1. Introduction
Neurological disorders are common, and predicted to affect one in six people [
1]. Scientists constantly work to find more effective methods of diagnosing and treating these devastating conditions. Animal subjects are invaluable in neuroscience as they allow researchers to induce diseases and their pathogenesis[
2]. Small animals, most commonly mice, led to the development of many important animal models[
3]. However, small animals lack the brain complexity and biological scale found in humans, resulting in many therapies failing to translate to human trials[
4][
5]. This issue led to an increased focus on developing large animal models. Animals such as sheep, pigs, and non-human primates resulted in more robust findings[
6]. Recently, minipigs have emerged at the forefront of neuroscience as they present multiple advantages over other large animals, such as relatively large gyrencephalic brain, similar to human brain anatomy, neurophysiological processes, and white-grey matter ratio (60:40)[
7]. Furthermore, minipig’s use presents considerably lower cost and fewer ethical issues[
8] when compared to alternatives.
Medical imaging, such as magnetic resonance (MR) imaging, provides researchers with non-invasive access to the brain. MRI produces higher-quality images compared to Computed Tomography without radiation risks, making it suitable for research. Medical image processing is a crucial part of research as it allows researchers to monitor their experiments and understand disease development. Processes such as image registration, skull stripping, tissue segmentation, and landmark detection are necessary for many experiments. However, many algorithms are created and optimized for MR analysis of human data and are not directly applicable or sufficiently sensitive to measure large-animal data. For example, due to differences in size and shape of the head, highly-successful tools such as SynthStrip[
9] fail on minipig images. This forces researchers into laborious, expensive, and error-prone manual processing. Therefore there is an urgent need for accurate and automated tools to analyze minipig data.
In this work, we propose PigSNIPE - a pipeline for the automated processing of minipig MR images, similar to software available for humans[
10]. This is an extension of our previous work[
11] and allows for image registration, AC-PC alignment, brain mask segmentation, skull stripping, tissue segmentation, caudate-putamen brain segmentation, and landmark detection in under two minutes. To the best of our knowledge, this is the first tool aimed at animal images. Our tool is open-source and can be found at
https://github.com/BRAINSia/PigSNIPE.
2. Data
In this study, we used two large-animal minipig datasets. The first (Germany) dataset consists of 106 scanning sessions from 33 Libechov minipigs[
12] collected using a 3T Philips Achieva scanner. Each scanning session contained single T1 and T2 weighted scans. T1w images were acquired at the acquisition resolution of
mm and spatial size of 450x576x576 voxels. The T2w images have a resolution of
mm and a spatial size of
voxels.
The second (Iowa) dataset was collected for a study investigating CLN2 gene mutation at the University of Iowa[
13]. The Iowa dataset consisted of 38 scanning sessions from 23 Yucatan minipigs using a 3.0T GE SIGNA Premier scanner. Many scanning sessions contain more than one T1w and T2w image resulting in a total of 178 T1w and 134 T2w images. Both T1w and T2w images were acquired at a resolution of
mm and spatial size of
voxels.
Based on our data, we created two training datasets. The first dataset was used for the training brainmasks models. As both Low-Resolution and High-Resolution brainmasks models work on either T1w or T2w images, we took advantage of all available data as shown in
Table 1. The second dataset was used to train Intracranial Volume, Gray-White Matter and CSF, and Caudate-Putamen Segmentation models, which require as input registered T1w and T2w images. We compute a cartesian product of all T1w and T2w images for each scanning session to maximize our training dataset to obtain all possible pairs. The range of all possible pairs per scanning session was between 1 and 28. Therefore to avoid the overrepresentation of some subjects, we limit the number of T1w-T2w pairs to four. The resulting training split is in
Table 2. For each dataset, we split the data by subjects into training, validation, and test sets (80%, 10%, 10% respectively) on the subject basis, to ensure data from the same animal appear only in one subset.
In addition to the original T1w and T2w scan data, we generated Inter-Cranial Volume (ICV) masks, Caudate-Putamen (CP) Segmentations, and Gray Matter-White Matter-Cerebrospinal Fluid (GWC) segmentations. The ICV masks serve as ground truth for Low-Resolution, High-Resolution, and ICV models. The CP segmentations were manually traced and used to train the CP segmentation model. The GWC masks were generated using Atropos software[
14] and are used in training the GWC model. The physical landmarks data consists of 19 landmarks used for training the landmark detection models. Detailed information on data and implementation of landmark detection can be found in our previous work[
11].
