Submitted:
15 January 2023
Posted:
19 January 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
- A state of art attention-based tab transformer model is presented to predict patients LoS using multiple modalities such as clinical features, patients’ demographics data, and X-ray reports.
- We present a framework where the result of machine learning based methods for LoS prediction can be analyzed with association mining rules and identify the cohort of risk factors affecting the LoS in hospitals.
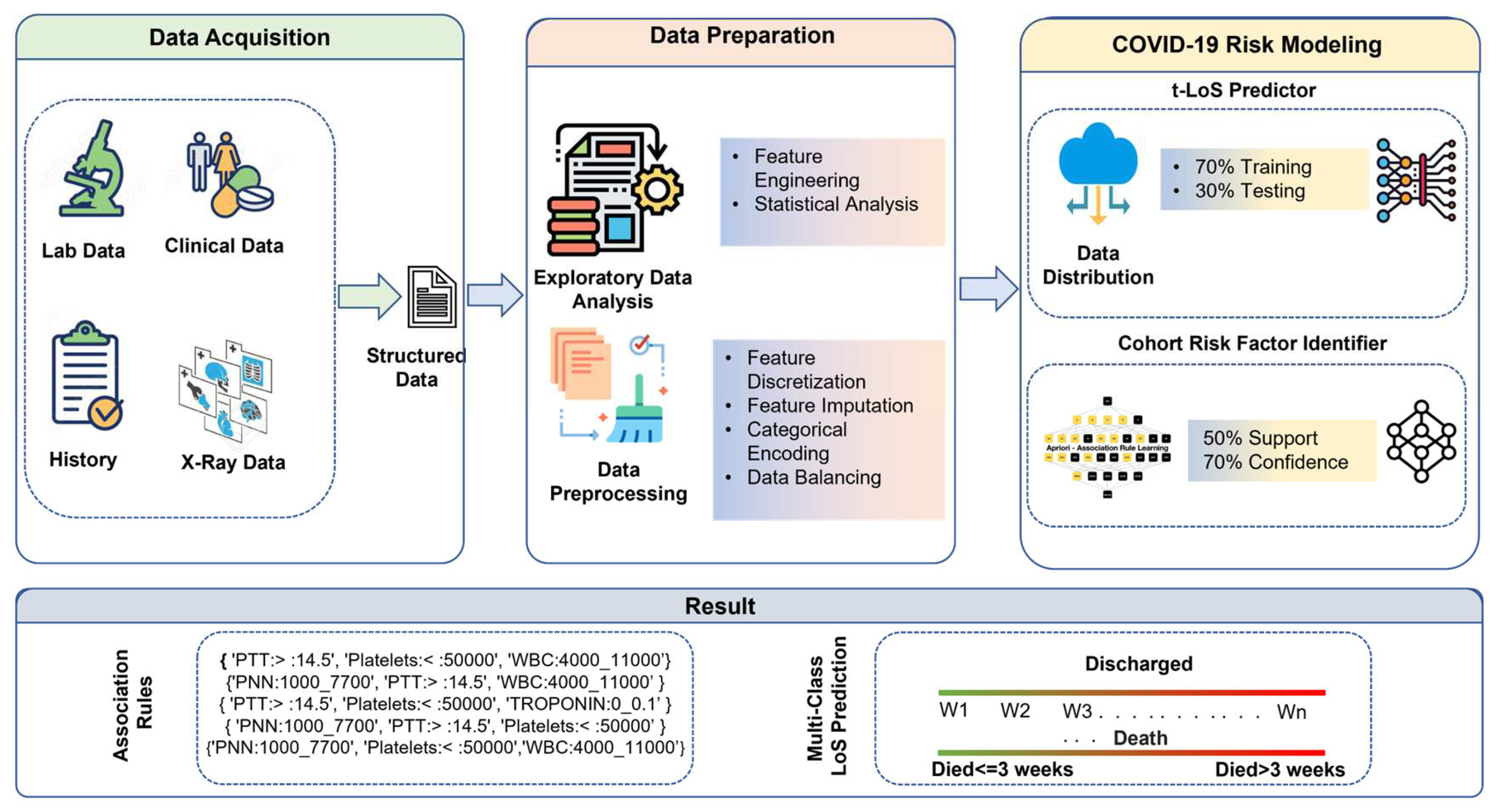
2. Materials and Methods
2.1 Data Acquisition
2.2. Data Preparation
2.2.1. Natural Binning:
2.2.2. Encoding of Categorical Features
2.2.3. LoS Category Creation
2.2.4. Data Balance with Respect to LoS
2.3 COVID-19 Risk Modeling
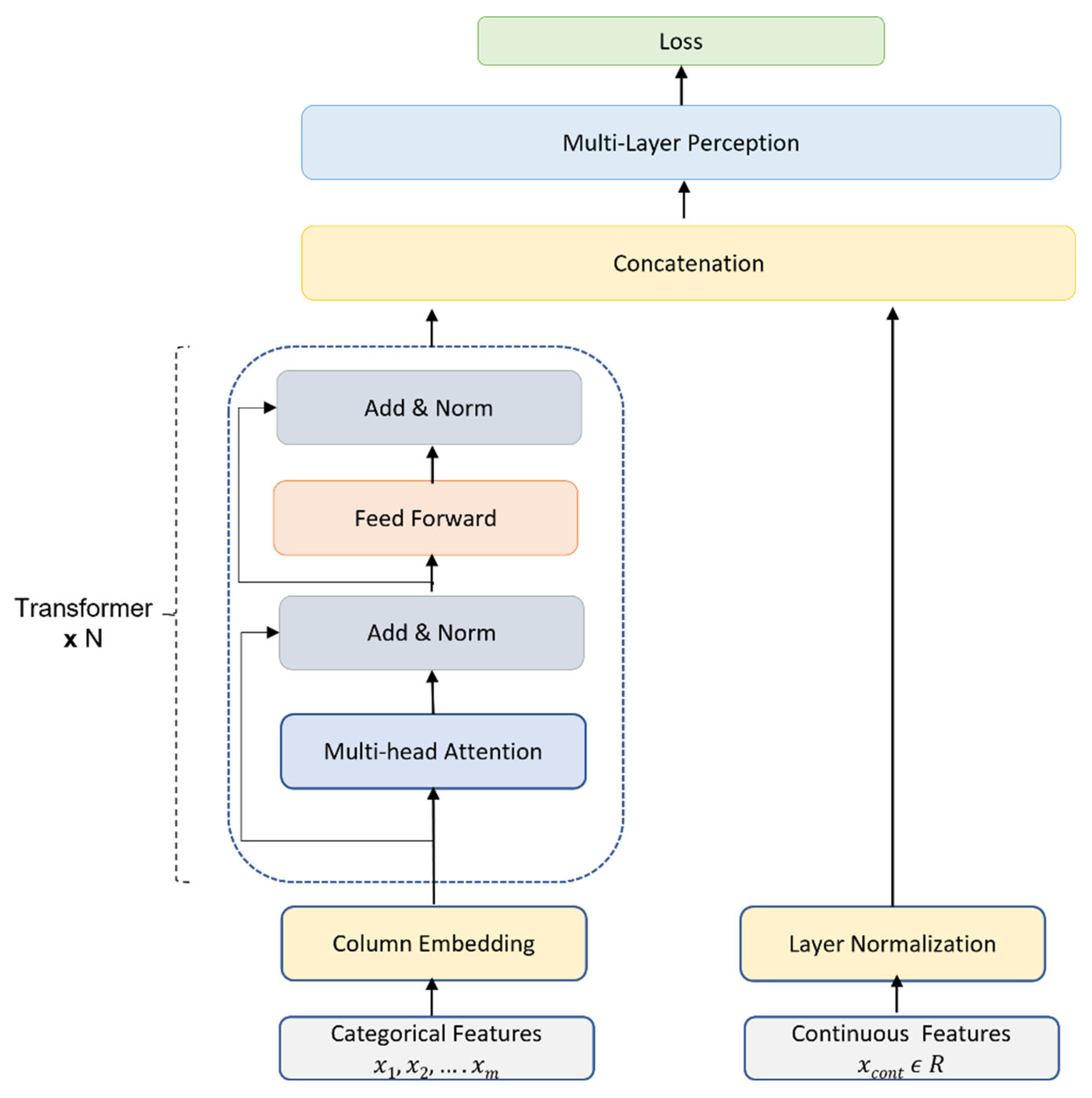
2.3.1 t-LoS Predictor
2.3.2. Cohort Risk Factors Identifier
3. Results and Evaluation
3.1. COVID-19 Risk Model Results
3.2. CRFI Results
3.2.1. CRFI for Discharged Patients’ Category
3.2.2. CRFI for Deceased Patients’ Category
4. Discussion
- Age appears to be a strong risk factor for COVID-19 severity and its outcomes. Statsenko et al. [20] performed detailed analysis and concluded that elderly patients with COVID-19 are more likely to progress to severe disease. The result of CRFI within deceased category consists of rules with age >=56 Years and <=73 Years while other age category rules are not frequently observed and found to be insignificant. In addition, the mining results on patients who stayed in hospital between three and four weeks contains 25% of the rules with age>=73. These observations validate the fact the age is correlated with COVD-19 severity and a significant factor in deciding LoS.
