Submitted:
02 February 2023
Posted:
03 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
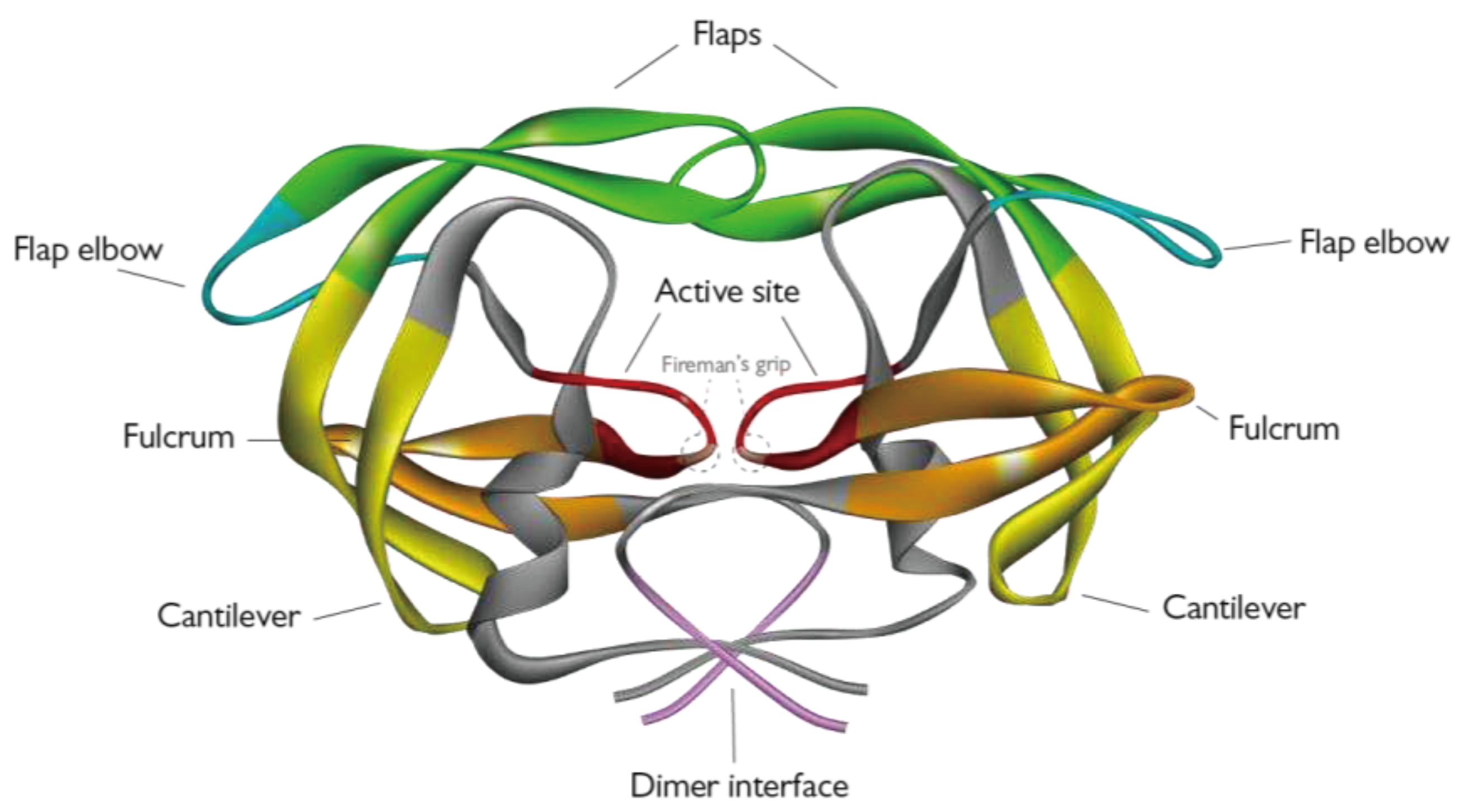
1.1. HIV-1 PR structure and function
1.2. Sequence specificity
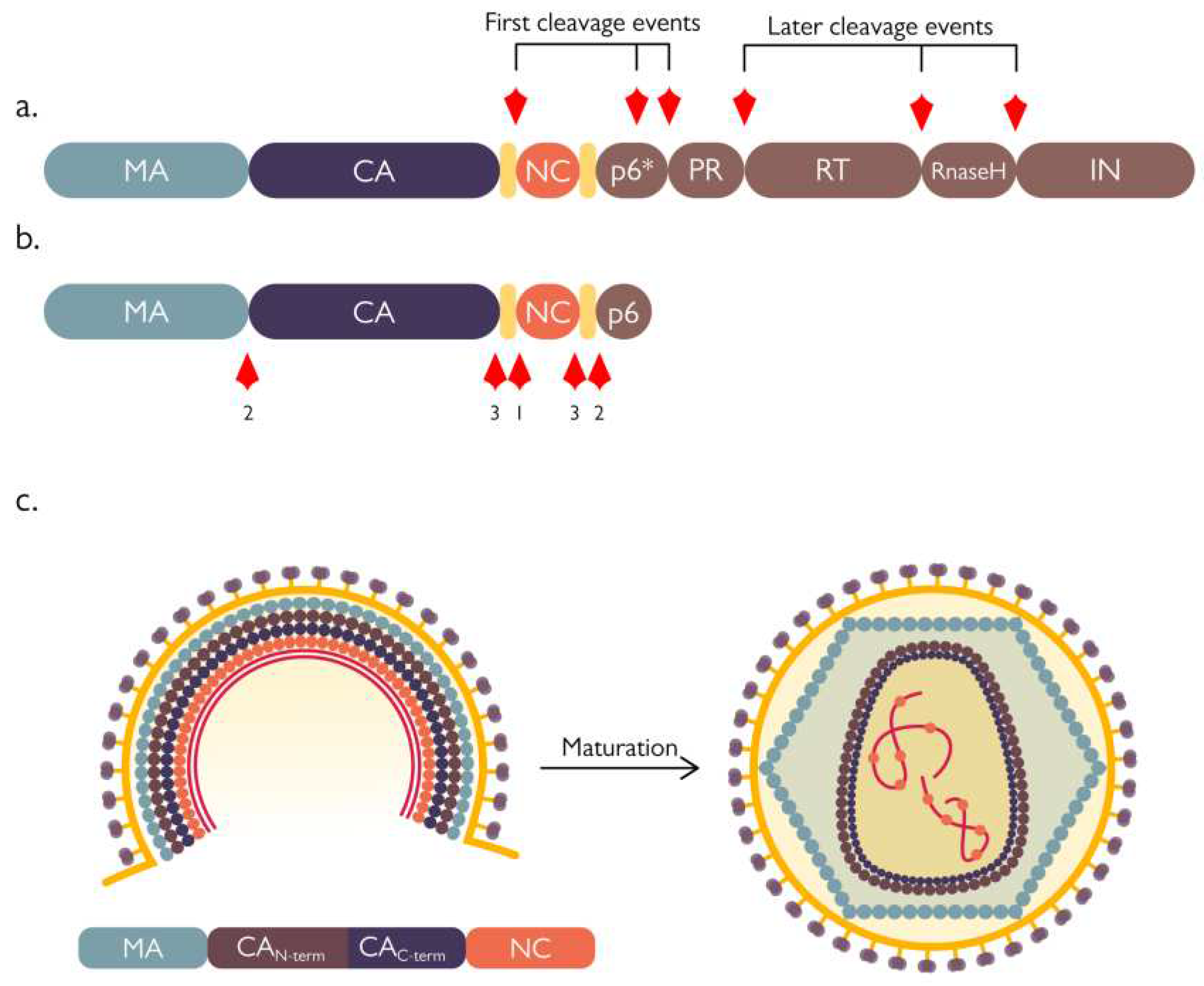
1.3. PR activation and virion maturation
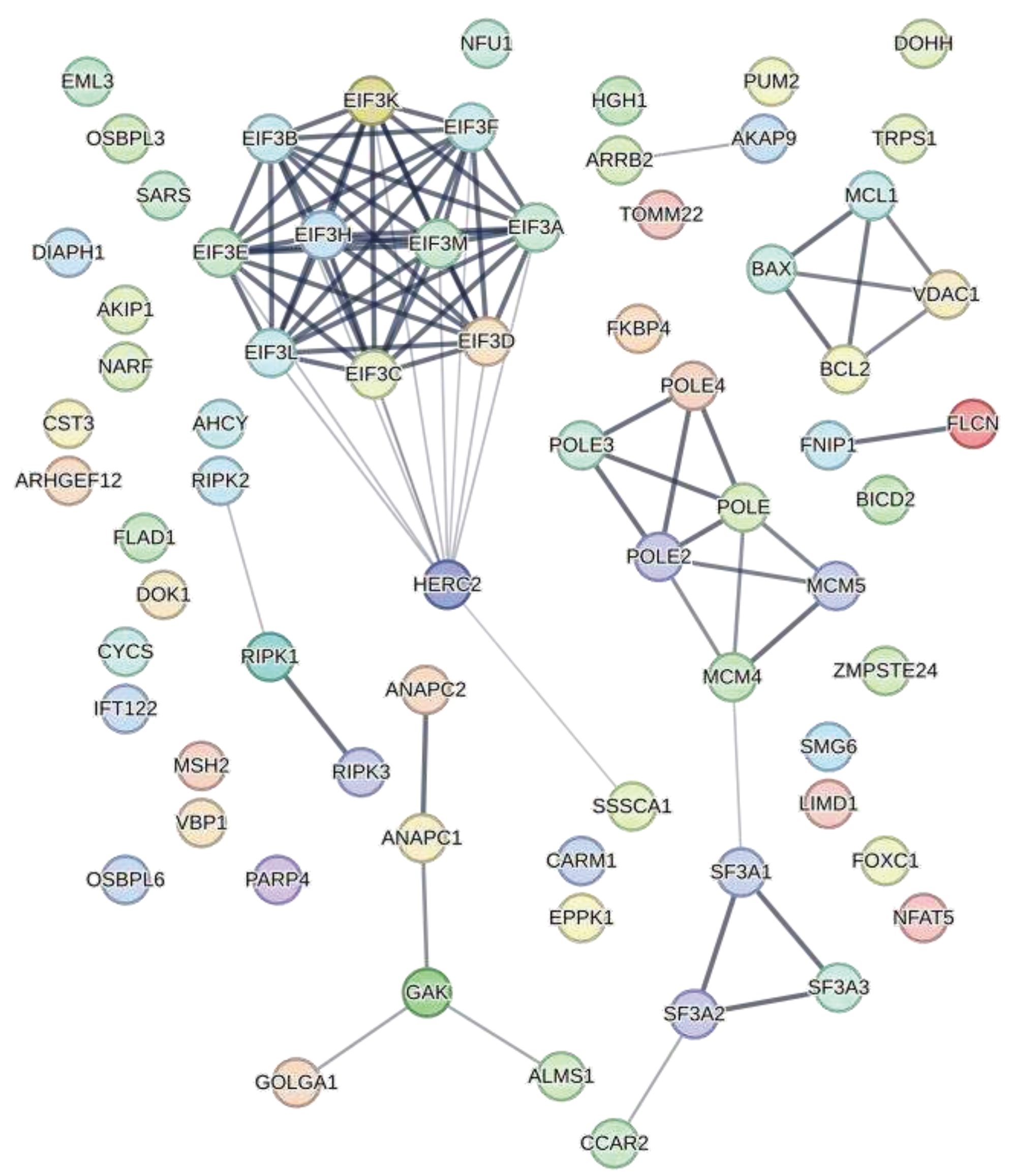
2. Cellular targets of HIV-1 Protease
| Protein | Cleavage sequence |
|---|---|
| HIV-1 cleavage sites | |
| Gag (MA-CA) | SQNY^PIVQ [17] |
| Gag (CA-p2) | ARVL^AEAM [17] |
| Gag (p2-NC) | ATIM^MQRG [17] |
| Gag (NC-p1) | RQAN^FLGK [17] |
| Gag (p1-p6) | PGNF^LQSR [17] |
| Gag-Pol (TF-PR) | SFNF^PQIT [17] |
| Gag-Pol (PR-RT) | TLNF^PISP [17] |
| Gag-Pol (RT-RH) | AETF^YVDG [17] |
| Gag-Pol (RH-IN) | RKIL^FLDG [17] |
| Host protein cleavage sites | |
| RIPK1 | PQVL^YQNN [23] |
| NDR1 | KDWV^FINY [24] |
| NDR2 | KDWV^FlNY [24] |
| proCaspase8 | PKVF^FIQA [25] |
| eIF4GI | KIIA^TVLM [26] |
| ATVL^MTED [26] | |
| RFSA^LQQA [26] | |
| eIF3d | RRNM^LQFN [22] |
| Bcl2 | RRDF^AEMS [27] |
| PABP | GFVS^FERH [28] |
| PRVM^STQR [28] | |
| GCN2 | GQDY^VETV [29] |
| NF-κB1 | HYGF^PTYG [30] |
| TBK1 | SNTL^VEMT [31] |
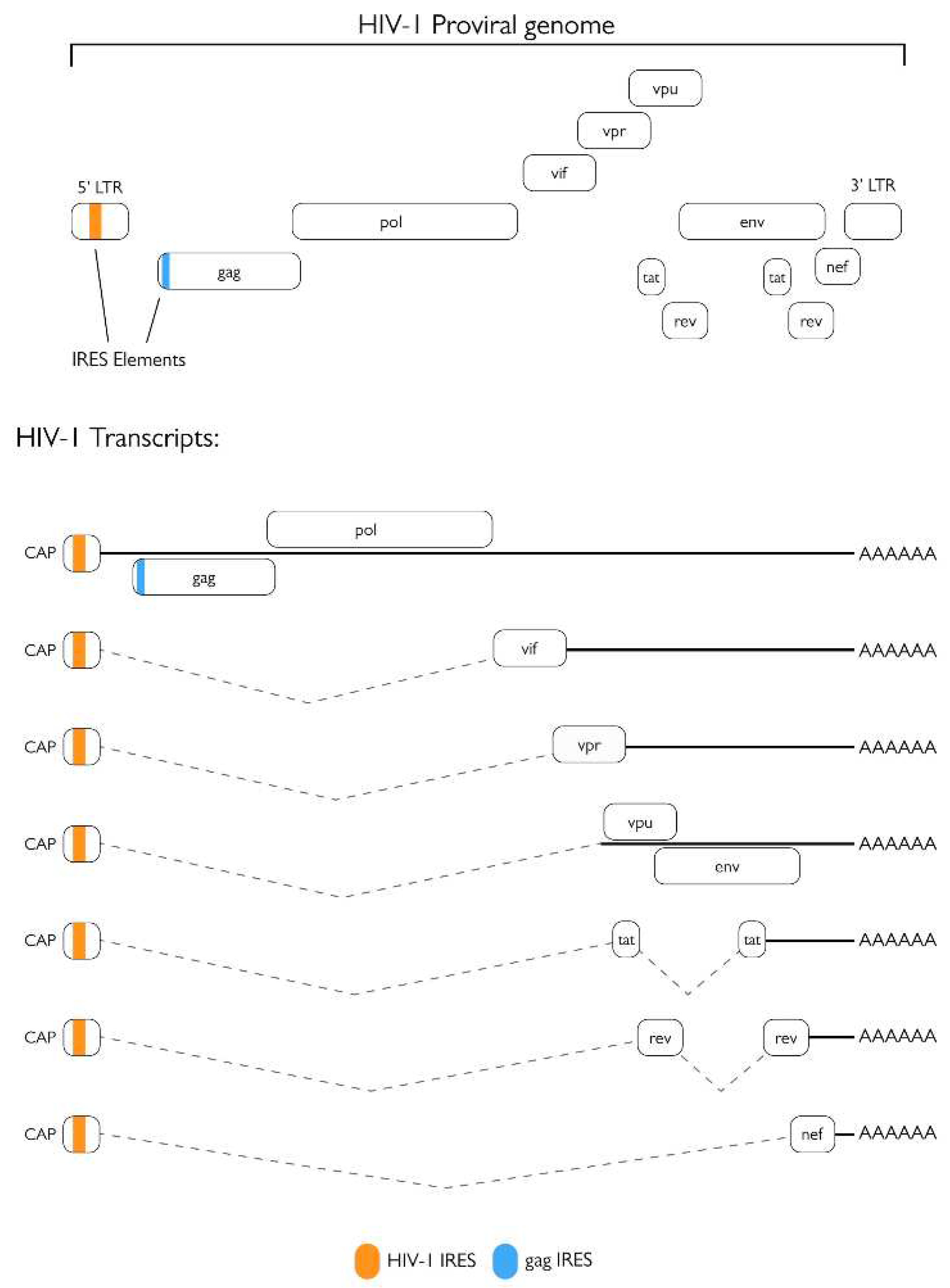
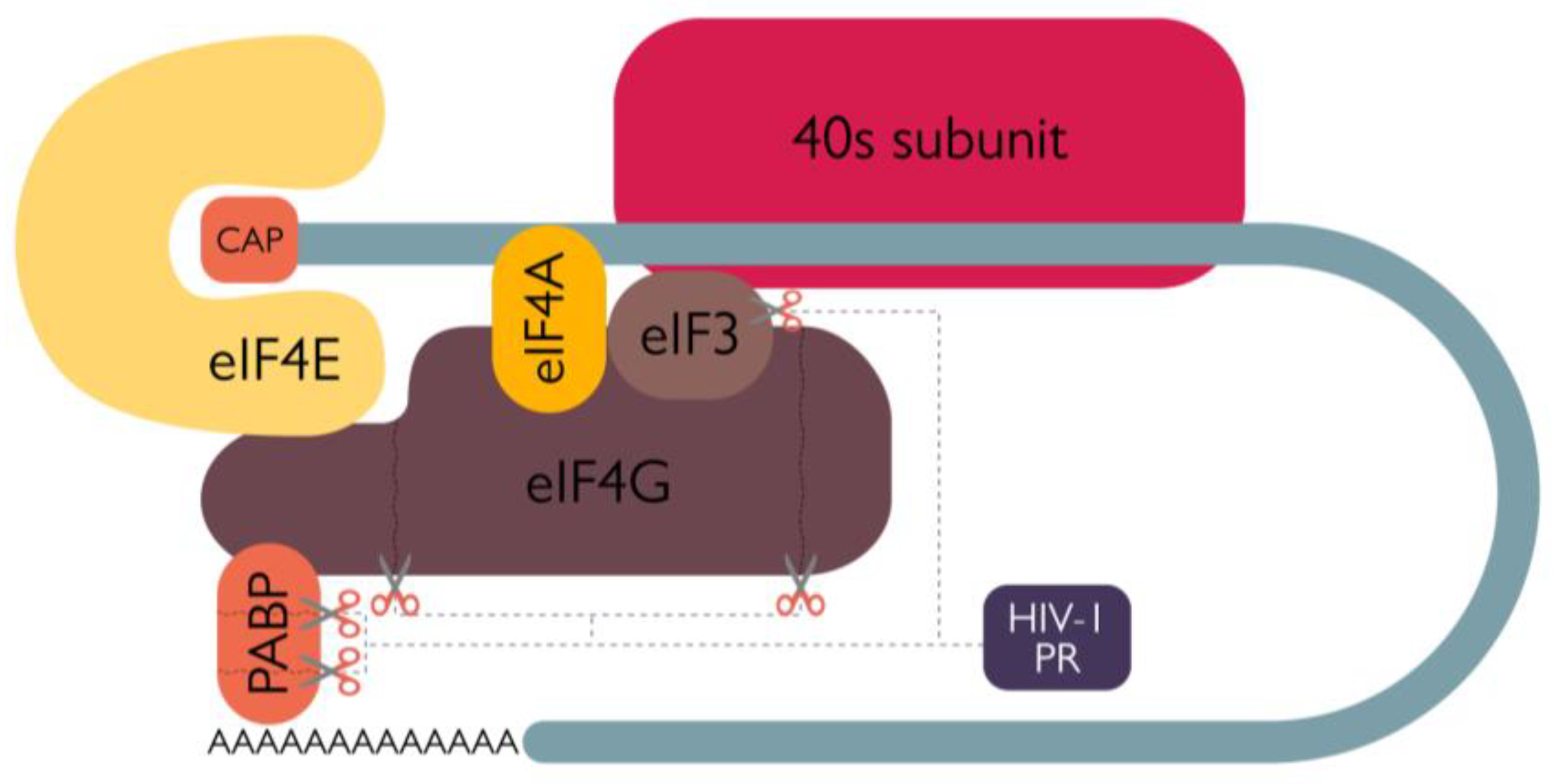
2.1. Role of the protease in protein synthesis modulation
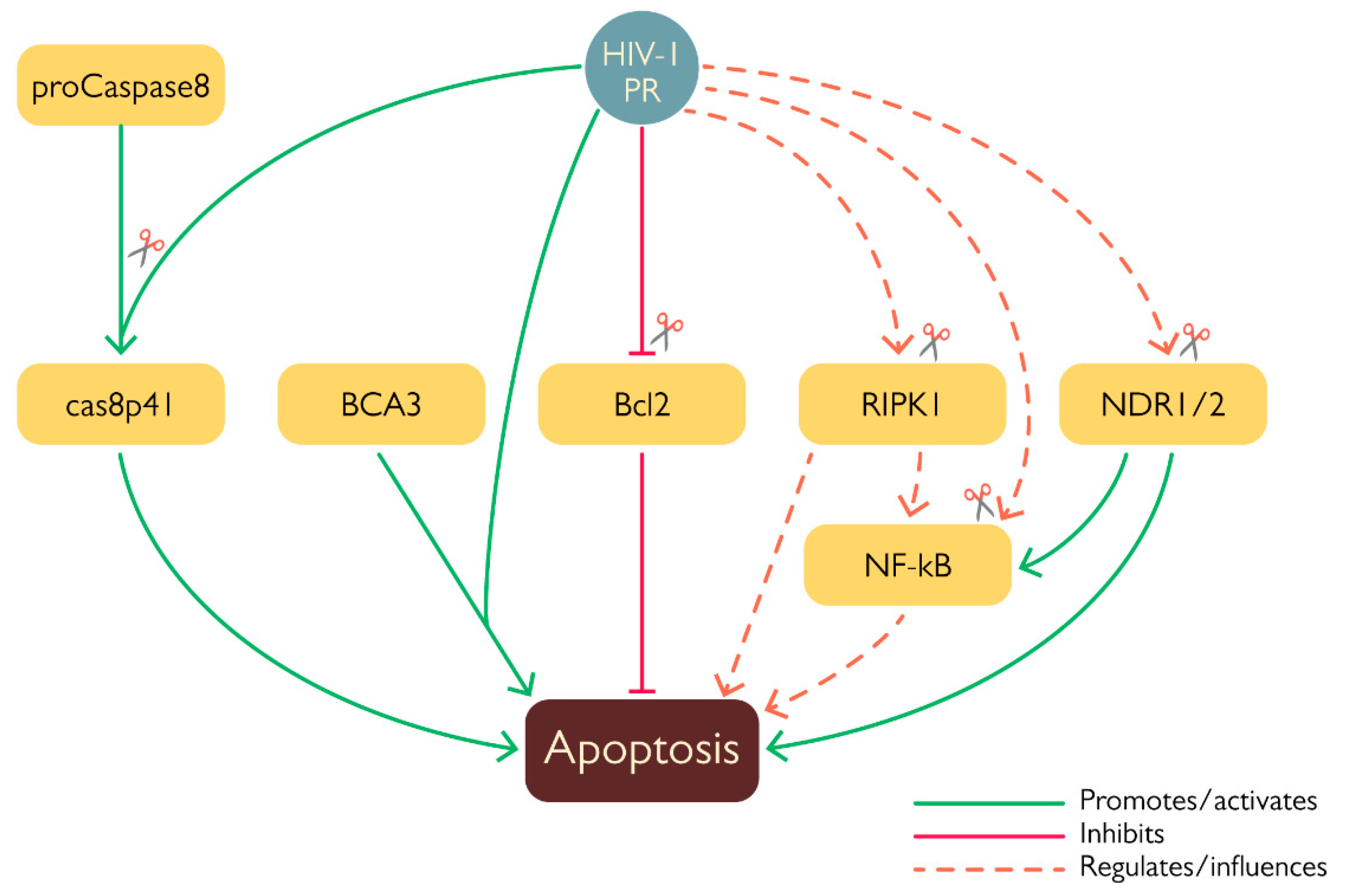
2.2. PR and apoptosis
2.3. Effect of PR on innate defenses
2.3. Cleavage of virion-incorporated restriction factors
2.4. PR-mediated cytoskeleton modulation
3. Conclusions
References
- Sharp, P.M.; Hahn, B.H. Origins of HIV and the AIDS Pandemic. Cold Spring Harb. Perspect. Med. 2011, 1. [Google Scholar] [CrossRef] [PubMed]
- IN DANGER: UNAIDS Global AIDS Update 2022; Geneva, 2022.
- Groopman, J.E. Zidovudine Intolerance. Rev. Infect. Dis. 1990, 12 Suppl 5, S500-6. [Google Scholar] [CrossRef]
- Larder, B.A.; Darby, G.; Richman, D.D. HIV with Reduced Sensitivity to Zidovudine (AZT) Isolated During Prolonged Therapy. Science (80-. ). 1989, 243, 1731–1734. [Google Scholar] [CrossRef] [PubMed]
- Hochhauser, D.; Harris, A.L. Drug Resistance. Br. Med. Bull. 1991, 47, 178–196. [Google Scholar] [CrossRef] [PubMed]
