1. Introduction
The innate immune system is the first line of host defense against multiple danger signals from pathogens or cellular damage. It encodes various host germline-encoded pattern recognition receptors (PRRs), which recognize pathogen-associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs)[
1,
2]. To fight infection and achieve homeostasis, the PRRs cause the activation of gene transcription or protease-dependent cytokine maturation, producing antiviral interferons (IFNs), proinflammatory cytokines, and chemokines [
1,
2]. Host antiviral innate immune responses are elicited during viral infections, and upon engagement by viral infections, cells use different PRRs to sense viral nucleic acids [
3]. DNA sensing PRRs are Toll-like receptor 9 (TLR9), cyclic GMP-AMP synthase (cGAS), stimulator of interferon genes (STING), absent in melanoma 2 (AIM2), and interferon gamma-inducible 16 (IFI16) [
4]. RNA sensing PRRs include endosomal TLR3, TLR7, TLR8, and cytosolic retinoic acid inducible gene-I (RIG-I), melanoma differentiation-associated gene 5 (MDA5), NOD-like receptor pyrin domain containing 3 (NLRP3) and nucleotide-binding and oligomerization domain containing 2 (NOD2) [
5].
The cGAS-STING pathway has been identified to be the important DNA-sensing machinery in innate immunity, against pathogen aggresion [
6,
7]. cGAS senses the presence of non-self and self DNA, and utilizes substrates ATP and GTP to catalyze the production of the second messenger cyclic GMP-AMP (2′3′-cGAMP) that then activates the signaling adaptor protein STING [
6,
7]. Activated STING recruits the downstream TANK-binding kinase 1 (TBK1) and TBK1 is auto-phosphorylated [
6,
8]. Then, the transcription factor IRF3, which is recruited by STING and phosphorylated by TBK1, translocates to the nucleus and induces antiviral type I IFNs and IFN stimulated genes (ISGs) [
6,
8]. Another transcription factor NF-κB is also activated by STING-TBK1 signaling and drives proinflammatory gene expressions [
6,
9].
Manganese (Mn) is required as an enzymatic cofactor in many physiologic processes, such as protein and energy metabolism, immune function, development, reproduction, neuronal regulation, and antioxidant defenses [
10,
11,
12]. Several canonical signaling pathways have been reported to be Mn-responsive, including ataxia telangiectasia mutated 2 (ATM), p53, phosphatidylinositol 3 kinase (PI3K), insulin and insulin-like growth factor-1 (IGF-1) pathways [
13,
14,
15]. However, attention has been recently paid to the Mn in the regulation of cGAS-STING pathway, which exert a potent host defense against DNA viruses [
16]. Mn
2+ was shown to increase the sensitivity of cGAS to double-stranded DNA (dsDNA) and its enzymatic activity. It also facilitates STING activity by boosting cGAMP-STING binding affinity [
16]. Further studies have revealed that Mn
2+ directly activated cGAS to induce a noncanonical catalytic synthesis of 2’3’-cGAMP, through similar overall conformation to dsDNA-activated cGAS [
17,
18].
Interestingly, recent publications have suggested the relevance of the cGAS-STING pathway in the process of RNA virus infections [
19,
20,
21,
22]. However, whether Mn
2+ plays a role in the cGAS-STING pathway mediated anti-RNA virus infections is still largely unclear. Moreover, how the Mn
2+ mediates its antiviral functions is not fully elucidated. In the current study, we found that Mn
2+ exerts a broad antiviral functions against various viruses including some RNA and DNA viruses, which is independent on the cGAS-STING pathway.
2. Materials and methods
2.1. Cells and viruses
Marc-145 cells and HEK293T cells were cultured in DMEM (HyClone Laboratories, Logan, UT, USA) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin solution at 37 °C with 5% CO
2. Porcine alveolar macrophages (3D4/21, ATCC CRL-2843) were grown in RPMI 1640 (Hyclone Laboratories, Logan, UT, USA) supplemented with 10% FBS with 1% penicillin–streptomycin solution, and maintained at 37 °C with 5% CO
2 in a humidified incubator. The viruses including a DNA virus Herpes Simplex Virus 1 (HSV1-GFP) as well as RNA viruses Vesicular Stomatitis Virus (VSV-GFP) and Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Viruses (HP-PRRSV XJ17-5-GFP and HP-PRRSV vaccine strain JXA1-R-GFP) were used as we previously reported [
22,
23,
24].
2.2. Cell treatments
The Marc-145 cells were pretreated with different concentrations of Mn2+ (MnCl2·4H2O) (Sigma-Aldrich, St. Louis, MO, USA) for 24 h and then infected with 0.1 multiplicity of infection (MOI) PRRSV XJ17-5 or JXA1-R. The 3D4/21 cells were also pretreated with Mn2+ (MnCl2·4H2O) for 24 h, and subsequently infected with 0.01 MOI HSV-1 or 0.001 MOI VSV. Additionally, 3D4/21 cells were transfected with cGAS agonist poly dA:dT (InvivoGen, Hong Kong, China) or STING agonist 2’3’-cGAMP (InvivoGen, Hong Kong, China) by using the transfection reagent Lipofectamine 2000 (ThermoFisher Scientific, Shanghai, China) and then exposed to Mn2+ treatment for 24 h.
