1. Introduction
Dermoscopy is a diagnostic imaging tool used by dermatologists to increase the reliability of skin disease diagnosis. Using huge datasets of dermoscopic images which are publicly available, a diversity of deep learning models have been dramatically developed to detect skin cancers from dermoscopic images [
1,
2].
On the other hand, the dermoscopy is not always available in local clinics due to the high cost of dermoscopes. Instead, lots of dermatologists exploit clinical skin images which can be acquired even using a general-purpose camera, in order not only to diagnose skin lesions directly from them but also to simplify the cumbersome process of choosing the best lesion from multiple lesions whose dermoscopic image should be finally taken for a dermatologist’s review.
Taking into account such benefits of clinical skin images as lower cost yet a wider field of view compared to dermoscopic images, a few machine learning models based on clinical skin images have been introduced for skin disease classification. Pacheco
et al. assessed four convolutional neural networks (CNNs) including GoogleNet, VGGNet, ResNet, and MobileNet, for skin cancer detection from a clinical image dataset they had collected [
3,
4]. The clinical image-based approach to skin disease detection is valuable to increase the possibility for telediagnosis and self-diagnosis that will be available without an expensive dermoscope before visiting a clinic.
Nonetheless, the clinical image-based approach to skin disease detection is technically challenging. It is likely to have worse performance than the dermoscopic image-based approach, since a clinical image is expected to have lower resolution and consequently to be less informative for accurate skin diagnosis than the corresponding dermoscopic image due to the lack of dermatologist-level microscopic features of skin lesions.
Reconstructing the dermoscopy-level high resolution skin image from a given clinical skin image can be taken into consideration as an efficient technical strategy to cope with the limitations of clinical image-based methods. As a substitute to genuine dermoscopic images, a synthetic dataset of dermoscopy-level super resolution (SR) images may be effectively exploited to train a machine learning model for skin disease detection. In addition, the methodology of clinical-to-dermoscopic reconstruction is practically instrumental for dermatologists to make better skin diagnosis without any dermoscope. It would also lead to the reduction in medical costs by referring to AI-based pre-diagnosis in advance before taking dermoscopic images of high cost.
The clinical-to-dermoscopic image generation can be understood as the problem of single image super resolution (SISR) by which a high resolution (HR) image is reconstructed from the corresponding low resolution (LR) image. Although SISR is an ill-posed problem which is intrinsically intractable, the most state-of-the-art deep learning models for SISR have achieved remarkable improvements in their performance [
5]. Since a super resolution model using deep convolutional networks (SRCNN) was introduced by Dong
et al. in 2014 [
6], a diverse of convolutional neural networks (CNNs) have been developed including the enhanced deep residual network (EDSR) [
7], wise-activated deep residual network (WDSR) [
8], residual dense network (RDN) [
9], large receptive field network (LRFNet) [
10], residual channel attention network (RCAN) [
11], cascading residual network (CARN) [
12], and information distillation network (IDN) [
13].
The other class of SISR networks has been developed based on the generative adversarial network (GAN). The pioneering SRGAN framework [
14] has been extended to ESRGAN [
15], cycle-in-cycle GAN [
16], and SRFeat [
17]. In contrast to the CNN-based methods which focus on pixel-level accuracy (measured by PSNR), the perceptual quality was more emphasized in the GAN-based methods. Both classes of SISR networks were effective to restore the local features, however they are still restrictive to recover global texture details with long-range dependency which cannot be easily inferred from neighboring pixels.
To make up for the weakness of existing networks, a novel SISR network based on vision transformers, so called the efficient super-resolution transformer (ESRT), was suggested by Lu
et al. in 2021, where a sequence of transformer blocks were added to restore the texture details of local regions referring the global information from distant regions after extracting high frequency features using the CNN backbone [
18].
Despite the rapid growth of SR technologies, few SISR networks have been reported to generate dermoscopic SR images from clinical LR skin images to the best of our knowledge. We evaluated a few representative SISR models including ESRT, and we found their limitations in recovering minute textures of skin lesions from LR skin images. Indeed, while ESRT is effective to preserve the long-range dependence of local features using transformers, there still exists a high risk that the perceptual quality and local texture details might be deteriorated since it is based on the
loss to minimize pixel-wise errors [
19].