- Tomasselli, A.G.; Thaisrivongs, S.; Heinrikson, R.L. Discovery and Design of HIV Protease Inhibitors as Drugs for Treatment of Aids; 1996; Volume 2, ISBN 1559386932. [Google Scholar]
- Collier, A.C.; Coombs, R.W.; Schoenfeld, D.A.; Bassett, R.L.; Timpone, J.; Baruch, A.; Jones, M.; Facey, K.; Whitacre, C.; McAuliffe, V.J.; et al. Treatment of Human Immunodeficiency Virus Infection with Saquinavir, Zidovudine, and Zalcitabine. AIDS Clinical Trials Group. N. Engl. J. Med. 1996, 334, 1011–1017. [Google Scholar] [CrossRef]
- Simon, V.; Ho, D.D.; Abdool Karim, Q. HIV/AIDS Epidemiology, Pathogenesis, Prevention, and Treatment. Lancet 2006, 368, 489–504. [Google Scholar] [CrossRef]
- Yang, H.; Nkeze, J.; Zhao, R.Y. Effects of HIV-1 Protease on Cellular Functions and Their Potential Applications in Antiretroviral Therapy. Cell Biosci. 2012, 2, 1. [Google Scholar] [CrossRef] [PubMed]
- Mager, P.P. The Active Site of HIV-1 Protease. Med. Res. Rev. 2001, 21, 348–353. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Kräusslich, H.G.; Müller, B. Retroviral Proteases and Their Roles in Virion Maturation. Virology 2015, 479–480, 403–417. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, J.; Chen, Z.; Wang, G.; Shao, Q.; Shi, J.; Zhu, W. Structural Insights into HIV-1 Protease Flap Opening Processes and Key Intermediates. RSC Adv. 2017, 7, 45121–45128. [Google Scholar] [CrossRef]
- Scott, W.R.P.; Schiffer, C.A. Curling of Flap Tips in HIV-1 Protease as a Mechanism for Substrate Entry and Tolerance of Drug Resistance. Structure 2000, 8, 1259–1265. [Google Scholar] [CrossRef] [PubMed]
- Trylska, J.; Tozzini, V.; Chang, C.E.A.; McCammon, J.A. HIV-1 Protease Substrate Binding and Product Release Pathways Explored with Coarse-Grained Molecular Dynamics. Biophys. J. 2007, 92, 4179–4187. [Google Scholar] [CrossRef] [PubMed]
- Laco, G.S. HIV-1 Protease Substrate-Groove: Role in Substrate Recognition and Inhibitor Resistance. Biochimie 2015, 118, 90–103. [Google Scholar] [CrossRef] [PubMed]
- Impens, F.; Timmerman, E.; Staes, A.; Moens, K.; Ariën, K.K.; Verhasselt, B.; Vandekerckhove, J.; Gevaert, K. A Catalogue of Putative HIV-1 Protease Host Cell Substrates. Biol. Chem. 2012, 393, 915–931. [Google Scholar] [CrossRef] [PubMed]