3. Materials and Methods
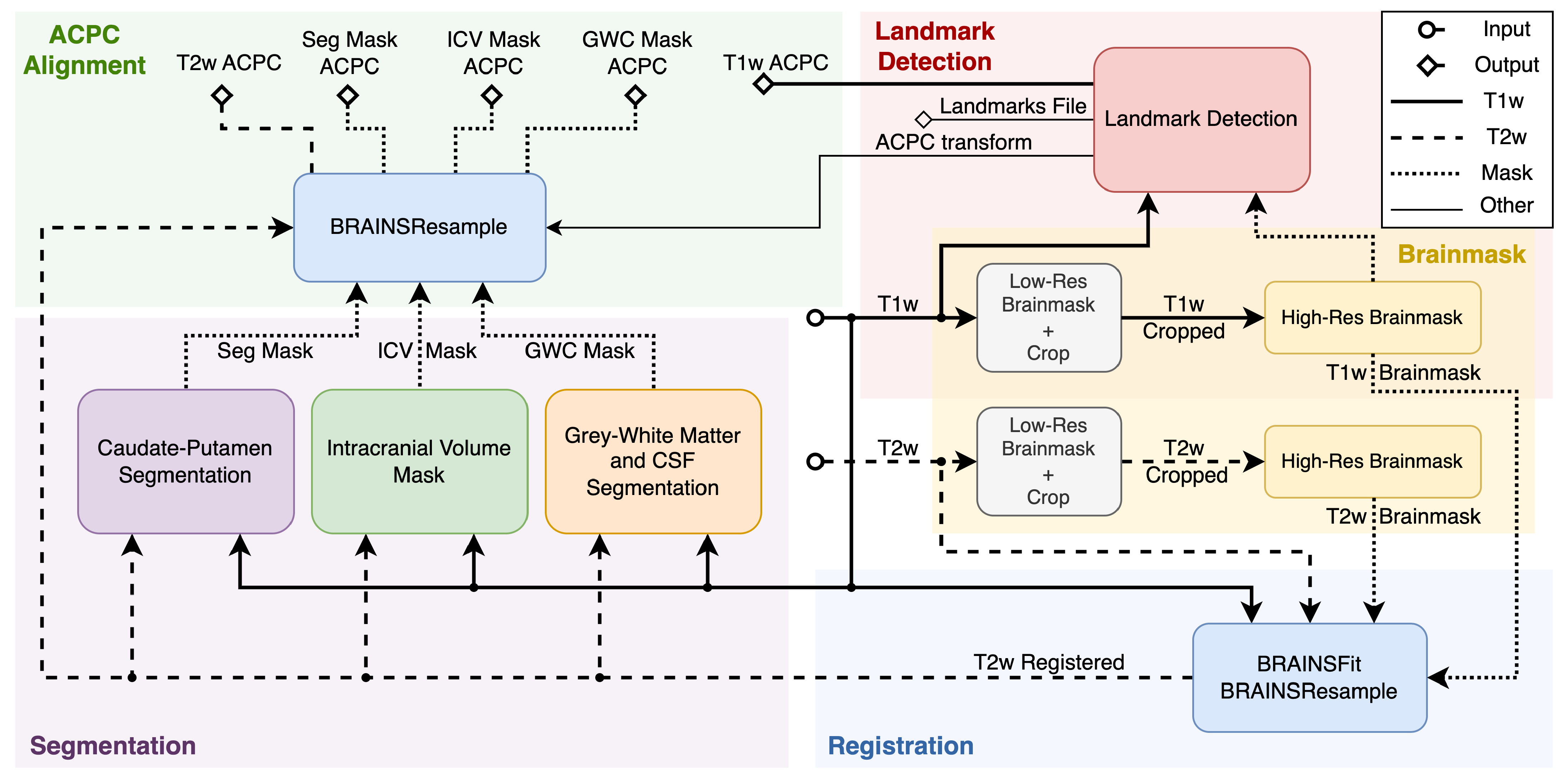
Figure 1.
Simplified view of the pipeline’s dataflow.
Figure 1.
Simplified view of the pipeline’s dataflow.
3.1. Pipeline Overview
Figure 1 displays the simplified view of the PigSNIPE pipeline. The pipeline’s inputs are T1w and T2w images in their original physical spaces. The starting step (Brainmask) is to compute image brainmasks as other processes require them. First, we compute a low-resolution brainmask to crop the original image around the brain. This step then allows for computing a high-resolution brainmask. Then, the original T1w and T2w images and their corresponding high-resolution brainmasks are used in the registration process (Registration step). The co-registered T1w and T2w images are used in the Segmentation part to compute Intracranial Volume mask (ICV), Gray Matter-White Matter-Cerebrospinal Fluid mask (GWC), and Caudate-Putamen segmentation (Seg). On the other side of the pipeline, the Landmark Detection step uses the T1w image with its high-resolution brainmask. This process produces ACPC transform, ACPC aligned T1w image, and computes 19 physical landmarks saved in both the original T1w and ACPC space. The last part of the pipeline is ACPC alignment. At this point, computed ICV, GWC, Seg, and T2w registered images are in the original T1w space. Therefore we can reuse the ACPC transform computed during the Landmark Detection process to resample the data into ACPC space.
3.2. Deep Learning Segmentation Model
All brainmask and segmentation are based on the same model architecture - 3D ResUNet architecture[
15]. Each model uses the Adam optimizer, a learning rate of 0.001, and the DiceCE loss, which combines Dice and Cross Entropy losses. The implementation leveraged existing MONAI[
16], PyTorch[
17], and Lightning[
18] libraries.
All models use five layers with 16, 32, 64, 128, and 256 channels per layer (with the Low-Resolution model being an exception and having 128 channels in the last layer) and three residual connections. The only variations between the models are image voxel size. In the Low-Resolution Model, we resample a 64x64x64 voxel region with 3mm isotropic spacing from the center of the image. This dramatically decreases the image size by over 95%, allowing for easy model training. All other models use an isotropic spacing of 0.5mm. The High-Resolution model uses a
region, the ICV and GWC model use a
region, and the Seg model uses a
voxel region. Those regions are resampled around the centroid of the Low-Resolution brainmask for High-Resolution Model, High-Res Brainmask for the ICV model, and ICV mask for GWC and Seg models. The centroids are computed using the ITK’s ImageMomentsCalculator[
19]. Additionally, to make the High-Resolution brainmask model robust to errors in the centroid position resulting from the inaccuracy of the Low-Resolution model, during training, we add a random vector to the computed centroid position.
For preprocessing, all models use intensity value scaling from -1.0 to 1.0 with truncation on 1% and 99% percentile and implement standard data augmentation techniques: random rotation, zoom, noise, and flip (except the Seg model). Additionally, for ICV, GWC, and Seg models, we pass both T1w and T2w images on two channels.
For post-processing during inference, we apply the FillHoles transform, which will fill all holes that could randomly occur in the predicted mask to all models. Additionally, all models except the GWC model use KeepLargestConnectedComponent, which removes noise outside the predicted mask.