In this paper, we propose a coarse-to-fine SR framework (CF-ESRT) based on efficient super resolution transformers for clinical-to-dermoscopic image reconstruction. In our proposed method, the coarse network based on the ESRT backbone is followed by the refinement network to further improve finer local textures and perceptual quality, and both networks were jointly trained in an end-to-end fashion [
20]. While only the
loss was used in ESRT, the perceptual loss and gradient loss were additionally applied to improve perceptual similarity and texture details (as difference gradient) between the predicted SR image and the corresponding ground truth.
In summary, our contribution is to show that such simple strategies as coarse-to-fine network architecture, perceptual and gradient loss were effective for the transformer-based SISR network ESRT to enhance local texture details as well as perceptual quality in SR skin images.
2. Materials and Methods
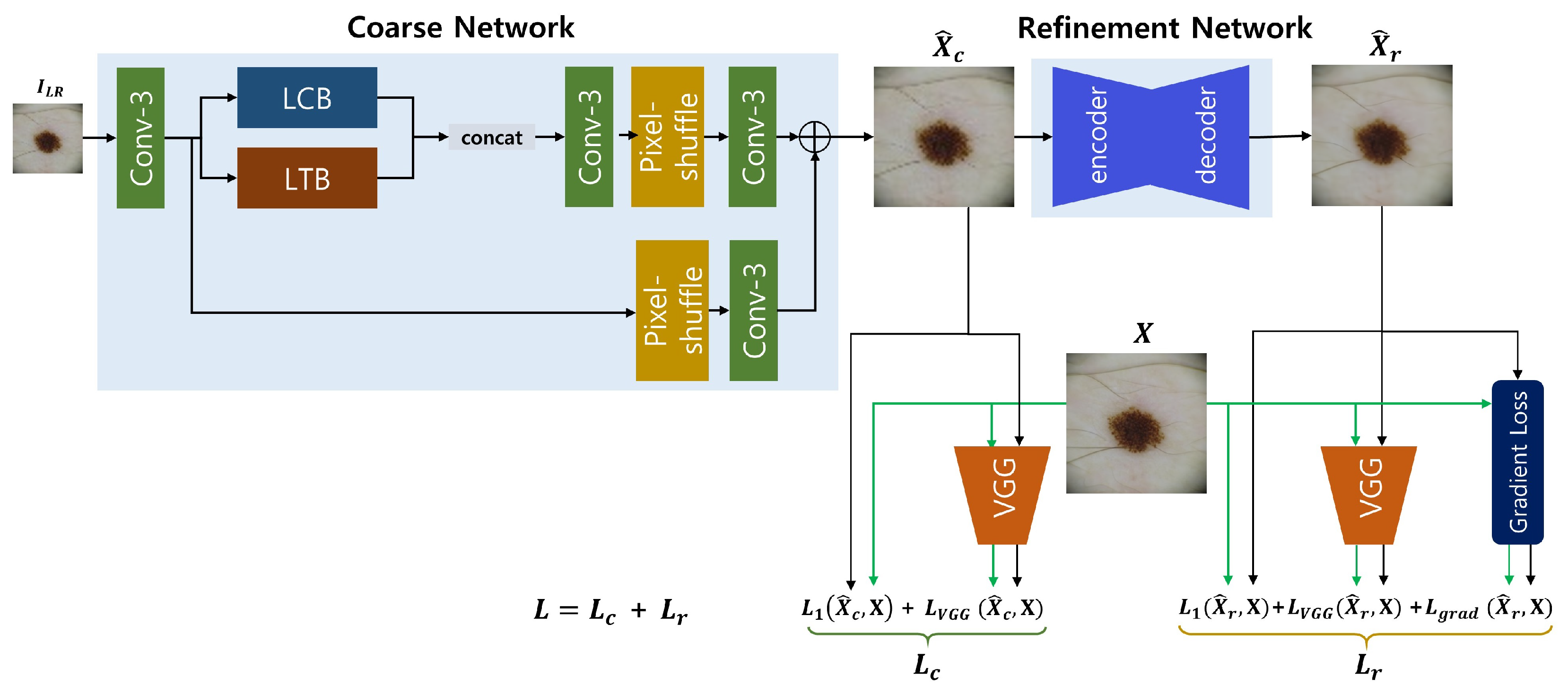
As illustrated in
Figure 1, the proposed super resolution framework for clinical-to-dermoscopic image reconstruction consists of a coarse network based on the ESRT backbone and a refinement network. The output image generated by the coarse network goes through the refinement network to achieve better perceptual quality and finer texture details. Both networks are trained in an end-to-end fashion by employing both perceptual and gradient losses to improve perceptual quality and minute texture details.