2.3. Western blot analysis
Proteins were extracted in radioimmunoprecipitation assay (RIPA) lysis buffer, mixed with 4 × loading buffer by 3:1 ratio, and boiled at 100°C for 5-10 min. The protein samples were separated on 10% SDS-PAGE gels, and then transferred to PVDF membranes. After blocking with 5% skim milk solution at room temperature (RT) for 60 min, membranes were incubated with individual primary antibodies at 4 °C overnight. The primary antibodies include anti-GFP (HT801-01, TransGen, Beijing, China), STING (19851-1-AP, ProteinTech, Wuhan, China), cGAS (sc-515777, Santa Cruz Biotechnology, Dallas, Texas, USA), IRF3 (11904S, CST, Boston, MA, USA), p-IRF3 (Ser396) (MA5-14947, ThermoFisher Scientific, Shanghai, China), TBK1 (3504S, CST, Boston, MA, USA), p-TBK1 (5483S, CST, Boston, MA, USA) and β-actin (5057, CST, Boston, MA, USA). Secondary antibody HRP-conjugated goat anti-mouse or rabbit IgG (Transgen Biotech, Beijing, China) was used to incubate with the membranes for 60 min at RT. Signals were detected using enhanced chemiluminescence (ECL) substrate (Tanon, Shanghai, China) and images were visualized by imaging system (Tanon, Shanghai, China).
2.4. Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIpure reagent (Aidlab, Beijing, China). The cDNA was synthesized using HiScript® 1st Strand cDNA Synthesis Kit (Vazyme, Nanjing, China). The target gene expressions were examined using ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) on a StepOne Plus real-time PCR system (Applied Biosystems, Foster City, CA, USA). The qPCR program was 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 1 min. β-actin served as an internal reference control. The relative mRNA levels were calculated using 2
−ΔΔCT method. For all the qPCR assays, an efficiency comprised between 90 and 110% was measured. The sequence of qPCR primers used in this study were listed in
Table S1.
2.5. CRISPR gRNA design and preparation of Knockout (KO) cells
The CRISPR gRNAs targeting porcine cGAS and monkey cGAS were designed using the web tool from Benchling (
www.benchling.com). For each gene, two gRNAs were chosen according to the predicted high scores, respectively, which are shown in
Table S2. The recombinant pX458-gRNA plasmids were obtained when the annealed gRNA encoding DNA sequences were cloned into the
Bbs I site of pX458-EGFP. Marc-145 cells or 3D4/21 cells were transfected with the corresponding recombinant pX458-gRNA plasmids using Lipofectamine 2000. At 24 h post transfection, the GFP positive cells were sorted by a FACS Aria SORP cell sorter (Becton Dickinson) and cultured in 96-well plates by limiting dilution for monoclonal growth. The individual cell clones were screened by PCR using primers shown in
Table S2. Briefly, the genomic PCR products were cloned into T vector with pClone007 versatile simple vector kit (TsingKe Biological Technology, Beijing, China). Base substitution, insertion and deletion (ins/del) mutations were analyzed after the sequencing of inserted fragments, and cGAS
-/- monkey Marc-145 cells and cGAS
-/- porcine macrophages (3D4/21) were each acquired (
Figure S1). In addition, the STING
-/- Marc-145 cells and STING
-/- 3D4/21 cells were previously obtained and have been used in our lab [
22,
25].
2.6. Virus tissue culture infectious dose 50 (TCID50) titrations
Marc-145 cells or 3D4/21 cells were seeded into 96-well plates, and then infected with 10-fold serial dilutions of various virus samples (Marc-145 cells for PRRSVs and 3D4/21 cells for VSV and HSV1). Next, the infected cell supernatants were replaced with fresh DMEM or RPMI 1640 containing 2% FBS, and the cells were monitored for the GFP fluorescence and cytopathic effects (CPE) characterized by cell clumping and shrinkage in Marc-145 cells or 3D4/21 cells after infections for 1-5 days. Finally, the viral titers were expressed as TCID50 and calculated using the method of Reed-Muench.
2.7. Dual-luciferase reporter promoter assay
293T cells were seeded in 96-well plates followed by transfection at next day. Cells were co-transfected with reporter plasmids, ISRE-Firefly luc (Fluc) or IFNβ-Fluc (10 ng/well) and Renilla luciferase (Rluc) reporters (0.2 ng/well), plus the indicated porcine cGAS and STING plasmids or vector control (10-30 ng/well) using Lipofectamine 2000. The total DNA per well was normalized with control vectors to 50 ng. Twenty-four hours post transfection, cells were treated with different concentrations of Mn2+ for another 24 h. Then, cells were harvested and luciferase activities were detected with the TransDetect Double-Luciferase Reporter Assay Kit (Vazyme, Nanjing, Jiangsu, China). The fold changes were calculated relative to control samples after Fluc normalization by corresponding Rluc.
2.8. Statistical analysis
The results were analyzed using the software GraphPad Prism 6.0 and expressed as the mean ± standard deviation (SD). Statistical analysis was conducted by one way ANOVA followed by Tukey’s post hoc test. The normality of the data distribution was assessed using Shapiro-Wilk test. The p value less than 0.05 was considered statistically significant.