3.3. Landmarks Detection
A fundamental step in neuroimage analysis is anatomical landmark detection. Landmarks have many uses in medical imaging, including morphometric analysis[
20], image-guided surgery[
21], or image registration[
22]. In addition, many analysis tools require landmarks to co-register different imaging sets.
The landmark detection is a two-stage process involving four deep reinforcement learning models[
23]. All models detect multiple landmarks simultaneously through the multi-agent, hard parameter sharing [
24][
25], deep Q-network [
26]. The models are optimized by Huber loss[
27], and Adam optimizer [
28], and controlled by an
-greedy exploration[
29] policy.
The first step is to crop the T1w image and compute the brain mask’s center of gravity, which is used for reinforcement learning initialization. In the first two models, we compute three landmarks allowing AC-PC alignment of the T1w image. After that, the image in the standard AC-PC space allows for accurate computation of the remaining 16 landmarks.
Detailed information about the models’ architecture, training, and results can be found in our previous work[
11].
3.4. Image Registration and AC-PC Alignment
Despite images from the same scanning session being taken minutes apart, many sources for possible image misalignment exist. For example, the animal might move during the scanning session resulting in poor alignment of the images. As ICV, GWC, and Seg models use both T1w and T2w images simultaneously; it is crucial to ensure proper image registration.
For all registrations, we use BRAINSFit and BRAINSResample algorithms from BRAINSTools package[
30]. The algorithms use high-resolution masks to ensure that the registration samples data points from the region of interest. By default, the pipeline uses Rigid registration with ResampleInPlace. This allows for image registration without interpolation errors. However, the user has the option to use the Rigid+Affine transform that allows for nonlinear transformation.
Additionally, the pipeline can transform all data into a standardized AC-PC (anterior commissure-posterior commissure) aligned space. This process combines the ACPC transform generated during the Landmark detection process and the T2w-to-T1w transform in native space to achieve T2w image in ACPC space. As all computed segmentations are in the T1w image, we can apply the ACPC transform to generate ACPC-aligned masks.
4. Results
4.1. Deep-Learning Model Accuracy
We evaluate the model performance using DICE[
31] and balanced Hausdorff distance[
32]. DICE is a similarity coefficient that measures the overlap over the union of two binary masks, and Hausdorff distance measures the average distance error between ground truth and predicted masks. To compute the metrics we used Evaluate Segmentation tool[
33] and TorchMetrics package[
34]
4.1.1. Brainmask
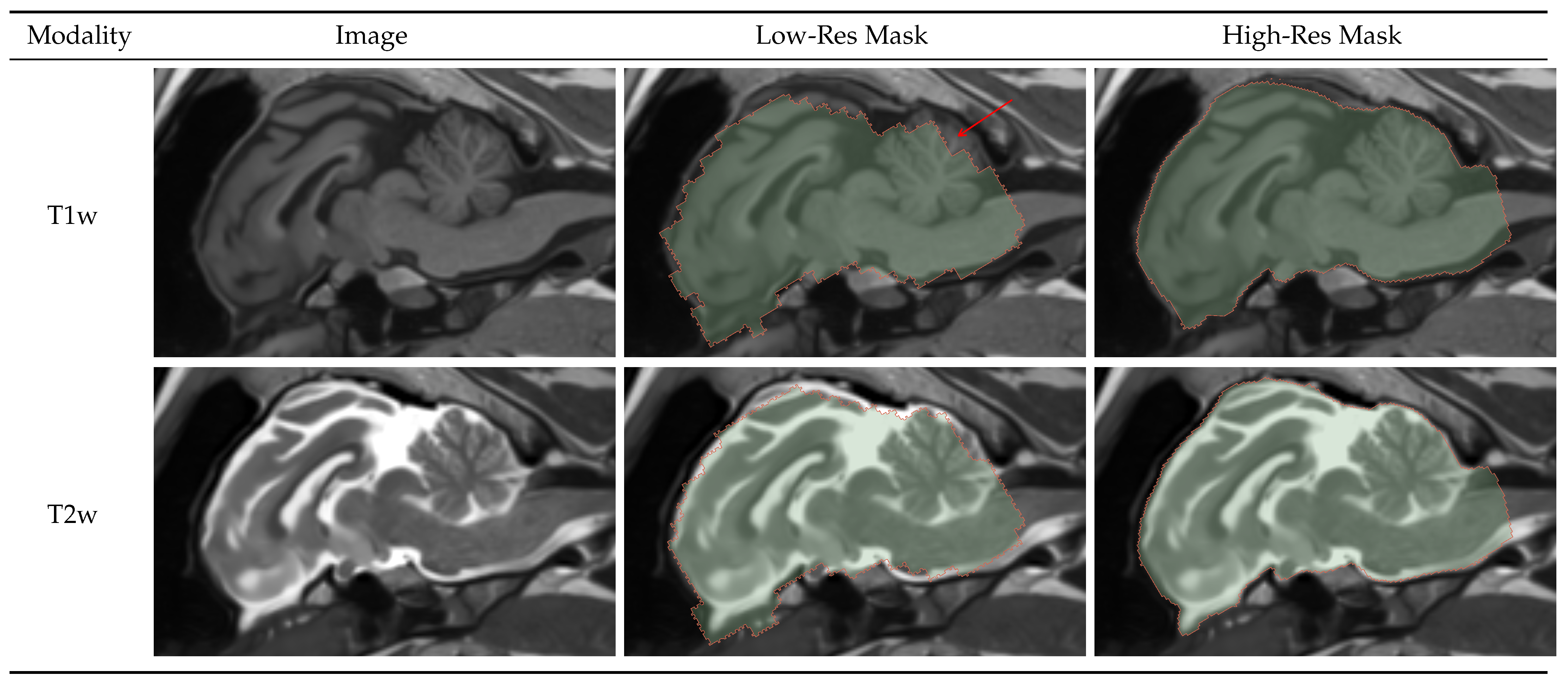
Figure 2.