- The detailed analysis on CRFI rules on the patients who stayed in hospital between 3 and 4 weeks showed that 43 % of the rules constitute either hypertension or diabetes, thus these comorbidities not only increase the LoS in hospitals but also leads to severe COVID leading to increased LoS in the hospital. Same was concluded by Adab et al., 2022 [21].
- The elevated level of D-dimers is an indicator and major risk factor for thrombosis (blood clotting) and increases the risk of medication and monitoring for a longer time [22]. We observed that for the people who discharged between 3 and 4 weeks CRFI results with D-dimers shows that 18 % of the rules have D- dimers value more than 500 ng/mL FEU, thus increasing their LoS. In addition, mining results on patients who stayed more than 3 weeks and died elevated D-Dimer values is present in 41% of the rules. This is also validated by the fact that the people who discharge within two weeks, CRFI results show only 4.5 % of the rules has D-Dimer value more than 500 ng/mL FEU and elevated D-Dimers values are not found to be significant on the CFRI results of patients who stayed less than one week.
- LDH is another factor that has an elevated level of more than 225 units/L in 23% of the rules based on CRFI results of patients discharged from hospital between 3 and 4 weeks.
- Wagner et al. [23] concluded that lymphocytes count is one of the prognostic factors in determining COVID-19 illness, and our CRFI results for patients who died after spending more than 3 weeks in hospitals found that all the rules with lymphocytes consist of values are between 500 and 1000 while its values are between 1000 and 4000, 86 % of the time for patients who discharged within two weeks. This again validate the fact the lower lymphocytes count is critical in determining COVID severity and LoS.
- In the start of Covid-19, medical community tried many treatments without much evidence. It is important to understand what medications based on lessons learned could be useful to treat infections caused by new strains of viruses as viable epidemic response strategies. Our study shows that drug such as Hydroxychloroquine, Favipiravir, reduces the patients LoS. CRFI results on patients who stayed less than a week in hospital show 51% of the rules consist of antibiotic medications, while who discharged in less than 2 weeks, 52 % of the rules consist of antiviral medication. These analysis shows that the usage of antiviral and antibiotic medication has reduced patient’s LoS.
5. Limitation and Future Directions
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- W.H., et al. (2021). Second round of the national pulse survey on continuity of essential health services during the COVID-19 pandemic: January-March 2021: interim report, 22 April 2021 (No. WHO/2019-nCoV/EHS_continuity/survey/2021.1). World Health Organization.
- Mathieu, E. (2022, December 28). Coronavirus (COVID-19) Hospitalizations. Our World in Data. Retrieved December 28, 2022, from https://ourworldindata.org/covid-hospitalizations.
- Bravata, D. M., Perkins, A. J., Myers, L. J., Arling, G., Zhang, Y., Zillich, A. J., ... & Keyhani, S. Association of intensive care unit patient load and demand with mortality rates in US Department of Veterans Affairs hospitals during the COVID-19 pandemic. JAMA network open 2021, 4(1), e2034266-e2034266. [CrossRef]
- Churpek, M. M., Wendlandt, B., Zadravecz, F. J., Adhikari, R., Winslow, C., & Edelson, D. P. Association between intensive care unit transfer delay and hospital mortality: a multicenter investigation. Journal of hospital medicine 2016, 11(11), 757-762. [CrossRef]
- Resar, R., Nolan, K., Kaczynski, D., & Jensen, K. Using real-time demand capacity management to improve hospitalwide patient flow. The Joint Commission Journal on Quality and Patient Safety 2011, 37(5), 217-AP3. [CrossRef]
- Weiss, A. J., & Elixhauser, A. (2014). Overview of hospital stays in the United States, 2012: statistical brief# 180.