- Lawal, M.M.; Sanusi, Z.K.; Govender, T.; Maguire, G.E.M.; Honarparvar, B.; Kruger, H.G. From Recognition to Reaction Mechanism: An Overview on the Interactions between HIV-1 Protease and Its Natural Targets. Curr. Med. Chem. 2018, 25, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.K.; Potempa, M.; Swanstrom, R. The Choreography of HIV-1 Proteolytic Processing and Virion Assembly. J. Biol. Chem. 2012, 287, 40867–40874. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Louis, J.M.; Aniana, A.; Suh, J.Y.; Clore, G.M. Visualizing Transient Events in Amino-Terminal Autoprocessing of HIV-1 Protease. Nature 2008, 455, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Agniswamy, J.; Sayer, J.M.; Weber, I.T.; Louis, J.M. Terminal Interface Conformations Modulate Dimer Stability Prior to Amino Terminal Autoprocessing of HIV-1 Protease. Biochemistry 2012, 51, 1041–1050. [Google Scholar] [CrossRef]
- Kaplan, A.H.; Swanstrom, R. Human Immunodeficiency Virus Type 1 Gag Proteins Are Processed in Two Cellular Compartments. Proc. Natl. Acad. Sci. U. S. A. 1991, 88, 4528–4532. [Google Scholar] [CrossRef]
- Jäger, S.; Cimermancic, P.; Gulbahce, N.; Johnson, J.R.; McGovern, K.E.; Clarke, S.C.; Shales, M.; Mercenne, G.; Pache, L.; Li, K.; et al. Global Landscape of HIV-Human Protein Complexes. Nature 2012, 481, 365–370. [Google Scholar] [CrossRef]
- Wagner, R.N.; Reed, J.C.; Chanda, S.K. HIV-1 Protease Cleaves the Serine-Threonine Kinases RIPK1 and RIPK2. Retrovirology 2015, 12, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Devroe, E.; Silver, P.A.; Engelman, A. HIV-1 Incorporates and Proteolytically Processes Human NDR1 and NDR2 Serine-Threonine Kinases. Virology 2005, 331, 181–189. [Google Scholar] [CrossRef]
- Nie, Z.; Bren, G.D.; Vlahakis, S.R.; Schimnich, A.A.; Brenchley, J.M.; Trushin, S.A.; Warren, S.; Schnepple, D.J.; Kovacs, C.M.; Loutfy, M.R.; et al. Human Immunodeficiency Virus Type 1 Protease Cleaves Procaspase 8 In Vivo. J. Virol. 2007, 81, 6947–6956. [Google Scholar] [CrossRef]
- Ventoso, I.; Blanco, R.; Perales, C.; Carrasco, L. HIV-1 Protease Cleaves Eukaryotic Initiation Factor 4G and Inhibits Cap-Dependent Translation. Proc. Natl. Acad. Sci. U. S. A. 2001, 98, 12966–12971. [Google Scholar] [CrossRef] [PubMed]