Result visualization for Low and High-Resolution models. Low-Resolution model robustly detects the brain allowing for image cropping and generating highly accurate high-resolution brainmask.
Figure 2.
Result visualization for Low and High-Resolution models. Low-Resolution model robustly detects the brain allowing for image cropping and generating highly accurate high-resolution brainmask.
Figure 2 shows a 2D sagittal slice of the T1w and T2w images and their corresponding low and high-resolution brainmasks. We can see that the low-resolution mask has very coarse boundaries and mistakes, such as missed part of the cerebellum indicated by the red arrow. Although the accuracy of the low-resolution mask is much worse than the high-resolution mask, it robustly finds the position of the brain. This allows for accurate image cropping and the generation of a highly accurate high-resolution mask.
Table 3 shows the DICE coefficient and balanced Average Hausdorff Distance (bAVD) results on the test set. We can see that the models perform well on both Germany and Iowa data. The bAVD for the low-resolution mask is, on average, 0.25 mm, which proves the robustness of the model’s ability to find the brain. The high-resolution model is highly accurate, with an average distance error of 0.04 mm and DICE of 0.97.
4.1.2. Intracranial Volume Mask
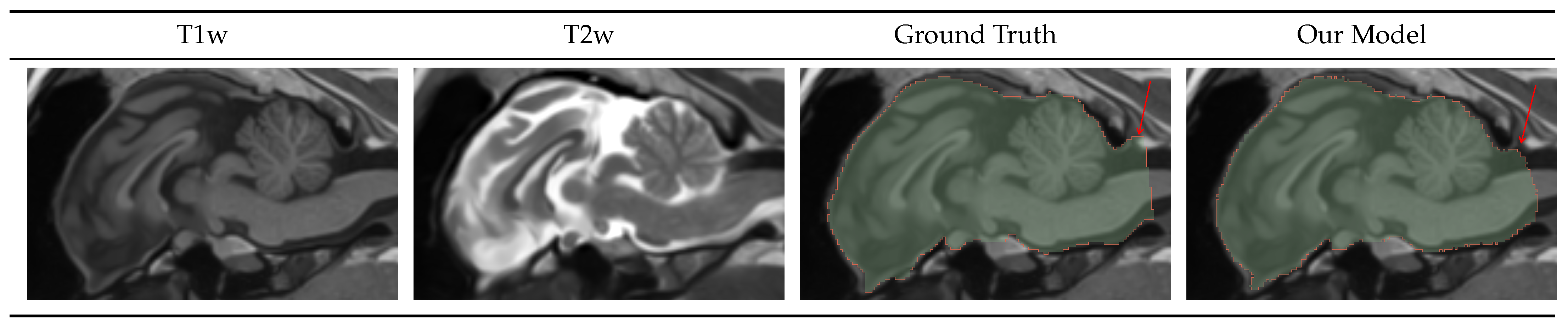
Figure 3.
Intracranial Volume Mask Visualization. Our model generates masks alike the ground truth labels, with the difference being the posterior cutoff point, as indicated by the red arrows.
Figure 3.
Intracranial Volume Mask Visualization. Our model generates masks alike the ground truth labels, with the difference being the posterior cutoff point, as indicated by the red arrows.
Figure 3 shows the input T1w and T2w image, the ground truth, and the predicted ICV mask. As seen in
Table 4 the ICV model is more accurate and consistent than the high-resolution model at the DICE of 0.98 and bAVD of 0.03 mm. Furthermore, those numbers are negatively impacted by the posterior mask cutoff differences, indicated by the red arrow.
4.1.3. Caudate-Putamen Segmentation Mask
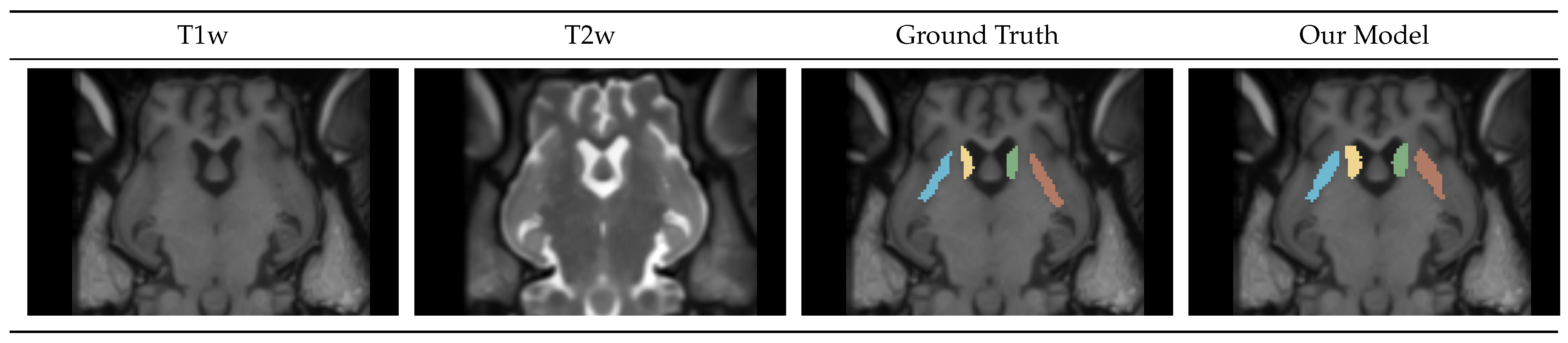
Figure 4.