2.1. Coarse network
The coarse network aims to reconstruct the coarse features of the input image. The basic architecture of the coarse network was adopted from ESRT whose main parts are the lightweight CNN backbone (LCB) and the lightweight transformer backbone (LTB) [
18]. In the original ESRT, both elements are sequentially connected where the potential SR features extracted from LCB is sent to LTB to restore local texture details which can be inferred from the other image regions using transformers.
We made two minor changes from the original network architecture of ESRT. First, the sequential pathway of LCB and LTB was parallelized, as shown in
Figure 1, assuming that both features from LCB and LTB had better be simultaneously used for SR image reconstruction. Second, the multi-level residual connection architecture of LCB was simplified by removing redundant residual connections. LCB comprises of a sequence of high preserving blocks (HPBs) to extract high frequency features. HPB is composed of a series of adaptive residual feature blocks (ARFBs) as well as high-frequency filtering module (HFM), that is designed to reduce computational costs by applying the reduction-expansion scheme (i.e., downsampling-upsampling) to several network levels. The conventional HPB in ESRT has multi-level residual connections in that HPB, its element ARFB, and ARFB’s sub-units have residual connections [
18]. Supposing that the multi-level residual connection architecture has no effects on extracting potential SR features which are valuable to improve the performance, we used a simplified HPB (sHPB) by removing the residual connection from the superior level of HPB.
LTB is composed of a series of efficient transformers (ETs) which are the encoder parts of the standard transformer [
18]. After a feature map is unfolded into a set of overlapping 2D patches in a similar manner as convolution, each patch is sent to the ETs after being flattened and all patches encoded by ETs are folded together to reconstruct the feature map.
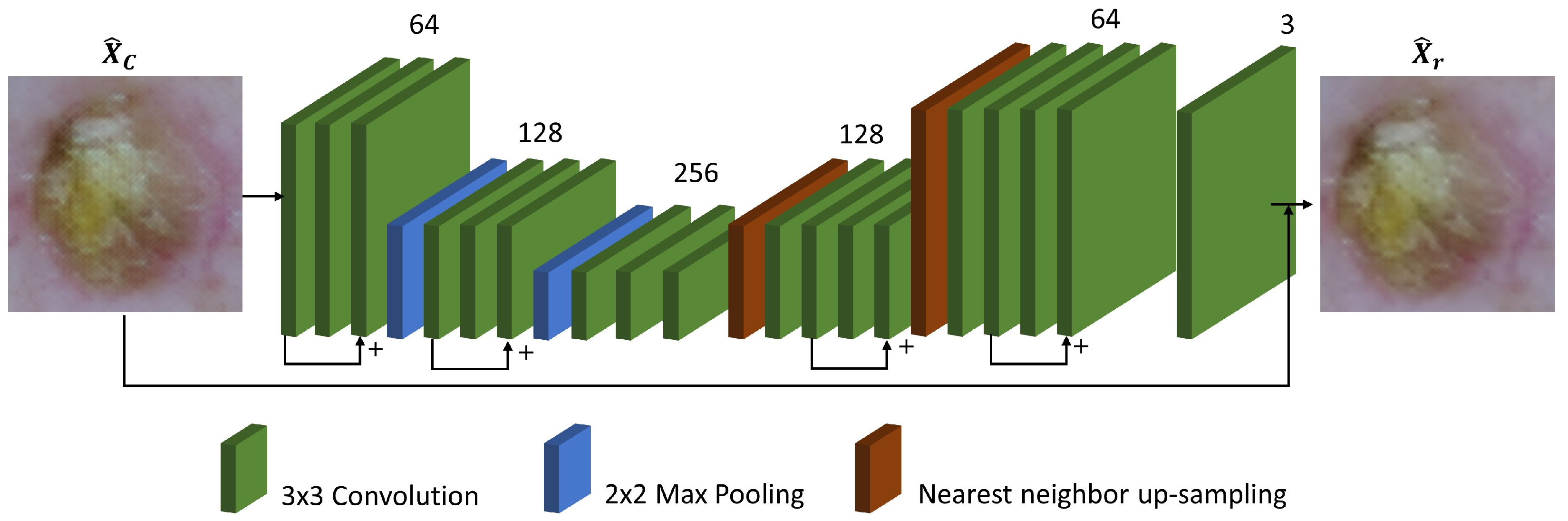
2.2. Refinement network
The refinement network aims to reduce the discrepancy in perceptual quality and texture details between the draft SR image predicted from the coarse network and the corresponding ground truth. As shown in
Figure 2, the refinement network has an encoder-decoder architecture with residual connections which may be beneficial to propagating information over layers, in a similar manner with the framework introduced by Kim
et al. [
20]. Max pooling with a 2 window and nearest neighbor interpolation were used for downsampling and upscaling respectively. All convolution filters are of size
and stride 1, and the number of channels was indicated for each convolution layer.