4. Discussion
The element Mn is critical for almost all forms of life [
10,
11,
12]. Cytosolic Mn
2+ has been reported to be involved in the dsDNA sensing activity of cGAS, and protects against DNA viruses [
16]. Recent studies have shown that Mn
2+ could directly activate cGAS, which was independent of dsDNA [
17,
18]. In addition, the overlapping mechanisms between the antiviral innate immunity developed against RNA and DNA viruses have been reviewed previously [
26]. Many RNA viruses of families
Flaviviridae,
Coronaviridae, and
Arteriviridae have been found to be associated with the cGAS-STING pathway [
19,
20,
21,
22,
27]. Likewise, in our study, we demonstrated that Mn
2+ exerts antiviral functions against a DNA virus (HSV-1) and some RNA viruses (PRRSV XJ17-5, PRRSV JXA1-R, and VSV) in a dose-dependent manner. The Mn
2+ exhibited a broad antiviral activity, which is similar to that mediated by the cGAS-STING pathway. At the first glance, it is logical to deduce that Mn
2+ exerts antiviral activity by acting on cGAS-STING signaling pathway. However, our further investigation revealed that Mn
2+ triggered antiviral activity is cGAS-STING independent, suggesting there is other cell mechanism which mediates the Mn
2+ antiviral functions.
Previously, Mn
2+ has been found to participate in the phosphorylation of p53 [
15,
28]. p53 is a tumor suppressor gene, and functions most commonly in cell cycle arrest, differentiation as well as apoptosis [
29]. Moreover, p53 has been reported to be involved the regulation of antiviral functions [
30,
31,
32]. For example, p53 overexpression represses HIV-1 long terminal repeat (LTR) transcriptional elongation via preventing the phosphorylation of serine 2 of the pol II C-terminal domain (CTD), resulting in the inhibition of HIV-1 transcription and replication [
30]. Influenza virus infection promotes the activation of the p53 pathway leading to apoptosis, and suppression of p53 activity contributes to influenza virus infection [
31]. Similarly, depletion of p53 was shown to promote porcine epidemic diarrhea virus (PEDV) infection susceptibility [
32]. p53 has robust antiviral immunity by activation of the IFN pathway and the induction of several antiviral proteins [
33,
34]. Miciak
et al have identified that p53 contributes to perpetuate IFN signaling through ISG-dependent positive feedback loops [
35]. Then, Hao
et al have discovered that p53 facilitates IFN signaling and secretion, and activates ISREs and ISG expression during viral infection [
32].
In addition, a recent report found that Mn
2+ alone activates phosphorylation of TBK1 with ATM involved, and enhances DNA- or RNA-mediated innate immune responses [
36]. TBK1 is the downstream mediator of multiple DNA sensors, such as Ku70, IFI16, cGAS, and DDX41 [
37]. Similarly, it is also involved in the signaling pathway of RNA sensors, such as RIG-I and MDA5 [
38,
39]. Here, our study demonstrates that Mn
2+ has antiviral functions against RNA viruses (PRRSVs, VSV) and a DNA virus (HSV-1) in cGAS
-/- and STING
-/- cells. Mn
2+ may initiate ATM-TBK1 signaling to exert its broad spectrum antiviral functions [
36]. Together, we speculate that Mn
2+ may induce activation of p53 and/or ATM-TBK1 to establish a powerfully antiviral state through cGAS-STING independent signaling pathways.
Nevertheless, we observed that the cGAS-STING signaling is indeed activated by the Mn
2+ treatment. This observation is consistent with the previous discovery that Mn
2+ suppressed virus replications through sensitizing both cGAS and STING [
16] and suggests that cGAS-STING is one of the cell machinery that mediates Mn
2+ antiviral functions. In our study, although Mn
2+ mediated antiviral functions via a cGAS-STING independent pathway, it was also identified to promote cGAS-STING signaling activity. Then, what is the relationship between cGAS-STING signaling and other cell machinery triggered by the Mn
2+? These cell machineries must be redundant, and together participate in the Mn
2+ triggered antiviral functions. However, the exact molecular mechanisms underlying Mn
2+ mediated antiviral functions deserve further exploration for a complete elucidation.
In summary, we demonstrated that Mn2+ inhibits PRRSV XJ17-5, PRRSV JXA1-R, VSV, and HSV-1 replications in a cGAS-STING independent manner. Our results reveal that Mn2+ may harbor multiple cellular mechanisms to exert its broad spectrum antiviral activity, and suggest that the Mn2+ has the potential to be used not only as an antiviral therapeutics but also as the immune adjuvant in some animal vaccines.
Figure 1.
Mn2+ exerts antiviral functions against PRRSV, VSV, and HSV-1. (A-D) Marc-145 cells were pretreated with Mn2+ (0, 100, 200, 500, 800, 1000 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV-GFP XJ17-5 (A and B) or HP-PRRSV-GFP JXA1-R (C and D) for 24 h and 48 h, respectively. (E-H) 3D4/21 cells were pretreated with Mn2+ (0, 50, 100, 200, 500, 800 μM) for 24 h, and then infected with 0.001 MOI VSV-GFP (E and F) or 0.01 MOI HSV-1-GFP (G and H) for 12 h. The GFP and β-actin protein levels were detected using Western blot analysis (A, C, E, G), and the virus titers in the supernatants were examined by TCID50 assay (B, D, F, H).