Caudate-Putamen Segmentation Mask Visualization. Mask generated by our model closely resembles the manually traced ground truth label.
Figure 4.
Caudate-Putamen Segmentation Mask Visualization. Mask generated by our model closely resembles the manually traced ground truth label.
Figure 4 shows a portion of an axial slice of the input T1w and T2w images, the Ground Truth manual segmentation, and our Model prediction. We can see that the boundaries of the predicted regions vary from the ground truth. This is because the Seg model tends to over-segment the regions.
Table 5 show the metrics for the Seg model performance on the test dataset. We can see that all regions have a similar DICE score of 0.8.
4.1.4. Gray-White-CSF Segmentation Mask
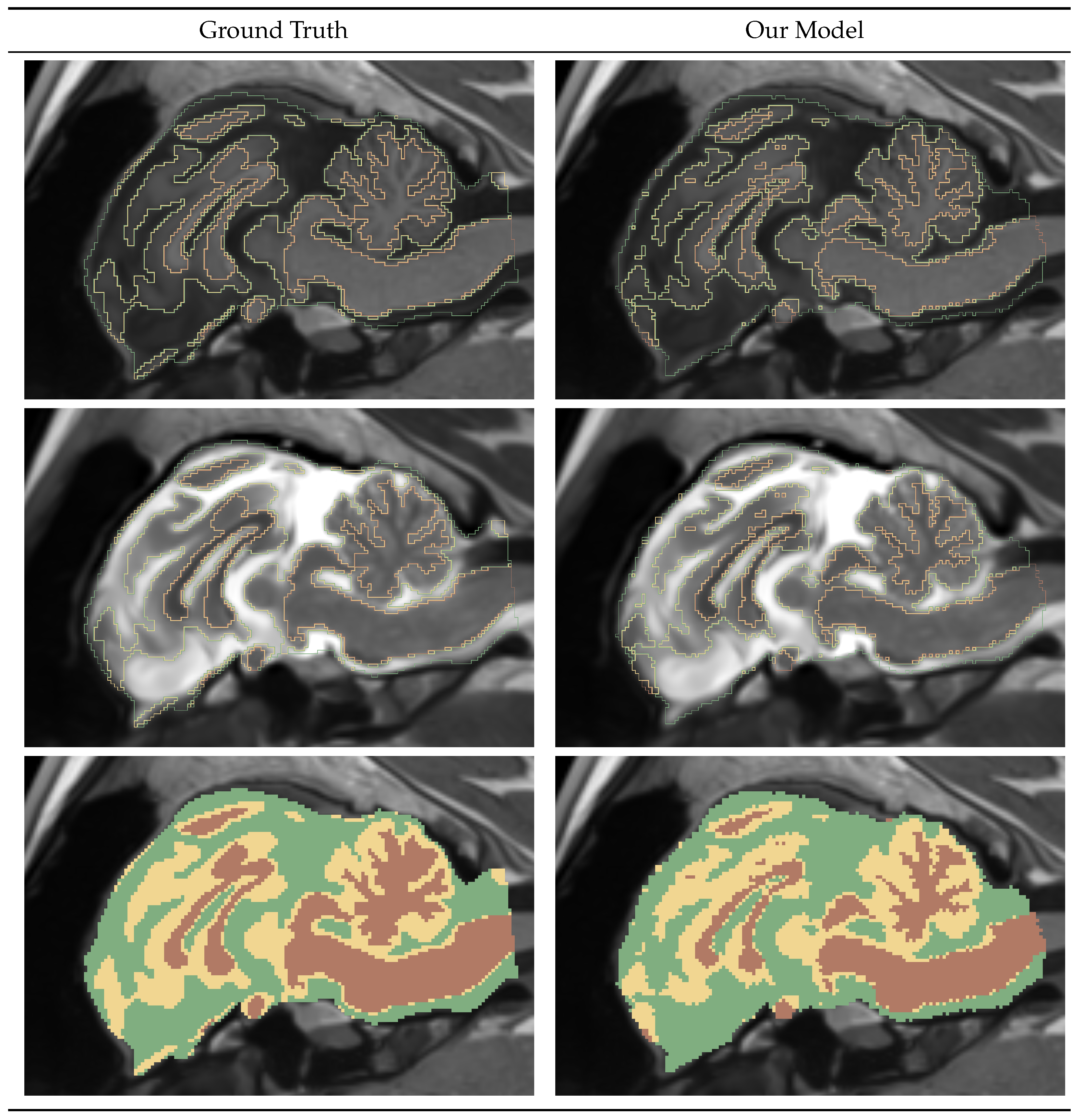
Figure 5.
Gray Matter-White Matter-Cerebrospinal Fluid Segmentation Mask Visualization.
Figure 5.
Gray Matter-White Matter-Cerebrospinal Fluid Segmentation Mask Visualization.
Figure 5 shows a sagittal slice of an example GWC model prediction. The left column shows Ground Truths, and the right column shows the predicted mask. The first two rows show masks outline overlayed on top of the input T1w and T2w images, and the last mask shows full masks. We can see differences in both mask predictions, especially in the cerebellum region.
Table 6 shows the DICE and bAVD results for the GWC model performance on the test dataset. We can see that consistently the CSF performance is lower and that the White Matter has the highest accuracy. We observe a low distance error of bAVD 0.05 mm for all masks.
4.2. Pipeline Performance
4.2.1. Runtime
The pipeline is designed for running single images. For example, on Iowa data with an image size 10Mb, the CPU pipeline executes in under two minutes. GPU usage can speed up this process by around 10-15 seconds. The largest components are the landmark detection process taking around a minute and the registration taking around 30 seconds.
The Germany data images are much bigger, with T2w images registered at over 30Mb and T1w images greater than 200Mb. The large file sizes negatively impact the pipeline performance, resulting in an execution time of 6 minutes. Half of that time is spent on registration and another 1.5-2 minutes on landmark detection.