- Strack, P.R.; Frey, M.W.; Rizzo, C.J.; Cordova, B.; George, H.J.; Meade, R.; Ho, S.P.; Corman, J.; Tritch, R.; Korant, B.D. Apoptosis Mediated by HIV Protease Is Preceded by Cleavage of Bcl-2. Proc. Natl. Acad. Sci. U. S. A. 1996, 93, 9571–9576. [Google Scholar] [CrossRef]
- Álvarez, E.; Castelló, A.; Menéndez-Arias, L.; Carrasco, L. HIV Protease Cleaves Poly(A)-Binding Protein. Biochem. J. 2006, 396, 219–226. [Google Scholar] [CrossRef] [PubMed]
- del Pino, J.; Jiménez, J.L.; Ventoso, I.; Castelló, A.; Muñoz-Fernández, M.Á.; de Haro, C.; Berlanga, J.J. GCN2 Has Inhibitory Effect on Human Immunodeficiency Virus-1 Protein Synthesis and Is Cleaved upon Viral Infection. PLoS One 2012, 7. [Google Scholar] [CrossRef]
- Rivière, Y.; Blank, V.; Kourilsky, P.; Israël, A. Processing of the Precursor of NF-ΚB by the HIV-1 Protease during Acute Infection. Nature 1991, 350, 625–626. [Google Scholar] [CrossRef] [PubMed]
- Jeremiah, S.S.; Miyakawa, K.; Matsunaga, S.; Nishi, M.; Kudoh, A.; Takaoka, A.; Sawasaki, T.; Ryo, A. Cleavage of TANK-Binding Kinase 1 by HIV-1 Protease Triggers Viral Innate Immune Evasion. Front. Microbiol. 2021, 12, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING V11: Protein-Protein Association Networks with Increased Coverage, Supporting Functional Discovery in Genome-Wide Experimental Datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Amorim, R.; Costa, S.M.; Cavaleiro, N.P.; da Silva, E.E.; da Costa, L.J. HIV-1 Transcripts Use IRES-Initiation under Conditions Where Cap-Dependent Translation Is Restricted by Poliovirus 2A Protease. PLoS One 2014, 9, e88619. [Google Scholar] [CrossRef] [PubMed]
- Castelló, A.; Franco, D.; Moral-López, P.; Berlanga, J.J.; Álvarez, E.; Wimmer, E.; Carrasco, L. HIV-1 Protease Inhibits Cap-and Poly(A)-Dependent Translation upon EIF4GI and PABP Cleavage. PLoS One 2009, 4. [Google Scholar] [CrossRef] [PubMed]
- Brasey, A.; Lopez-Lastra, M.; Ohlmann, T.; Beerens, N.; Berkhout, B.; Darlix, J.-L.; Sonenberg, N. The Leader of Human Immunodeficiency Virus Type 1 Genomic RNA Harbors an Internal Ribosome Entry Segment That Is Active during the G2/M Phase of the Cell Cycle. J. Virol. 2003, 77, 3939–3949. [Google Scholar] [CrossRef] [PubMed]