- Luo, L., Lian, S., Feng, C., Huang, D., & Zhang, W. (2017, March). Data mining-based detection of rapid growth in length of stay on COPD patients. In 2017 IEEE 2nd International Conference on Big Data Analysis (ICBDA) (pp. 254-258). IEEE.
- Dogu, E., Albayrak, Y. E., & Tuncay, E. Length of hospital stay prediction with an integrated approach of statistical-based fuzzy cognitive maps and artificial neural networks. Medical & Biological Engineering & Computing 2021, 59(3), 483-496. [CrossRef]
- Kulkarni, H., Thangam, M., & Amin, A. P. Artificial neural network-based prediction of prolonged length of stay and need for post-acute care in acute coronary syndrome patients undergoing percutaneous coronary intervention. European Journal of Clinical Investigation 2021, 51(3), e13406. [CrossRef]
- Dan, T., Li, Y., Zhu, Z., Chen, X., Quan, W., Hu, Y., ... & Cai, H. (2020, December). Machine learning to predict ICU admission, ICU mortality and survivors’ length of stay among COVID-19 patients: toward optimal allocation of ICU resources. In 2020 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (pp. 555-561). IEEE.
- Vekaria, B., Overton, C., Wiśniowski, A., Ahmad, S., Aparicio-Castro, A., Curran-Sebastian, J., ... & Elliot, M. J. Hospital length of stay for COVID-19 patients: Data-driven methods for forward planning. BMC Infectious Diseases 2021, 21(1), 1-15. [CrossRef]
- Zebin, T., & Chaussalet, T. J. (2019, July). Design and implementation of a deep recurrent model for prediction of readmission in urgent care using electronic health records. In 2019 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB) (pp. 1-5). IEEE.
- Johnson, A. E., Pollard, T. J., Shen, L., Lehman, L. W. H., Feng, M., Ghassemi, M., ... & Mark, R. G. MIMIC-III, a freely accessible critical care database. Scientific data 2016, 3(1), 1–9. [CrossRef]
- Harerimana, G., Kim, J. W., & Jang, B. A deep attention model to forecast the Length of Stay and the in-hospital mortality right on admission from ICD codes and demographic data. Journal of Biomedical Informatics 2021, 118, 103778. [CrossRef]
- Rajkomar, A., Oren, E., Chen, K., Dai, A. M., Hajaj, N., Hardt, M., ... & Dean, J. Scalable and accurate deep learning with electronic health records. NPJ digital medicine 2018, 1(1), 1–10. [CrossRef]
- Chawla, N. V., Bowyer, K. W., Hall, L. O., & Kegelmeyer, W. P. SMOTE: synthetic minority over-sampling technique. Journal of artificial intelligence research 2002, 16, 321–357. [CrossRef]
- Huang, X., Khetan, A., Cvitkovic, M., & Karnin, Z. (2020). Tabtransformer: Tabular data modeling using contextual embeddings. arXiv preprint arXiv:2012.06678.
- Borgelt, C., & Kruse, R. (2002). Induction of association rules: Apriori implementation. In Compstat (pp. 395-400). Physica, Heidelberg.
- GitHub – Covid19_research. (n.d.). Retrieved December 28, 2022, from https://github.com/smileslab/Covid19_research/tree/main/Association_Mining.
- Statsenko, Y., Al Zahmi, F., Habuza, T., Almansoori, T. M., Smetanina, D., Simiyu, G. L., ... & Al Koteesh, J. Impact of Age and Sex on COVID-19 Severity Assessed From Radiologic and Clinical Findings. Frontiers in cellular and infection microbiology 2022, 1395. [CrossRef]
- Adab, P., Haroon, S., O’Hara, M. E., & Jordan, R. E. Comorbidities and covid-19. bmj 2022, 377.