4.2.2. Memory and Hardware
The overall memory requirement to support full pipeline capabilities is 10GB. The majority of that are the Reinforcement Learning models taking 9GB. The GPUs used for training the models were NVIDIA GTX8000 and NVIDIA GV100, and running the pipeline was executed on Intel Xeon with 56 cores and 72 threads. However, all processes except the registration execute using a single core.
5. Discussion
We present MinipigBrain, a Deep Learning based pipeline for minipig MRI processing. Trained on heterogeneous datasets, the pipeline can perform multiple operations commonly used in human neurodegenerative research for application in minipig models. The pipeline is constructed to be applicable on similarly shaped large animal quadrupeds (i.e. sheep, goat, etc.), but lack of access to those data prevented validation across species.
5.1. Deep Learning Models
Splitting the initial brainmask process into low and high-resolution masks was necessary to extract a region of interest from the original image space. The low-resolution masks proved robust and sufficiently accurate for preparing the data for the subsequent processes.
The Caudate-Putamen segmentation model accuracy is worse than the high-resolution and ICV models, which produce highly precise masks. The caudate and putamen are hard to distinguish, small regions. Even minor errors in segmenting such small regions lead to a higher impact on the DICE metric. Additionally, we predict that the worse quality of T2w images in the Germany dataset harmed the training process, and we are investigating ways to improve the model’s performance.
The evaluation of the tissue classification model is more complicated. The Ground Truth masks used for Gray Matter-White Matter-Cerebrospinal Fluid segmentation were generated using the Atropos tool. That tool is aimed at human data rather than minipigs and may have produced suboptimal results. During visual inspection, we found many mistakes in the ground truth images. Unfortunately, we did not have the available resources to manually fix those labels manually. This negatively impacted the evaluation of our model as the DICE metric only measures the overlap between the masks and not the quality of the masks. However, despite the suboptimal labels, during the training process the model was able to generalize and in many cases, produced arguably better results than the "ground truth" masks.
5.2. Future Work
To improve the performance of segmentation and tissue classification, we plan to experiment with transfer learning. By leveraging a large human dataset and non-linear transformation, we expect to improve the models’ accuracy.
Additionally, we plan to add batch execution capability to the pipeline. Loading a model onto the GPU takes more time than inference time. We have nine different Deep Learning models, and being able to execute images from different subjects in a batch should significantly speed up the execution time.
Author Contributions
Conceptualization, M.B. and H.J.J.; methodology, M.B.; software, M.B.; validation, M.B. and H.J.J.; formal analysis, M.B.; investigation, M.B.; resources, H.J.J. and J.C.S.; data curation, M.B. and K.K.; writing—original draft preparation, M.B.; writing—review and editing, M.B. and H.J.J.; visualization, M.B.; supervision, H.J.J.; project administration, H.J.J. and J.C.S.; funding acquisition, H.J.J. and J.C.S.. All authors have read and agreed to the published version of the manuscript.
Funding
Funding for data collection of the Iowa datasets was provided by Noah’s Hope and Hope 4 Bridget Foundations. Imaging data collection was conducted on an MRI instrument funded by NIH 1S10OD025025-01. Funding for the collection of the german imaging data was provided by the CHDI Foundation and partners who support the work of the George-Huntington Institute and thus provided additional funding for the initial collection of these datasets.
Institutional Review Board Statement
Germany Data: All research and animal care procedures used to collect this data were approved by the Landesamt für Natur und Umweltschutz, Nordrhein-Westfalen, Germany [84-02.04.2011.A160][
12].
Iowa Data: All procedures used to collect this data were approved by the Institutional Animal Care and Use Committees (IACUC) of the University of Iowa and Exemplar Genetics in accordance with regulations[
13].
Data Availability Statement
Not applicable.
Acknowledgments
For their work on the german dataset, we want to acknowledge Dr. Ralf Reilmann, who secured funding and oversaw the methodological development of MR imaging protocols, and Robin Schubert, who worked on data acquisition and processing. In addition, we would like to thank Dr. Jill Weimer - principal investigator of Noah’s Hope, for her help securing funding for the Iowa dataset collection. Finally, we would also like to thank Alexander B. Powers for his help on the Reinforcement Learning implementation for landmark detection and Jerzy Twarowski and Ariel Wooden for their support and figure design critique.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| T1w |
T1-weighted MRI sequence |
| T2w |
T2-weighted MRI sequence |
| CSF |
Cerebrospinal Fluid |
| GWC |
Gray Matter-White Matter-CSF |
| Seg |
Caudate-Putamen Segmentation |
| bAVD |
balanced Average Hausdorff Distance |
References
-
Neurological disorders : public health challenges; World Health Organization: Geneva, 2006.