- Vallejos, M.; Carvajal, F.; Pino, K.; Navarrete, C.; Ferres, M.; Huidobro-Toro, J.P.; Sargueil, B.; López-Lastra, M. Functional and Structural Analysis of the Internal Ribosome Entry Site Present in the MRNA of Natural Variants of the HIV-1. PLoS One 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Buck, C.B.; Shen, X.; Egan, M.A.; Pierson, T.C.; Walker, C.M.; Siliciano, R.F. The Human Immunodeficiency Virus Type 1 Gag Gene Encodes an Internal Ribosome Entry Site. J. Virol. 2001, 75, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Plank, T.D.M.; Whitehurst, J.T.; Kieft, J.S. Cell Type Specificity and Structural Determinants of IRES Activity from the 5’ Leaders of Different HIV-1 Transcripts. Nucleic Acids Res. 2013, 41, 6698–6714. [Google Scholar] [CrossRef]
- Baboonian, C.; Dalgleish, A.; Bountiff, L.; Gross, J.; Oroszlan, S.; Rickett, G.; Smith-Burchnell, C.; Troke, P.; Merson, J. HIV-1 Proteinase Is Required for Synthesis of pro-Viral DNA. Biochem. Biophys. Res. Commun. 1991, 179, 17–24. [Google Scholar] [CrossRef]
- Jacobsen, H.; Ahlborn-laake, L.; Gugel, R.; Mous, J.A.N. Progression of Early Steps of Human Immunodeficiency Virus Type 1 Replication in the Presence of an Inhibitor of Viral Protease A-I. 1992, 66, 5087–5091.
- Nagy, K.; Young, M.; Baboonian, C.; Merson, J.; Whittle, P.; Oroszlan, S. Antiviral Activity of Human Immunodeficiency Virus Type 1 Protease Inhibitors in a Single Cycle of Infection: Evidence for a Role of Protease in the Early Phase. J. Virol. 1994, 68, 757–765. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Manchester, M.; Smith, T.; Yang, Y.L.; Swanstrom, R. Conditional Human Immunodeficiency Virus Type 1 Protease Mutants Show No Role for the Viral Protease Early in Virus Replication. J. Virol. 1996, 70, 5840–5844. [Google Scholar] [CrossRef]
- Uchida, H.; Maeda, Y.; Mitsuya, H. HIV-1 Protease Does Not Play a Critical Role in the Early Stages of HIV-1 Infection. Antiviral Res. 1997, 36, 107–113. [Google Scholar] [CrossRef]