Figure 1.
Mn2+ exerts antiviral functions against PRRSV, VSV, and HSV-1. (A-D) Marc-145 cells were pretreated with Mn2+ (0, 100, 200, 500, 800, 1000 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV-GFP XJ17-5 (A and B) or HP-PRRSV-GFP JXA1-R (C and D) for 24 h and 48 h, respectively. (E-H) 3D4/21 cells were pretreated with Mn2+ (0, 50, 100, 200, 500, 800 μM) for 24 h, and then infected with 0.001 MOI VSV-GFP (E and F) or 0.01 MOI HSV-1-GFP (G and H) for 12 h. The GFP and β-actin protein levels were detected using Western blot analysis (A, C, E, G), and the virus titers in the supernatants were examined by TCID50 assay (B, D, F, H).
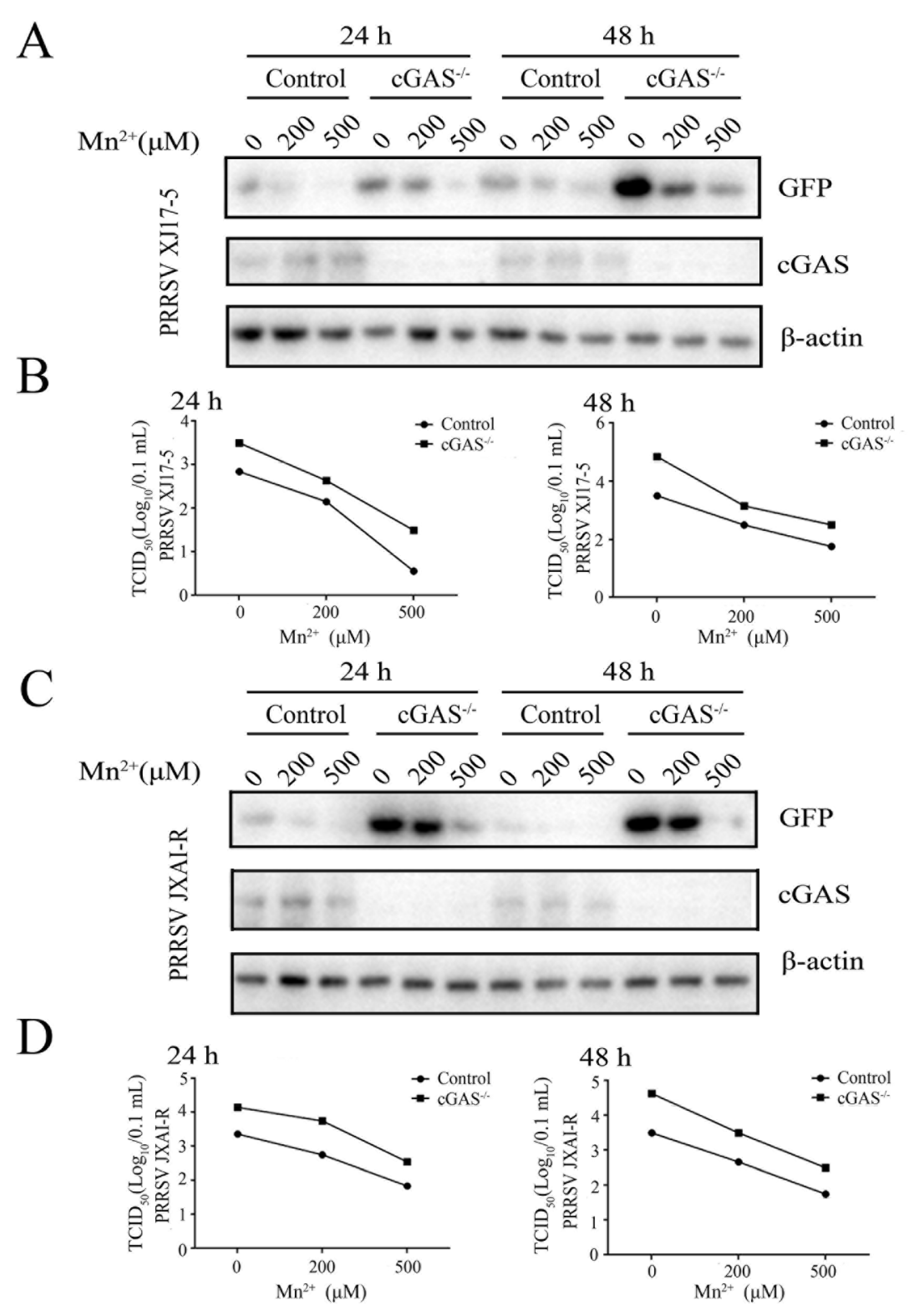
Figure 2.
Mn2+ mediates antiviral functions against PRRSV in the cGAS-independent manner. The cGAS-/- Marc-145 cells and normal Marc-145 control cells were pretreated with Mn2+ (0, 200, 500 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV XJ17-5 (A and B) or 0.1 MOI PRRSV JXA1-R (C and D) for 24 h and 48 h, respectively. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-cGAS and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
Figure 2.