- Lehmann, A., Prosch, H., Zehetmayer, S., Gysan, M. R., Bernitzky, D., Vonbank, K., ... & Gompelmann, D. Impact of persistent D-dimer elevation following recovery from COVID-19. PLoS One 2021, 16(10), e0258351. [CrossRef]
- Wagner, J., DuPont, A., Larson, S., Cash, B., & Farooq, A. Absolute lymphocyte count is a prognostic marker in Covid-19: a retrospective cohort review. International Journal of Laboratory Hematology 2020, 42(6), 761-765. [CrossRef]
| Dataset Source | Description | Features Frequency |
|---|---|---|
| General | Contain general information such as demographic data (gender, age, and ethnicity), epidemiological data (date of admission, date of death) and comorbidities such as hypertension, diabetes, COPD etc. | 68 |
| Lab Data | Contain elements related to blood test such as WBC count, PNN, Lymphocyte’s count, Hemoglobin, Platelets, Creatinine, ALT LDH, FERRITIN, D-DIMER, CRP, PROCALCITONIN, TROPONIN, Pro-BNP, PTT, Vitamin D and IL6 |
17 |
| X-Ray Data | Contain elements related to X-Ray such as presence of consolidation, presence of ground glass, and opacities bilateral or unilateral | 4 |
| Patient Characteristics | Details: % Patients | ||
|---|---|---|---|
| General | Demographic | Gender | Female: 49.7 %; Male: 50.3 % |
| Age: Mean Median IQR |
58.8 Years 60 Years 26.7 Years |
||
| Egypt: 2% | |||
| Filipino: 1.3% | |||
| Iraq: .32 % | |||
| Nationality | Saudi Arabia: 95.7 % | ||
| Sudan: .36 % | |||
| United Kingdom: .32 % | |||
| Comorbidities | Diabetes | 69.2% | |
| Hypertension | 64.3% | ||
| Heart Ischemic | 17.2% | ||
| Heart Failure | 5.0% | ||
| Cardiomyopathies | 1.3% | ||
| COPD | 2.0% | ||
| Heart Failure | 4.9% | ||
| Lung Interstitial Disease | 0.3% | ||
| Bronchial Asthma | 15.0% | ||
| Cerebrovascular | 4.2% | ||
| Neurologic (Dementia) | 4.2% | ||
| Cirrhosis | 1.3% | ||
| HIV | 0.0% | ||
| Liver Disease | 2.0% | ||
| Shortness of Breath | 85.7% | ||
| Others | Psychiatric History | 1.3% | |
| End Stage Renal | 11.0% | ||
| Hemodialysis | 4.5% | ||
| Cancer | 6.0% | ||
| Solid Organ Transplant | 5.5% | ||
| Hematopoietic Cell Transplant | 0.0% | ||
| Smoker | 0.3% | ||
| Pregnancy | 5.0% | ||
| Sick Cell | 0.3% | ||
| Obesity | 5.5% | ||
| Fever | 55.0% | ||
| Hemoptysis | 1.0% | ||
| Diarrhea | 11.0% | ||
| Cough | 72.0% | ||
| Headache | 7.5% | ||
| Abdominal Pain | 8.0% | ||
| Myalgia | 11.0% | ||
| Loss of Smell or Taste | 8.0% | ||
| Temperature | 100.0% | ||
| Respiratory Rate | 13.6% | ||
| Pulse | 100.0% | ||
| Nausea or vomiting | 8.0% | ||
| Diastolic BP | 100.0% | ||
| Systolic BP | 100.0% | ||
| Chest pain | 4.0% | ||
| Lab Parameters | LDH | 100.0% | |
| Glassgow | 100.0% | ||
| PaCO2 | |||
| HCO3 | |||
| PaO2 | |||
| Ph | |||
| Lymphocytes | |||
| PaO2 | |||
| WBC | |||
| ALT | |||
| PTT | |||
| D-Dimer | |||
| Platelets | |||
| WBC | |||
