P.X. Lim, J.Z. Lee, B.J. Tan, N. Shyh-Chang, E.H. Lim, W.T. Lim, D.S. Tan, E.H. Tan, B.C. Tai, R.A. Soo, W.L. Tam, and B. Lim, Tumour-initiating cell-specific miR-1246 and miR-1290 expression converge to promote non-small cell lung cancer progression. Nature communications 7 (2016) 11702. [CrossRef]
- T.A. Farazi, S.A. Juranek, and T. Tuschl, The growing catalog of small RNAs and their association with distinct Argonaute/Piwi family members. Development 135 (2008) 1201-14. [CrossRef]
- J.S. Mattick, and I.V. Makunin, Non-coding RNA. Human molecular genetics 15 Spec No 1 (2006) R17-29. [CrossRef]
- V. Ambros, B. Bartel, D.P. Bartel, C.B. Burge, J.C. Carrington, X. Chen, G. Dreyfuss, S.R. Eddy, S. Griffiths-Jones, M. Marshall, M. Matzke, G. Ruvkun, and T. Tuschl, A uniform system for microRNA annotation. Rna 9 (2003) 277-9. [CrossRef]
- J. Li, B. Wu, J. Xu, and C. Liu, Genome-wide identification and characterization of long intergenic non-coding RNAs in Ganoderma lucidum. PloS one 9 (2014) e99442. [CrossRef]
- P. Xu, S.Y. Vernooy, M. Guo, and B.A. Hay, The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Current biology : CB 13 (2003) 790-5. [CrossRef]
- E. Huntzinger, and E. Izaurralde, Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature reviews. Genetics 12 (2011) 99-110. [CrossRef]
- A. Nascimento, D.R. Valadao de Souza, R. Pessoa, A.J. Pietrobon, Y. Nukui, J. Pereira, J. Casseb, A.C. Penalva de Oliveira, P. Loureiro, A.J. da Silva Duarte, P.B. Clissa, and S.S. Sanabani, Global expression of noncoding RNome reveals dysregulation of small RNAs in patients with HTLV-1-associated adult T-cell leukemia: a pilot study. Infectious agents and cancer 16 (2021) 4. [CrossRef]
- M. Shimoyama, Diagnostic criteria and classification of clinical subtypes of adult T-cell leukaemia-lymphoma. A report from the Lymphoma Study Group (1984-87). British journal of haematology 79 (1991) 428-37. [CrossRef]
- W. Si, J. Shen, H. Zheng, and W. Fan, The role and mechanisms of action of microRNAs in cancer drug resistance. Clinical epigenetics 11 (2019) 25. [CrossRef]
- P.B. Clissa, R. Pessôa, K.F. Ferraz, D.R. de Souza, and S.S. Sanabani, Data on global expression of non-coding RNome in mice gastrocnemius muscle exposed to jararhagin, snake venom metalloproteinase. Data Brief 9 (2016) 685-688. [CrossRef]
- D.R. Valadao de Souza, R. Pessoa, A. Nascimento, Y. Nukui, J. Pereira, J. Casseb, A.C. Penalva de Oliveira, A.J. da Silva Duarte, P.B. Clissa, and S.S. Sanabani, Small RNA profiles of HTLV-1 asymptomatic carriers with monoclonal and polyclonal rearrangement of the T-cell antigen receptor gamma-chain using massively parallel sequencing: A pilot study. Oncology letters 20 (2020) 2311-2321. [CrossRef]
- Y. Yamada, M. Tomonaga, H. Fukuda, S. Hanada, A. Utsunomiya, M. Tara, M. Sano, S. Ikeda, K. Takatsuki, M. Kozuru, K. Araki, F. Kawano, M. Niimi, K. Tobinai, T. Hotta, and M. Shimoyama, A new G-CSF-supported combination chemotherapy, LSG15, for adult T-cell leukaemia-lymphoma: Japan Clinical Oncology Group Study 9303. British journal of haematology 113 (2001) 375-82. [CrossRef]
- A. Bazarbachi, F. Suarez, P. Fields, and O. Hermine, How I treat adult T-cell leukemia/lymphoma. Blood 118 (2011) 1736-45. [CrossRef]