Mn2+ mediates antiviral functions against PRRSV in the cGAS-independent manner. The cGAS-/- Marc-145 cells and normal Marc-145 control cells were pretreated with Mn2+ (0, 200, 500 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV XJ17-5 (A and B) or 0.1 MOI PRRSV JXA1-R (C and D) for 24 h and 48 h, respectively. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-cGAS and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
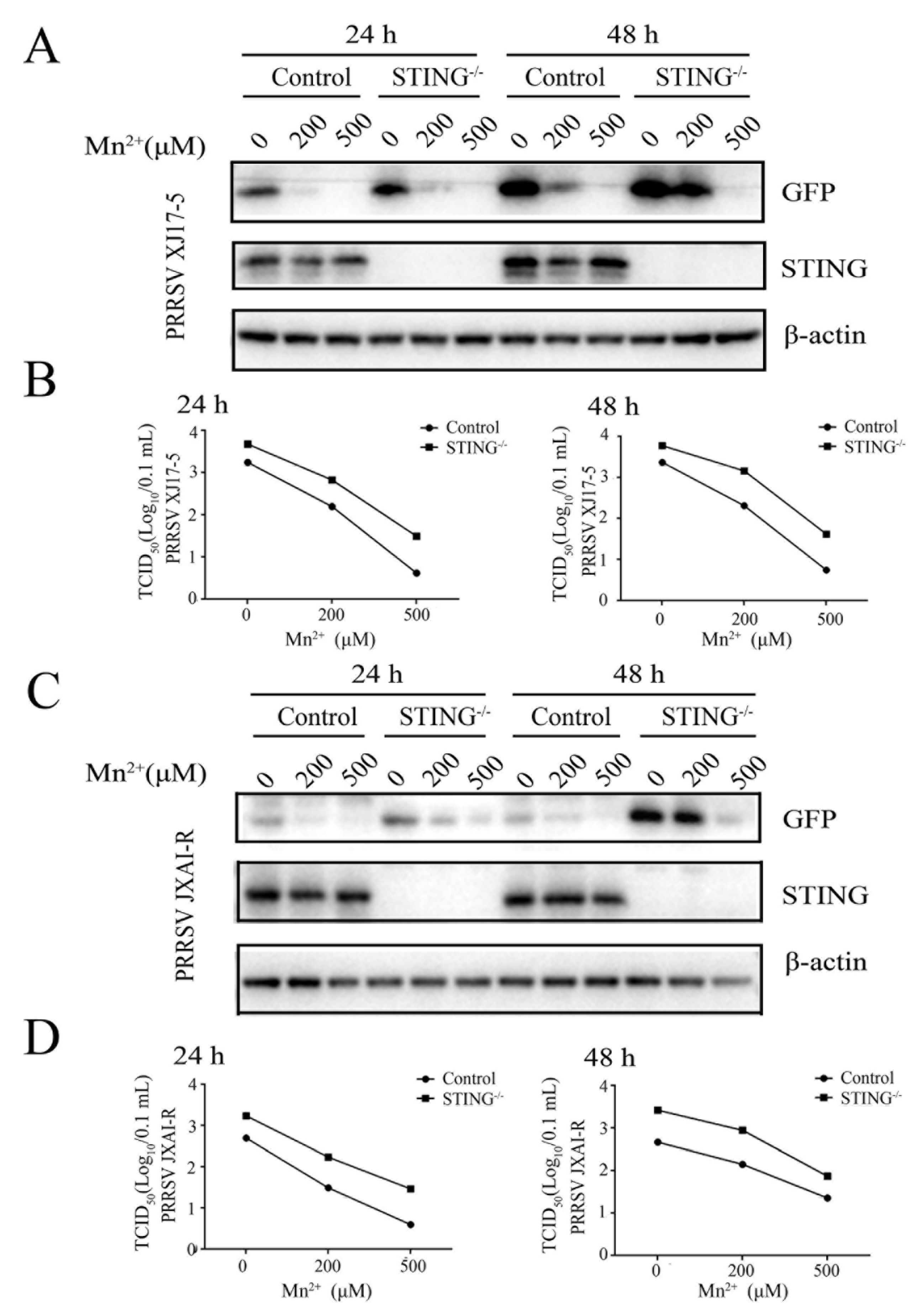
Figure 3.
Mn2+ mediates antiviral functions against PRRSV in the STING-independent manner. The STING-/- Marc-145 cells and normal Marc-145 control cells were pretreated with Mn2+ (0, 200, 500 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV XJ17-5 (A and B) or 0.1 MOI PRRSV JXA1-R (C and D) for 24 h and 48 h, respectively. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-STING and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
Figure 3.