2.3. Loss function
The loss function
for the coarse network consists of the
loss reflecting pixel-wise errors and the perceptual loss reflecting VGG16 feature domain errors, as given in the equation
where
is an input image,
is the output image from the coarse network,
is a constant and
is a feature map from the third convolution layer at the fifth block in VGG16 [
21].
On the other hand, the loss function
for the refinement network consists of not only the
loss and the perceptual loss but also the gradient loss reflecting errors in high frequency details, which is given as
where
is the output image from the refinement network, and both
and
are constants. The gradient loss
is given as
where
and
denote image gradients in horizontal and vertical directions respectively. Then, the entire coarse-to-fine framework is trained in an end-to-end fashion with the total loss
[
22].
3. Results
3.1. Implementation and datasets
The dermoscopic image dataset ISIC2019 of 25,331 images was used as a training dataset, and 20% of the dataset was used for quantitative evaluation [
23]. The clinical skin image dataset PAD-UFES-20 was used for qualitative evaluation [
4].
Our method was implemented in Pytorch by extending the original ESRT, and trained on 8 NVIDIA RTX A6000 with the mini-batch size of 112. The learning rate was initially set to be 0.0002 and reduced in half every 20 epochs. The Adam optimizer was utilized to optimize the training process [
24]. The proposed network was compared with bicubic interpolation, SRCNN [
6], SRGAN [
14], and ESRT [
18].