- H. Mesrian Tanha, M. Mojtabavi Naeini, S. Rahgozar, A. Moafi, and M.A. Honardoost, Integrative computational in-depth analysis of dysregulated miRNA-mRNA interactions in drug-resistant pediatric acute lymphoblastic leukemia cells: an attempt to obtain new potential gene-miRNA pathways involved in response to treatment. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 37 (2016) 7861-72. [CrossRef]
- S. Cuninghame, R. Jackson, and I. Zehbe, Hypoxia-inducible factor 1 and its role in viral carcinogenesis. Virology 456-457 (2014) 370-83. [CrossRef]
- A. Bazarbachi, M.E. El-Sabban, R. Nasr, F. Quignon, C. Awaraji, J. Kersual, L. Dianoux, Y. Zermati, J.H. Haidar, O. Hermine, and H. de The, Arsenic trioxide and interferon-alpha synergize to induce cell cycle arrest and apoptosis in human T-cell lymphotropic virus type I-transformed cells. Blood 93 (1999) 278-83. [CrossRef]
- M.E. El-Sabban, R. Nasr, G. Dbaibo, O. Hermine, N. Abboushi, F. Quignon, J.C. Ameisen, F. Bex, H. de The, and A. Bazarbachi, Arsenic-interferon-alpha-triggered apoptosis in HTLV-I transformed cells is associated with tax down-regulation and reversal of NF-kappa B activation. Blood 96 (2000) 2849-55. [CrossRef]
- R. Nasr, A. Rosenwald, M.E. El-Sabban, B. Arnulf, P. Zalloua, Y. Lepelletier, F. Bex, O. Hermine, L. Staudt, H. de The, and A. Bazarbachi, Arsenic/interferon specifically reverses 2 distinct gene networks critical for the survival of HTLV-1-infected leukemic cells. Blood 101 (2003) 4576-82. [CrossRef]
- Y.R. Fauzi, S. Nakahata, S. Chilmi, T. Ichikawa, P. Nueangphuet, R. Yamaguchi, T. Nakamura, K. Shimoda, and K. Morishita, Antitumor effects of chloroquine/hydroxychloroquine mediated by inhibition of the NF-kappaB signaling pathway through abrogation of autophagic p47 degradation in adult T-cell leukemia/lymphoma cells. PloS one 16 (2021) e0256320. [CrossRef]
- E. Cesarman, A. Chadburn, G. Inghirami, G. Gaidano, and D.M. Knowles, Structural and functional analysis of oncogenes and tumor suppressor genes in adult T-cell leukemia/lymphoma shows frequent p53 mutations. Blood 80 (1992) 3205-16. [CrossRef]
- S. Nishimura, N. Asou, H. Suzushima, T. Okubo, T. Fujimoto, M. Osato, H. Yamasaki, L. Lisha, and K. Takatsuki, p53 gene mutation and loss of heterozygosity are associated with increased risk of disease progression in adult T cell leukemia. Leukemia 9 (1995) 598-604.
- A. Sakashita, T. Hattori, C.W. Miller, H. Suzushima, N. Asou, K. Takatsuki, and H.P. Koeffler, Mutations of the p53 gene in adult T-cell leukemia. Blood 79 (1992) 477-80. [CrossRef]
- N.M. Hassan, L.A. Refaat, G.N. Ismail, M. Abdellateif, S.A. Fadel, and R.S. AbdelAziz, Diagnostic, prognostic and predictive values of miR-100 and miR-210 in pediatric acute lymphoblastic Leukemia. Hematology 25 (2020) 405-413. [CrossRef]
- X.J. Li, X.Q. Luo, B.W. Han, F.T. Duan, P.P. Wei, and Y.Q. Chen, MicroRNA-100/99a, deregulated in acute lymphoblastic leukaemia, suppress proliferation and promote apoptosis by regulating the FKBP51 and IGF1R/mTOR signalling pathways. British journal of cancer 109 (2013) 2189-98. [CrossRef]
- D.M. Garcia, D. Baek, C. Shin, G.W. Bell, A. Grimson, and D.P. Bartel, Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other microRNAs. Nature structural & molecular biology 18 (2011) 1139-46. [CrossRef]
- T. Ikezoe, C. Nishioka, K. Bandobashi, Y. Yang, Y. Kuwayama, Y. Adachi, T. Takeuchi, H.P. Koeffler, and H. Taguchi, Longitudinal inhibition of PI3K/Akt/mTOR signaling by LY294002 and rapamycin induces growth arrest of adult T-cell leukemia cells. Leukemia research 31 (2007) 673-82. [CrossRef]