Mn2+ mediates antiviral functions against PRRSV in the STING-independent manner. The STING-/- Marc-145 cells and normal Marc-145 control cells were pretreated with Mn2+ (0, 200, 500 μM) for 24 h, and then infected with 0.1 MOI HP-PRRSV XJ17-5 (A and B) or 0.1 MOI PRRSV JXA1-R (C and D) for 24 h and 48 h, respectively. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-STING and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
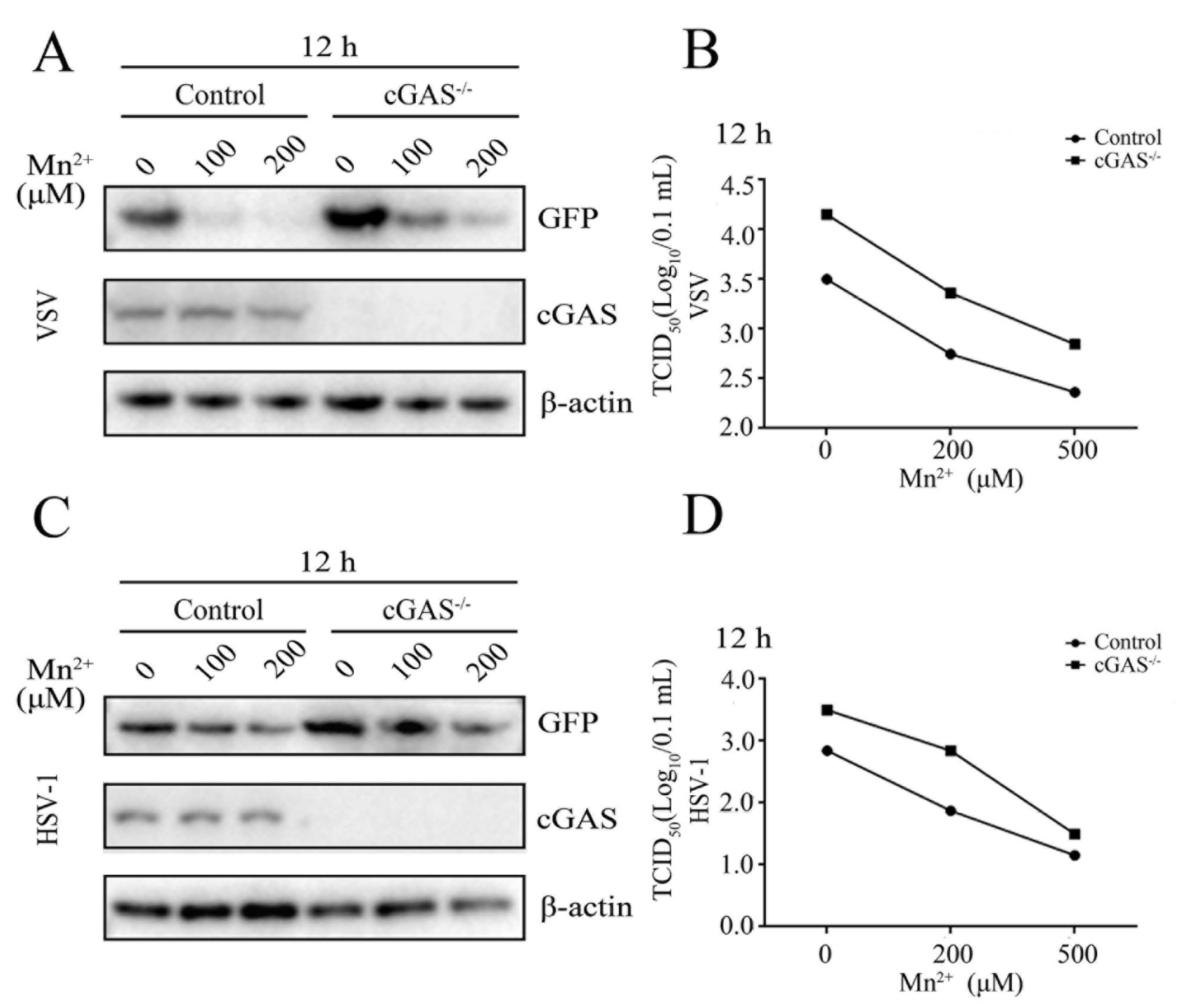
Figure 4.
Mn2+ mediates antiviral functions against VSV and HSV-1 in the cGAS-independent manner. (A-B) cGAS-/- 3D4/21 cells and normal 3D4/21 control cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and then infected with 0.001 MOI VSV for 12 h. (C-D) cGAS-/- 3D4/21 cells and normal 3D4/21 cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and subsequently infected with 0.01 MOI HSV-1 for 12 h. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-cGAS and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
Figure 4.