- Stefanidou, M.; Herrera, C.; Armanasco, N.; Shattock, R.J. Saquinavir Inhibits Early Events Associated with Establishment of HIV-1 Infection: Potential Role for Protease Inhibitors in Prevention. Antimicrob. Agents Chemother. 2012, 56, 4381–4390. [Google Scholar] [CrossRef] [PubMed]
- Monette, A.; Valiente-Echeverría, F.; Rivero, M.; Cohen, E. ´ A.A.; Lopez-Lastra, M. Dual Mechanisms of Translation Initiation of the Full-Length HIV-1 MRNA Contribute to Gag Synthesis. PLoS One 2013, 8, 68108. [Google Scholar] [CrossRef] [PubMed]
- Vichalkovski, A.; Gresko, E.; Cornils, H.; Hergovich, A.; Schmitz, D.; Hemmings, B.A. NDR Kinase Is Activated by RASSF1A/MST1 in Response to Fas Receptor Stimulation and Promotes Apoptosis. Curr. Biol. 2008, 18, 1889–1895. [Google Scholar] [CrossRef] [PubMed]
- Gougeon, M.L. Apoptosis as an HIV Strategy to Escape Immune Attack. Nat. Rev. Immunol. 2003, 3, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Baum, E.Z.; Bebernitz, G.A.; Gluzman, Y. Isolation of Mutants of Human Immunodeficiency Virus Protease Based on the Toxicity of the Enzyme in Escherichia Coli. Proc. Natl. Acad. Sci. U. S. A. 1990, 87, 5573–5577. [Google Scholar] [CrossRef] [PubMed]
- M’Barek, N. Ben; Audoly, G.; Raoult, D.; Gluschankof, P. HIV-2 Protease Resistance Defined in Yeast Cells. Retrovirology 2006, 3. [Google Scholar] [CrossRef] [PubMed]
- Konvalinka, J.; Litterst, M.A.; Welker, R.; Kottler, H.; Rippmann, F.; Heuser, A.M.; Kräusslich, H.G. An Active-Site Mutation in the Human Immunodeficiency Virus Type 1 Proteinase (PR) Causes Reduced PR Activity and Loss of PR-Mediated Cytotoxicity without Apparent Effect on Virus Maturation and Infectivity. J. Virol. 1995, 69, 7180–7186. [Google Scholar] [CrossRef] [PubMed]
- Rumlová, M.; Křížová, I.; Keprová, A.; Hadravová, R.; Doležal, M.; Strohalmová, K.; Pichová, I.; Hájek, M.; Ruml, T. HIV-1 Protease-Induced Apoptosis. Retrovirology 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Rumlová, M.; Křížová, I.; Zelenka, J.; Weber, J.; Ruml, T. Does BCA3 Play a Role in the HIV-1 Replication Cycle? Viruses 2018, 10. [Google Scholar] [CrossRef]