- Moon, C. New Insights into and Emerging Roles of Animal Models for Neurological Disorders. International Journal of Molecular Sciences 2022, 23. [Google Scholar] [CrossRef]
- Leung, C.; Jia, Z. Mouse Genetic Models of Human Brain Disorders. Frontiers in Genetics 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- McGarry, A.; McDermott, M.; Kieburtz, K.; de Blieck, E.A.; Beal, F.; Marder, K.; Ross, C.; Shoulson, I.; Gilbert, P.; Mallonee, W.M.; et al. A randomized, double-blind, placebo-controlled trial of coenzyme Q10 in Huntington disease. Neurology 2016, 88, 152–159. [Google Scholar] [CrossRef]
- Hersch, S.M.; Schifitto, G.; Oakes, D.; Bredlau, A.L.; Meyers, C.M.; Nahin, R.; and, H.D.R. The CREST-E study of creatine for Huntington disease. Neurology 2017, 89, 594–601. [Google Scholar] [CrossRef]
- Eaton, S.L.; Wishart, T.M. Bridging the gap: large animal models in neurodegenerative research. Mammalian Genome 2017, 28, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Ardan, T.; Baxa, M.; Levinská, B.; Sedláčková, M.; Nguyen, T.D.; Klíma, J.; Juhás, Š.; Juhásová, J.; Šmatlíková, P.; Vochozková, P.; Motlík, J.; Ellederová, Z. Transgenic minipig model of Huntington’s disease exhibiting gradually progressing neurodegeneration. Disease Models & Mechanisms, 2019. [Google Scholar] [CrossRef]
- VODIČKA, P.; SMETANA, K.; DVOŘÁNKOVÁ, B.; EMERICK, T.; XU, Y.Z.; OUREDNIK, J.; OUREDNIK, V.; MOTLÍK, J. The Miniature Pig as an Animal Model in Biomedical Research. Annals of the New York Academy of Sciences 2005, 1049, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Hoopes, A.; Mora, J.S.; Dalca, A.V.; Fischl, B.; Hoffmann, M. SynthStrip: skull-stripping for any brain image. NeuroImage 2022, 260, 119474. [Google Scholar] [CrossRef]
- Pierson, R.; Johnson, H.; Harris, G.; Keefe, H.; Paulsen, J.; Andreasen, N.; Magnotta, V. Fully automated analysis using BRAINS: autoWorkup. NeuroImage 2011, 54, 328–36. [Google Scholar] [CrossRef]
- Brzus, M.; Powers, A.B.; Knoernschild, K.S.; Sieren, J.C.; Johnson, H.J. Multi-agent reinforcement learning pipeline for anatomical landmark detection in minipigs. In Proceedings of the Medical Imaging 2022: Image Processing; Colliot, O.; Išgum, I., Eds. International Society for Optics and Photonics, SPIE, 2022, Vol. 12032, p. 1203229. [CrossRef]
- Schubert, R.; Frank, F.; Nagelmann, N.; Liebsch, L.; Schuldenzucker, V.; Schramke, S.; Wirsig, M.; Johnson, H.; Kim, E.Y.; Ott, S.; Hölzner, E.; Demokritov, S.O.; Motlik, J.; Faber, C.; Reilmann, R. Neuroimaging of a minipig model of Huntington’s disease: Feasibility of volumetric, diffusion-weighted and spectroscopic assessments. Journal of Neuroscience Methods 2016, 265, 46–55, Current Methods in Huntington’s Disease Research. [Google Scholar] [CrossRef]
- Swier, V.J.; White, K.A.; Johnson, T.B.; Sieren, J.C.; Johnson, H.J.; Knoernschild, K.; Wang, X.; Rohret, F.A.; Rogers, C.S.; Pearce, D.A.; Brudvig, J.J.; Weimer, J.M. A Novel Porcine Model of CLN2 Batten Disease that Recapitulates Patient Phenotypes. Neurotherapeutics 2022, 19, 1905–1919. [Google Scholar] [CrossRef]
- Avants, B.B.; Tustison, N.J.; Wu, J.; Cook, P.A.; Gee, J.C. An Open Source Multivariate Framework for n-Tissue Segmentation with Evaluation on Public Data. Neuroinformatics 2011, 9, 381–400. [Google Scholar] [CrossRef] [PubMed]
- Kerfoot, E.; Clough, J.; Oksuz, I.; Lee, J.; King, A.P.; Schnabel, J.A. Left-Ventricle Quantification Using Residual U-Net. Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges; Pop, M., Sermesant, M., Zhao, J., Li, S., McLeod, K., Young, A., Rhode, K., Mansi, T., Eds.; Springer International Publishing: Cham, 2019; pp. 371–380. [Google Scholar]
- Consortium, T.M. Project MONAI, 2020. [CrossRef]
- Paszke, A.; Gross, S.; Massa, F.; Lerer, A.; Bradbury, J.; Chanan, G.; Killeen, T.; Lin, Z.; Gimelshein, N.; Antiga, L.; Desmaison, A.; Kopf, A.; Yang, E.; DeVito, Z.; Raison, M.; Tejani, A.; Chilamkurthy, S.; Steiner, B.; Fang, L.; Bai, J.; Chintala, S. PyTorch: An Imperative Style, High-Performance Deep Learning Library. Advances in Neural Information Processing Systems; Wallach, H.; Larochelle, H.; Beygelzimer, A.; d’Alché-Buc, F.; Fox, E.; Garnett, R., Eds. Curran Associates, Inc., 2019, Vol. 32.
- Falcon, W.; Borovec, J.; Wälchli, A.; Eggert, N.; Schock, J.; Jordan, J.; Skafte, N.; Ir1dXD.; Bereznyuk, V.; Harris, E.; et al. PyTorchLightning/pytorch-lightning: 0.7.6 release, 2020. [CrossRef]
- Johnson, H.J.; McCormick, M.M.; Ibanez, L. Template:The ITK Software Guide Book 1: Introduction and Development Guidelines-Volume 1, 2015.
- Chollet, M.B.; Aldridge, K.; Pangborn, N.; Weinberg, S.M.; DeLeon, V.B. Landmarking the Brain for Geometric Morphometric Analysis: An Error Study. PLoS ONE 2014, 9, e86005. [Google Scholar] [CrossRef] [PubMed]
- da Silva, Jr, E. B.; Leal, A.G.; Milano, J.B.; da Silva, Jr, L.F.M.; Clemente, R.S.; Ramina, R. Image-guided surgical planning using anatomical landmarks in the retrosigmoid approach. Acta Neurochir. (Wien) 2010, 152, 905–910. [Google Scholar] [CrossRef]
- Visser, M.; Petr, J.; Müller, D.M.J.; Eijgelaar, R.S.; Hendriks, E.J.; Witte, M.; Barkhof, F.; van Herk, M.; Mutsaerts, H.J.M.M.; Vrenken, H.; de Munck, J.C.; De Witt Hamer, P.C. Accurate MR Image Registration to Anatomical Reference Space for Diffuse Glioma. Frontiers in Neuroscience 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- François-Lavet, V.; Henderson, P.; Islam, R.; Bellemare, M.G.; Pineau, J. An Introduction to Deep Reinforcement Learning. CoRR 2018, abs/1811.12560, [1811.12560].