Mn2+ mediates antiviral functions against VSV and HSV-1 in the cGAS-independent manner. (A-B) cGAS-/- 3D4/21 cells and normal 3D4/21 control cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and then infected with 0.001 MOI VSV for 12 h. (C-D) cGAS-/- 3D4/21 cells and normal 3D4/21 cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and subsequently infected with 0.01 MOI HSV-1 for 12 h. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-cGAS and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
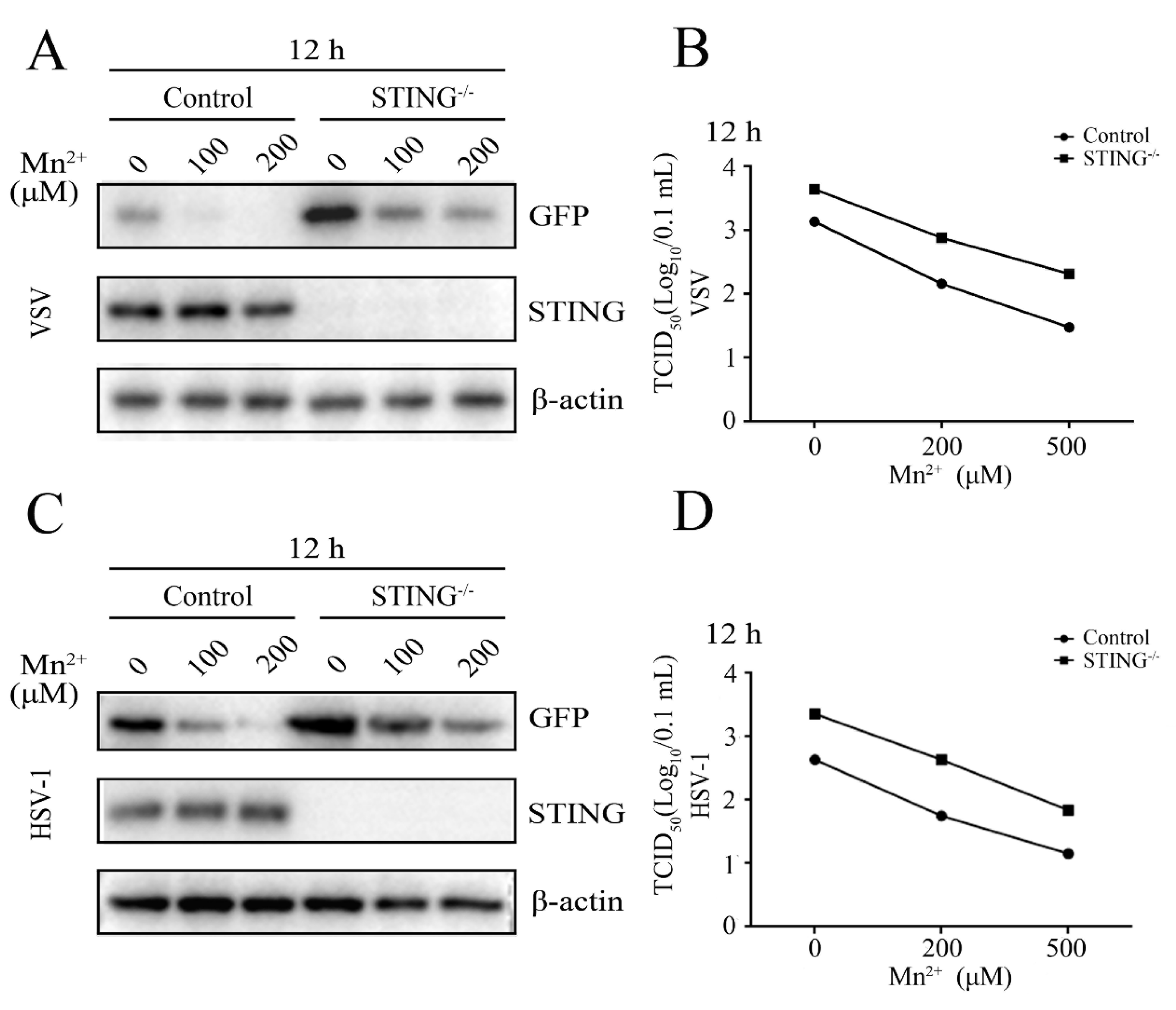
Figure 5.
Mn2+ mediates antiviral functions against VSV and HSV-1 in the STING-independent manner. (A-B) STING-/- 3D4/21 cells and normal 3D4/21 control cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and then infected with 0.001 MOI VSV for 12 h. (C-D) STING-/- 3D4/21 cells and normal 3D4/21 cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and subsequently infected with 0.01 MOI HSV-1 for 12 h. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-STING and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
Figure 5.
Mn2+ mediates antiviral functions against VSV and HSV-1 in the STING-independent manner. (A-B) STING-/- 3D4/21 cells and normal 3D4/21 control cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and then infected with 0.001 MOI VSV for 12 h. (C-D) STING-/- 3D4/21 cells and normal 3D4/21 cells were pretreated with Mn2+ (0, 100, 200 μM) for 24 h, and subsequently infected with 0.01 MOI HSV-1 for 12 h. The cell samples were collected and subjected to Western blot analysis using anti-GFP, anti-STING and anti-β-actin (A and C). The virus titers in the supernatants were detected using TCID50 assay (B and D).