In the first phase of training, both coarse and refinement networks are jointly trained in an end-to-end fashion. As a training image dataset, each dermoscopic image was downsampled into the size of
and used as ground truth. The corresponding input LR images of
were generated by downsampling the ground truth images using the bicubic method. In the second phase, the coarse network is frozen but only the refinement network is additionally trained using image patches of
split from original dermoscopic images as ground truth. Unlike the first phase of training, the perceptual loss was excluded from the refinement loss
defined in equation
2.
3.2. Quantitative and qualitative evaluations
The peak signal-to-noise ratio (PSNR) and structural similarity (SSIM) were used to assess the quantitative performance, while the Frechet Inception Distance (FID) and the learned perceptual image patch similarity (LPIPS) were used to assess the perceptual quality [
25,
26].
In
Table 1, the pixel-wise errors (PSNR) and the structural similarity (SSIM) in the proposed method were similar to SRCNN and ESRT. On the other hand, the perception-related metrics FID and LPIPS were significantly improved using the proposed coarse-to-fine ESRT network.
Indeed,
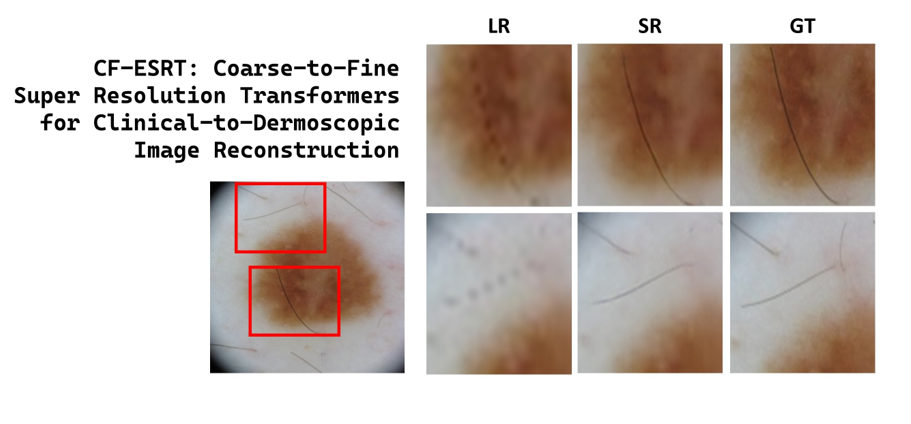
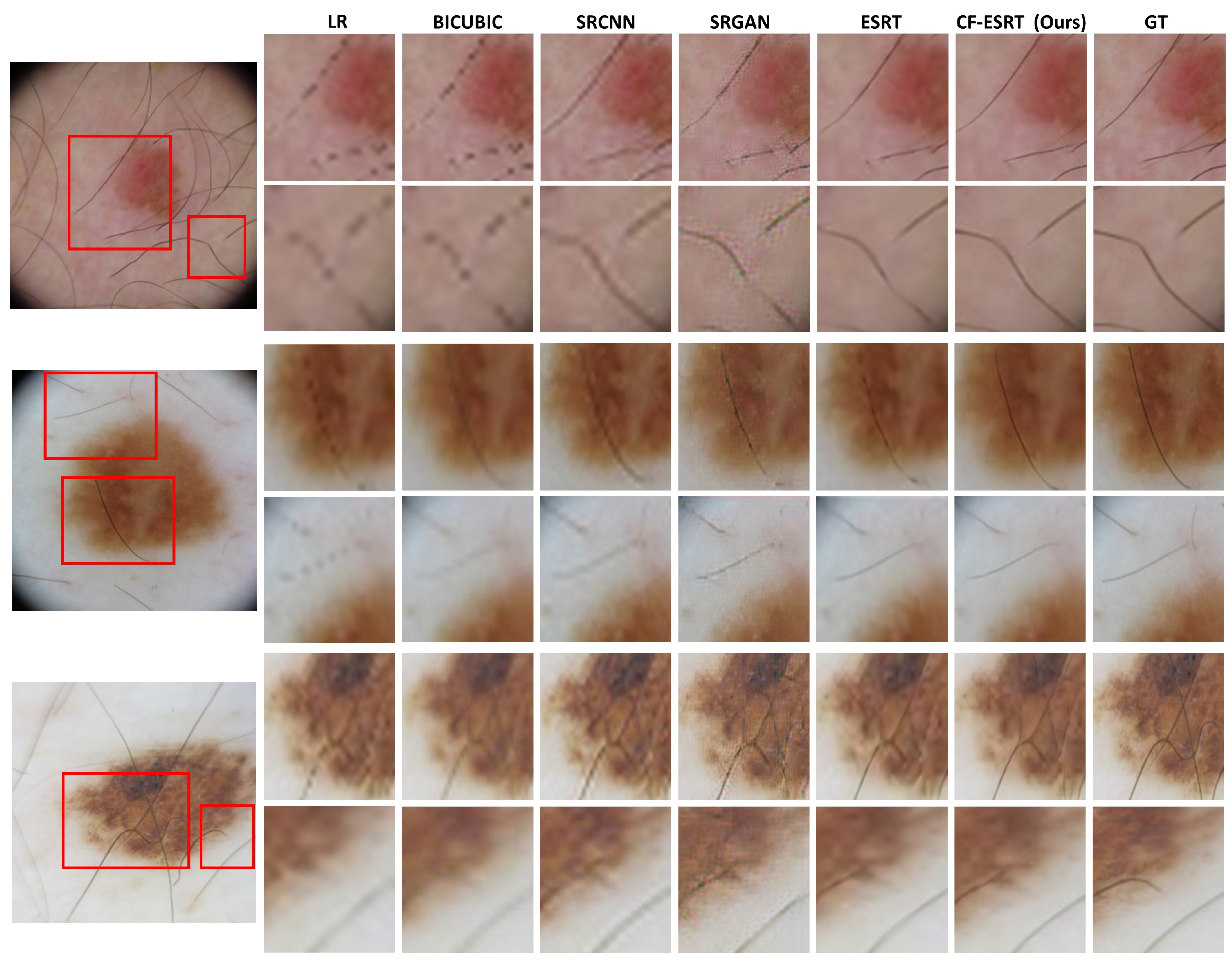
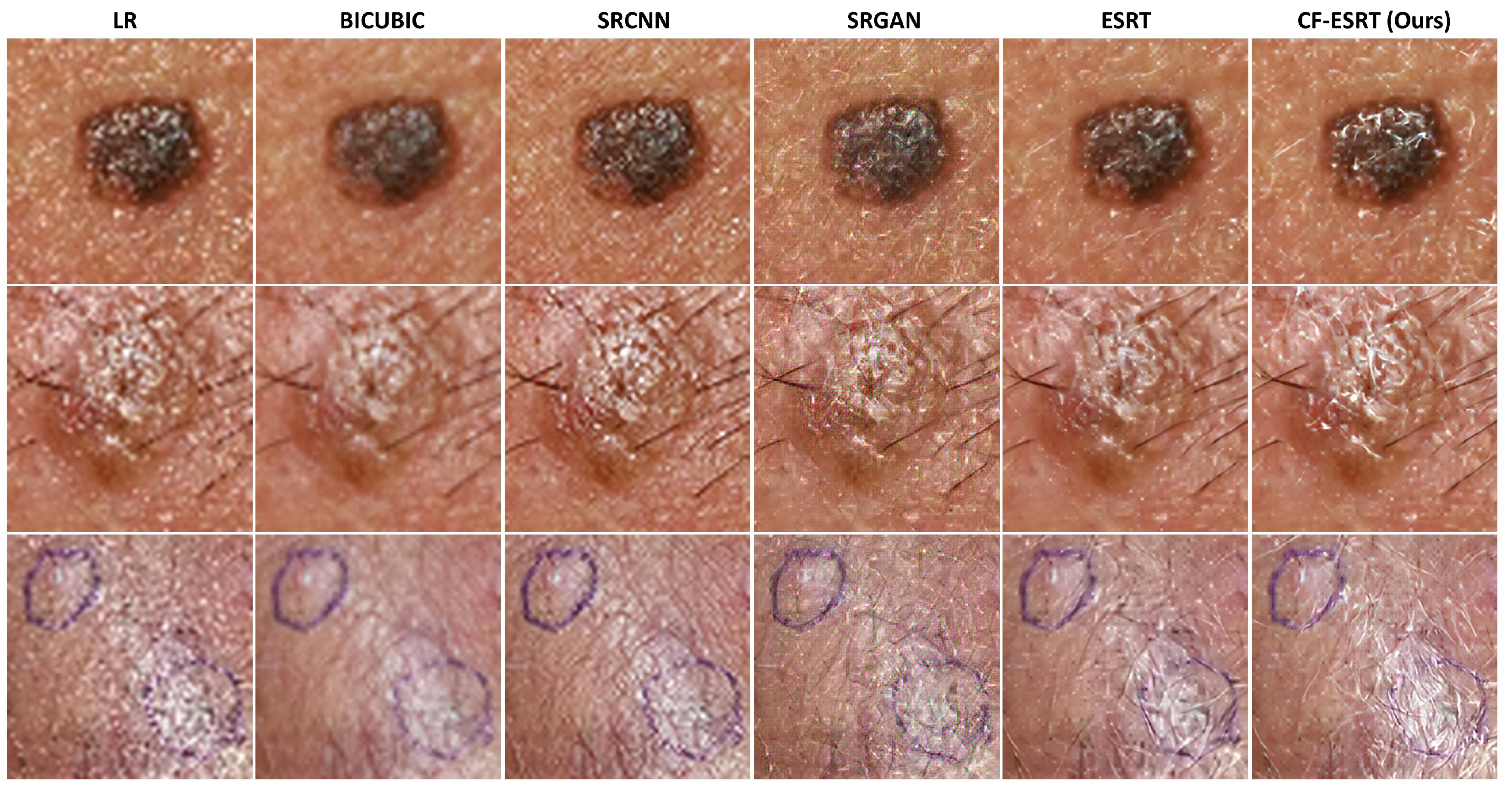
Figure 3 shows the visual comparison of SR skin images which were reconstructed from the LR skin images downsampled from dermoscopic images in ISIC2019.