| Hemoglobin | |||
| CRP | |||
| Ferritin | |||
| AST | |||
| Pro BNP | |||
| PROCALCITONI | |||
| TROPONIN | |||
| Vitamin D | |||
| IL6 | |||
| Blood Group | |||
| INR | |||
| Fibrinogen | |||
| PNN | |||
| Antiviral | |||
| Antibiotic | |||
| Anticoagulant | |||
| Medications | Immunomodulators | 80.0% | |
| Presence of Consolidation | 98.0% | ||
| Presence of Ground Glass Opacities | 92.0% | ||
| Bilateral or Unilateral | 87.0% | ||
| X-Ray | 72.0% | ||
| Patients’ Feature | Optimized Binning Interval |
|---|---|
| Age | <=37 Years |
| >=38 Years and <=55 Years | |
| >=56 Years and <=73 Years | |
| >=74 Years | |
| pH | {<= 7.35; 7.35 - 7.45; >7.45} |
| PaO2 | {<=80 mm Hg; >80 mm Hg} |
| PaCO2 | {<= 35 mm Hg; 35 mmHg- 45mm Hg; > 45 mm Hg} |
| HCO3 | {<=21 mEq/L); 21 mEq/L) - 27 mEq/L)} |
| Temperature | {<= 36 °C; 37.6 °C - 38.6 °C; > 38.6 °C} |
| Respiratory Rate | {<=12 bpm; 12 bpm - 20 bpm; 20 bpm - 28 bpm} |
| Pulse | {<= 79 bpm; 79 bpm - 95 bpm; 95 bpm -111 bpm; 111 bpm - 134 bpm; 134 beats per minute - 185 bpm} |
| Systolic Blood Pressure | {<= 90 mmHg; 90 mmHg - 130 mmHg} |
| Diastolic Blood Pressure | {<= 60 mmHg; 60 mmHg - 90 mmHg} |
| Glasgow | {<4; 4-8; 8-12; 12-14; > 14;} |
| WBC | {<=4000 /µL; 4000 /µL -11000 /µL; > 4000 /µL} |
| PNN | {<=500 mm3; 500 mm3 - 1000 mm3; 1000 mm3 - 7700 mm3;7700 mm3 - 15000 mm3} |
| Lymphocytes | {<=500 cells/µL; 500 cells/µL -1000 cells/µL; 1000 cells/µL -4000 cells/µL; >4000 cells/µL} |
| Hemoglobin | {<= 8 g/dl; 8 g/dl - 10 g/dl; 10 g/dl - 12 g/dl} |
| Platelets | {<=50000 /µL; 50000 /µL -150000/ µL; 150000 / µL - 450000 / µL} |
| Creatinine | {<= 59 mg/dL; 59 mg/dL - 104 mg/dL; 104 mg/dL - 250 mg/dL; 250 mg/dL - 500 mg/dL} |
| ALT | 1 U/L -41 U/L;>41 U/L |
| LDH | {<= 135 IU/L; 135 IU/L - 225 IU/L} |
| FERRITIN | {<= 792; 792 -1976; 1976 - 4374; 4374 - 7627;7627 - 159000} |
| D_DIMER | {0 ng/mL-500ng/mL; >500ng/mL} |
| CRP | {<= 6 mg/L; 6 mg/L - 100 mg/L; >100 mg/L} |
| PROCALCITONIN | {<=0.25 ng/mL; 0.25 ng/mL - 0.5 ng/mL; > 0.5 ng/mL} |
| TROPONIN | {<=0.1 ng/mL;>0.1 ng/mL} |
| ProBNP | {<=12 pg/mL; 12 pg/mL -5 pg/mL; 5 pg/mL - 450 pg/mL} |
| PTT | {<= 11.5; 11.5 -14.5} |
| Vitamin D | {<=50 nmol/L; 50 nmol/L - 250 nmol/L} |
| IL6 | {<=37.5 pg/ml; >37.5 pg/ml} |
| Classes | LoS in Hospital | Patient Frequency Original |
Patients Frequency After SMOTE -N |
|---|---|---|---|
| Deceased | Less than or equal 3 weeks | 36 | 36 |
| Greater than 3 weeks | 24 | 36 | |
| Discharge | Less than or equal to 1 week | 84 | 84 |
| Greater than 1 week and less than 2 weeks | 79 | 84 | |
| Greater than 2 weeks and less than 3 weeks | 37 | 84 | |
| Greater than 3 weeks and less than 4 weeks | 12 | 84 | |
| Greater than 4 weeks | 36 | 84 |
| Classifiers | Discharge Dataset | Deceased Dataset | ||||||
|---|---|---|---|---|---|---|---|---|