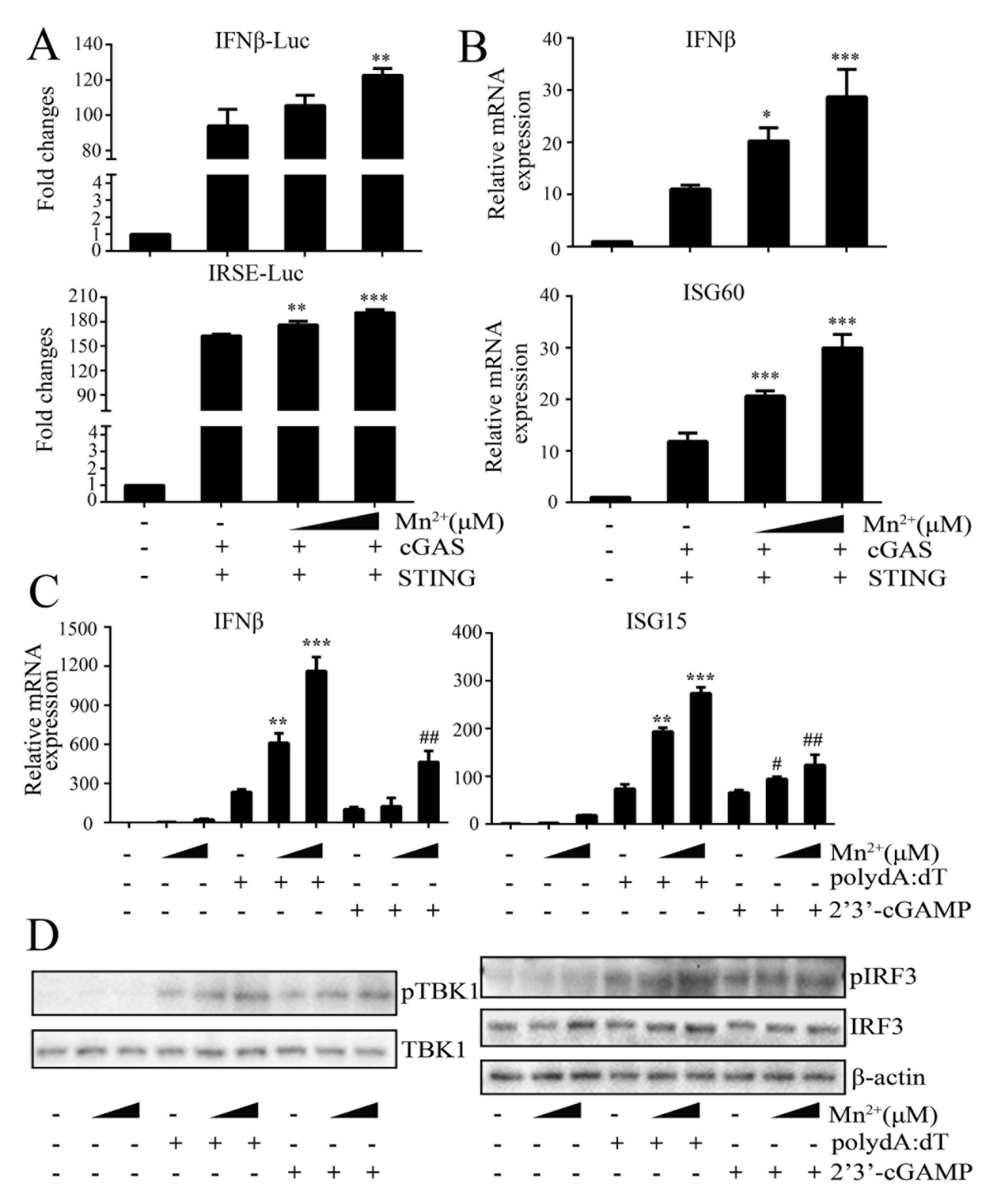
Figure 6.
Mn2+ promotes cGAS-STING signaling activity. (A) HEK293T cells in 96-well plates were con-transfected with 20 ng cGAS-HA and 10 ng STING-GFP, plus 10 ng IFNβ-luc or ISRE-luc and 0.2 ng pRL-TK plasmid for 24 h. Cells were then treated with Mn2+ (0, 100, 200 μM) for 24 h, and the luciferase activities were examined. (B) HEK293T cells in 24-well plates were co-transfected with 400 ng cGAS-HA and 400 ng STING-GFP plasmids for 24 h. Cells were then exposed to Mn2+ (0, 100, 200 μM) for another 24 h, and subjected to RT-qPCR for downstream gene detection. (C-D) 3D4/21 cells in 24-well plates were transfected with polydA:dT (1 μg/mL) or 2’3’-cGAMP (2 μg/mL) for 8 h using lipofectiamine 2000. Twenty-four hours post transfection, cells were incubated with Mn2+ (0, 100, 200 μM) for another 24 h, and then downstream gene expressions were detected using RT-qPCR (C) and Western blot analysis with the indicated antibodies (D). * or #, p < 0.05. ** or ##, p < 0.01. ***, p < 0.001.
Figure 6.
Mn2+ promotes cGAS-STING signaling activity. (A) HEK293T cells in 96-well plates were con-transfected with 20 ng cGAS-HA and 10 ng STING-GFP, plus 10 ng IFNβ-luc or ISRE-luc and 0.2 ng pRL-TK plasmid for 24 h. Cells were then treated with Mn2+ (0, 100, 200 μM) for 24 h, and the luciferase activities were examined. (B) HEK293T cells in 24-well plates were co-transfected with 400 ng cGAS-HA and 400 ng STING-GFP plasmids for 24 h. Cells were then exposed to Mn2+ (0, 100, 200 μM) for another 24 h, and subjected to RT-qPCR for downstream gene detection. (C-D) 3D4/21 cells in 24-well plates were transfected with polydA:dT (1 μg/mL) or 2’3’-cGAMP (2 μg/mL) for 8 h using lipofectiamine 2000. Twenty-four hours post transfection, cells were incubated with Mn2+ (0, 100, 200 μM) for another 24 h, and then downstream gene expressions were detected using RT-qPCR (C) and Western blot analysis with the indicated antibodies (D). * or #, p < 0.05. ** or ##, p < 0.01. ***, p < 0.001.