- Ruder, S. An Overview of Multi-Task Learning in Deep Neural Networks. ArXiv 2017, abs/1706.05098.
- Caruana, R. Multitask Learning. Machine Learning 1997, 28, 41–75. [Google Scholar] [CrossRef]
- Watkins, C.J.C.H.; Dayan, P. Q-learning. Machine Learning 1992, 8, 279–292. [Google Scholar] [CrossRef]
- Huber, P.J. Robust Estimation of a Location Parameter. The Annals of Mathematical Statistics 1964, 35, 73–101. [Google Scholar] [CrossRef]
- Kingma, D.P.; Ba, J. Adam: A Method for Stochastic Optimization. 3rd International Conference on Learning Representations, ICLR 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings; Bengio, Y.; LeCun, Y., Eds., 2015.
- Thrun, S. Efficient Exploration In Reinforcement Learning. Technical Report CMU-CS-92-102, Carnegie Mellon University, Pittsburgh, PA, 1992.
- BRAINSTools. https://github.com/BRAINSia/BRAINSTools.
- Dice, L.R. Measures of the Amount of Ecologic Association Between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Aydin, O.U.; Taha, A.A.; Hilbert, A.; Khalil, A.A.; Galinovic, I.; Fiebach, J.B.; Frey, D.; Madai, V.I. On the usage of average Hausdorff distance for segmentation performance assessment: hidden error when used for ranking. European Radiology Experimental 2021, 5. [Google Scholar] [CrossRef] [PubMed]
- Taha, A.A.; Hanbury, A. Metrics for Evaluating 3D Medical Image Segmentation: analysis, selection, and tool. BMC Medical Imaging 2015, 15, 29. [Google Scholar] [CrossRef]
- Detlefsen, N.S.; Borovec, J.; Schock, J.; Jha, A.H.; Koker, T.; Liello, L.D.; Stancl, D.; Quan, C.; Grechkin, M.; Falcon, W. TorchMetrics - Measuring Reproducibility in PyTorch. Journal of Open Source Software 2022, 7, 4101. [Google Scholar] [CrossRef]
Table 1.
Training dataset Low-Resolution and High-Resolution Brainmask models.
Table 1.
Training dataset Low-Resolution and High-Resolution Brainmask models.
| |
Germany Dataset |
Iowa Dataset |
Total |
| Number of |
Images |
Subjects |
Images |
Subjects |
Images |
Subjects |
| Training |
164 |
26 |
243 |
18 |
407 |
44 |
| Validation |
30 |
4 |
39 |
3 |
69 |
7 |
| Test |
18 |
3 |
30 |
2 |
48 |
5 |
Table 2.
Training dataset for Segmentation models.
Table 2.
Training dataset for Segmentation models.
| |
Germany Dataset |
Iowa Dataset |
Total |
| Number of |
Image Pairs |
Subjects |
Image Pairs |
Subjects |
Image Pairs |
Subjects |
| Training |
78 |
26 |
113 |
18 |
191 |
44 |
| Validation |
15 |
4 |
19 |
3 |
34 |
7 |
| Test |
8 |
3 |
14 |
2 |
22 |
5 |
Table 3.
Accuracy Evaluation for Low-Resolution and High-Resolution Brainmask models.
Table 3.
Accuracy Evaluation for Low-Resolution and High-Resolution Brainmask models.
| |
Low-Resolution |
High-Resolution |
| Dataset |
DICE |
bAVD |
DICE |
bAVD |
| Iowa |
0.89 |
0.21 |
0.97 |
0.03 |
| Germany |
0.88 |
0.32 |
0.97 |
0.06 |
| Total |
0.88 |
0.25 |
0.97 |
0.04 |
Table 4.
DICE and balanced average Hausdorff distance results for Intracranial Volume Model.
Table 4.
DICE and balanced average Hausdorff distance results for Intracranial Volume Model.
| Dataset |
DICE |
bAVD |
| Iowa |
0.97 |
0.03 |
| Germany |
0.98 |
0.03 |
| Total |
0.98 |
0.03 |
Table 5.
DICE and balanced average Hausdorff distance results for Caudate-Putamen Segmentation Model.
Table 5.
DICE and balanced average Hausdorff distance results for Caudate-Putamen Segmentation Model.
| Dataset |
Left Caudate |
Right Caudate |
Left Putamen |
Right Putamen |
Global bAVD |
| Germany |
0.82 |
0.79 |
0.79 |
0.81 |
0.29 |
| Iowa |
0.80 |
0.83 |
0.78 |
0.80 |
0.27 |
| Total |
0.81 |
0.82 |
0.78 |
0.80 |
0.28 |
Table 6.
Gray Matter-White Matter-Cerebrospinal Fluid Segmentation Results.
Table 6.
Gray Matter-White Matter-Cerebrospinal Fluid Segmentation Results.
| Dataset |
Gray Matter |
White Matter |
CSF |
Global bAVD |
| Germany |
0.79 |
0.89 |
0.71 |
0.06 |
| Iowa |
0.81 |
0.89 |
0.78 |
0.05 |
| Total |
0.80 |
0.89 |
0.76 |
0.05 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).