- Cartier, C.; Hemonnot, B.; Gay, B.; Bardy, M.; Sanchiz, C.; Devaux, C.; Briant, L. Active CAMP-Dependent Protein Kinase Incorporated within Highly Purified HIV-1 Particles Is Required for Viral Infectivity and Interacts with Viral Capsid Protein. J. Biol. Chem. 2003, 278, 35211–35219. [Google Scholar] [CrossRef]
- Baichwal, V.R.; Baeuerle, P.A. Apoptosis: Activate NF-ΚB or Die? Curr. Biol. 1997, 7, R94–R96. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Ong, N.; An, H.; Zheng, Y. The Emerging Roles of NDR1/2 in Infection and Inflammation. Front. Immunol. 2020, 11, 534. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Ma, X.; Cheng, H.; Jiang, W.; Xu, X.; Zhang, Y.; Zhang, Y.; Guo, Z.; Yu, Y.; Xu, H.; et al. Stk38 Protein Kinase Preferentially Inhibits TLR9-Activated Inflammatory Responses by Promoting MEKK2 Ubiquitination in Macrophages. Nat. Commun. 2015 61 2015, 6, 1–11. [Google Scholar] [CrossRef]
- Ma, X.; Wang, D.; Li, N.; Gao, P.; Zhang, M.; Zhang, Y. Hippo Kinase NDR2 Inhibits IL-17 Signaling by Promoting Smurf1-Mediated MEKK2 Ubiquitination and Degradation. Mol. Immunol. 2019, 105, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, Q.; Wu, C.; Li, H.; Shou, J.; Yang, Y.; Gu, M.; Ma, C.; Lin, W.; Zou, Y.; et al. Downregulated NDR1 Protein Kinase Inhibits Innate Immune Response by Initiating an MiR146a-STAT1 Feedback Loop. Nat. Commun. 2018 91 2018, 9, 1–16. [Google Scholar] [CrossRef]
- Solis, M.; Nakhaei, P.; Jalalirad, M.; Lacoste, J.; Douville, R.; Arguello, M.; Zhao, T.; Laughrea, M.; Wainberg, M.A.; Hiscott, J. RIG-I-Mediated Antiviral Signaling Is Inhibited in HIV-1 Infection by a Protease-Mediated Sequestration of RIG-I. J. Virol. 2011, 85, 1224–1236. [Google Scholar] [CrossRef]
- Jurczyszak, D.; Zhang, W.; Terry, S.N.; Kehrer, T.; Bermúdez González, M.C.; McGregor, E.; Mulder, L.C.F.; Eckwahl, M.J.; Pan, T.; Simon, V. HIV Protease Cleaves the Antiviral M6A Reader Protein YTHDF3 in the Viral Particle. PLOS Pathog. 2020, 16, e1008305. [Google Scholar] [CrossRef]