| F1 | Accuracy | Precision | Recall | F1 | Accuracy | Precision | Recall | |
| LR | 0.74 | 0.73 | 0.77 | 0.74 | 0.68 | 0.68 | 0.7 | 0.73 |
| RF | 0.73 | 0.71 | 0.76 | 0.72 | 0.68 | 0.68 | 0.7 | 0.73 |
| DT | 0.65 | 0.65 | 0.68 | 0.65 | 0.62 | 0.64 | 0.64 | 0.66 |
| AB | 0.62 | 0.61 | 0.63 | 0.62 | 0.61 | 0.64 | 0.61 | 0.62 |
| GB | 0.54 | 0.52 | 0.61 | 0.53 | 0.50 | 0.5 | 0.6 | 0.6 |
| TabT* | 0.92 | 0.73 | 0.83 | 0.93 | 0.84 | 0.77 | 0.75 | .98 |
| Dataset Type | LoS Category |
Association Rules |
|---|---|---|
| Discharged Dataset | LoS ≤ 1 Week | {Anticoagulant, Cough, Antibiotics, Antiviral} |
| {Cough, LDH> 225, Antibiotics, Antiviral} | ||
| {Anticoagulant, SOB, Immunomodulators, LDH> 225, Antibiotics, Platelets< 50000} | ||
| {PaO2 (0 to 80), Anticoagulant, SOB, LDH>225, Antibiotics} | ||
|
LoS >1 Week AND ≤ 2 Weeks |
{Fever, DIMER (0 to 500), Immunomodulators, Antibiotics, Temperature (36 to 37.6)} | |
| {PaO2(0 to 80), CSA_Fever:0, Immunomodulators, LDH> 225, Antibiotics, Antiviral} | ||
| {Anticoagulant, Fever, FERRITIN< 792, Immunomodulators, Glasgow> 14, Platelets<50000} | ||
| {Anticoagulant, Fever, SOB, HTN, Glasgow> 14, Antiviral} | ||
|
LoS > 2 Weeks AND LoS ≤ 3 Weeks |
{Fever, DIMER> 500, LDH> 225, Antiviral} | |
| {Anticoagulant, Fever, HTN, Diastolic BP (60 to 90), Antiviral} | ||
| {Anticoagulant, Fever, HTN, Immunomodulators, Diastolic BP (60 to 90)} | ||
| {CRP (6 to 100), Fever, LDH> 225, Antiviral} | ||
|
LoS > 3 Weeks AND LoS ≤ 4 Weeks |
{Anticoagulant, Lymphocytes (1000 to 4000), Antibiotics, Respiratory Rate (20 to 28), PNN (1000 to 7700)} | |
| {Anticoagulant, HTN, Immunomodulators, Lymphocytes (1000 to 4000), Antibiotics, Respiratory Rate (20 to 28), Antiviral} | ||
| {HTN, Immunomodulators, Lymphocytes (1000 to 4000), Antibiotics, Respiratory Rate (20 to 28), Antiviral} | ||
| {Anticoagulant, Immunomodulators, Lymphocytes (1000 to4000), PNN (1000 to 7700)} | ||
| LoS ≥ 4 Weeks | {Immunomodulators, Platelets< 50000, Antiviral, abnormal X-Ray} | |
| {Antibiotics, PTT> 14.5, Platelets< 50000} | ||
| {Anticoagulant, Immunomodulators, Antiviral} | ||
| {PNN (1000 to 7700), PTT> 14.5, Platelets< 50000, Antiviral} | ||
| Deceased dataset | LoS <=3 Weeks | {SOB, Antibiotics, PTT> 14.5, Platelets<50000, TROPONIN (0 to 0.1)} |
| {SOB, Antibiotics, Glasgow> 14, PNN:1000_7700, Antiviral} | ||
| {LDH> 225, Diastolic BP (60 to 90), Glasgow:> :14, Antiviral} | ||
| {PaO2(0 to 80), Cough, Antibiotics, Glasgow> 14, Platelets< 50000} | ||
| LoS >3 Weeks | {ALT (0 to 41), Diabetes, HTN, Immunomodulators, Antibiotics} | |
| {Ph (7.35 to 7.45), ALT (0 to 41), HTN, Immunomodulators, Antibiotics, Platelets < 50000} | ||
| {ALT (0 to 41), SOB, HTN, Immunomodulators, Antibiotics, PTT> 14.5} | ||
| {ALT (0 to 41), SOB, HTN, Immunomodulators, Glasgow> 14, Antiviral} |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