- A. Caccamo, A. Magri, D.X. Medina, E.V. Wisely, M.F. Lopez-Aranda, A.J. Silva, and S. Oddo, mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging cell 12 (2013) 370-80. [CrossRef]
- R.S. Almeida, E.S.M. Costa, L.L. Coutinho, R. Garcia Gomes, F. Pedrosa, J.D. Massaro, E.A. Donadi, and N. Lucena-Silva, MicroRNA expression profiles discriminate childhood T- from B-acute lymphoblastic leukemia. Hematological oncology 37 (2019) 103-112. [CrossRef]
- P. Roth, A. Keller, J.D. Hoheisel, P. Codo, A.S. Bauer, C. Backes, P. Leidinger, E. Meese, E. Thiel, A. Korfel, and M. Weller, Differentially regulated miRNAs as prognostic biomarkers in the blood of primary CNS lymphoma patients. European journal of cancer 51 (2015) 382-90. [CrossRef]
- R. Montalbo, L. Izquierdo, M. Ingelmo-Torres, J.J. Lozano, D. Capitan, A. Alcaraz, and L. Mengual, Prognostic value of circulating microRNAs in upper tract urinary carcinoma. Oncotarget 9 (2018) 16691-16700. [CrossRef]
- X. Cheng, P. Kan, Z. Ma, Y. Wang, W. Song, C. Huang, and B. Zhang, Exploring the potential value of miR-148b-3p, miR-151b and miR-27b-3p as biomarkers in acute ischemic stroke. Bioscience reports 38 (2018). [CrossRef]
- L. Liu, S. Wang, R. Chen, Y. Wu, B. Zhang, S. Huang, J. Zhang, F. Xiao, M. Wang, and Y. Liang, Myc induced miR-144/451 contributes to the acquired imatinib resistance in chronic myelogenous leukemia cell K562. Biochemical and biophysical research communications 425 (2012) 368-73. [CrossRef]
- J. Jin, Y. Wang, Y. Xu, X. Zhou, Y. Liu, X. Li, and J. Wang, MicroRNA-144 regulates cancer cell proliferation and cell-cycle transition in acute lymphoblastic leukemia through the interaction of FMN2. The journal of gene medicine 19 (2017). [CrossRef]
- Y. Liu, X. Wang, J. Jiang, Z. Cao, B. Yang, and X. Cheng, Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Molecular immunology 48 (2011) 1084-90. [CrossRef]
- T. Wang, F. Wu, and D. Yu, miR-144/451 in hematopoiesis and beyond. ExRNA 1 (2019) 16. [CrossRef]
- J. Iqbal, G. Wright, C. Wang, A. Rosenwald, R.D. Gascoyne, D.D. Weisenburger, T.C. Greiner, L. Smith, S. Guo, R.A. Wilcox, B.T. Teh, S.T. Lim, S.Y. Tan, L.M. Rimsza, E.S. Jaffe, E. Campo, A. Martinez, J. Delabie, R.M. Braziel, J.R. Cook, R.R. Tubbs, G. Ott, E. Geissinger, P. Gaulard, P.P. Piccaluga, S.A. Pileri, W.Y. Au, S. Nakamura, M. Seto, F. Berger, L. de Leval, J.M. Connors, J. Armitage, J. Vose, W.C. Chan, L.M. Staudt, P. Lymphoma Leukemia Molecular Profiling, and T.c.L.P. the International Peripheral, Gene expression signatures delineate biological and prognostic subgroups in peripheral T-cell lymphoma. Blood 123 (2014) 2915-23. [CrossRef]
- Jurkovicova, M. Magyerkova, L. Kulcsar, M. Krivjanska, V. Krivjansky, A. Gibadulinova, I. Oveckova, and M. Chovanec, miR-155 as a diagnostic and prognostic marker in hematological and solid malignancies. Neoplasma 61 (2014) 241-51. [CrossRef]
- K. Ishihara, D. Sasaki, K. Tsuruda, N. Inokuchi, K. Nagai, H. Hasegawa, K. Yanagihara, and S. Kamihira, Impact of miR-155 and miR-126 as novel biomarkers on the assessment of disease progression and prognosis in adult T-cell leukemia. Cancer epidemiology 36 (2012) 560-5. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).