- Ebrahimi, D.; Richards, C.M.; Carpenter, M.A.; Wang, J.; Ikeda, T.; Becker, J.T.; Cheng, A.Z.; McCann, J.L.; Shaban, N.M.; Salamango, D.J.; et al. Genetic and Mechanistic Basis for APOBEC3H Alternative Splicing, Retrovirus Restriction, and Counteraction by HIV-1 Protease. Nat. Commun. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Engeland, C.E.; Oberwinkler, H.; Schumann, M.; Krause, E.; Muller, G.A.; Krausslich, H.-G. The Cellular Protein Lyric Interacts with HIV-1 Gag. J. Virol. 2011, 85, 13322–13332. [Google Scholar] [CrossRef] [PubMed]
- Yamada, E.; Yoshikawa, R.; Nakano, Y.; Misawa, N.; Koyanagi, Y.; Sato, K. Impacts of Humanized Mouse Models on the Investigation of HIV-1 Infection: Illuminating the Roles of Viral Accessory Proteins in Vivo. Viruses 2015, 7, 1373–1390. [Google Scholar] [CrossRef]
- Ye, X.; Zhang, Y.; Xu, Q.; Zheng, H.; Wu, X.; Qiu, J.; Zhang, Z.; Wang, W.; Shao, Y.; Xing, H.Q. HIV-1 Tat Inhibits EAAT-2 through AEG-1 Upregulation in Models of HIV-Associated Neurocognitive Disorder. Oncotarget 2017, 8, 39922–39934. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.P.; Koyuncu, O.O.; Enquist, L.W. Subversion of the Actin Cytoskeleton during Viral Infection. Nat. Rev. Microbiol. 2011, 9, 427–439. [Google Scholar] [CrossRef]
- Honer, B.; Shoeman, R.L.; Traub, P. Human Immunodeficiency Virus Type 1 Protease Microinjected into Cultured Human Skin Fibroblasts Cleaves Vimentin and Affects Cytoskeletal and Nuclear Architecture. J. Cell Sci. 1991, 100, 799–807. [Google Scholar] [CrossRef] [PubMed]
- Shoeman, R.L.; Sachse, C.; Höner, B.; Mothes, E.; Kaufmann, M.; Traub, P. Cleavage of Human and Mouse Cytoskeletal and Sarcomeric Proteins by Human Immunodeficiency Virus Type 1 Protease: Actin, Desmin, Myosin, and Tropomyosin. Am. J. Pathol. 1993, 142, 221–230. [Google Scholar] [PubMed]
- Adams, L.D.; Tomasselli, A.G.; Robbins, P.; Moss, B.; Heinrikson, R.L. HIV-1 Protease Cleaves Actin During Acute Infection of Human T-Lymphocytes. AIDS Res. Hum. Retroviruses 1992, 8, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Naghavi, M.H. The Host Cytoskeleton: A Key Regulator of Early HIV-1 Infection. FEBS J. 2022. [Google Scholar] [CrossRef]
- Matarrese, P.; Malorni, W. Human Immunodeficiency Virus (HIV)-1 Proteins and Cytoskeleton: Partners in Viral Life and Host Cell Death. Cell Death Differ. 2005 121 2005, 12, 932–941. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