Figure 4 shows SR skin images reconstructed from real clinical images in PAD-UFES-20. In both results, our proposed method accurately recovers minute texture details of skin lesions as well as global structure while the other state-of-the-art methods tend to produce either blurry or erroneous results.
We also conducted a survey on visual quality of 10 SR images from 15 users. Users were asked to give a grade between 1 and 5 on how much realistic the given image is.
Table 1 shows that the mean opinion score (MOS) of the proposed network was prominently superior to the other methods.
3.3. Ablation study
We analyzed the effect of perceptual and gradient losses on the model performance. As shown in
Table 2, PSNR and SSIM were slightly decreased but both FID and LPIPS were significantly improved by using the full loss including perceptual and gradient losses defined in
Section 2.3, compared to the case of using
loss only. This indicates that both perceptual and gradient losses have a significant impact on improving the perceptual quality of the generated SR images.
4. Conclusion
We proposed an extension of the efficient super resolution transformers (ESRT) for clinical-to-dermoscopic image reconstruction by adding the refinement network to the original ESRT and enforcing the perceptual and gradient losses. The proposed coarse-to-fine ESRT network exhibited a significant improvement in perceptual quality metrics.
Limitations of the proposed method deserve to be mentioned. First, the quantitative performance of the proposed network was not significantly improved as shown in PSNR and SSIM. Despite the outstanding enhancement of perceptual quality, the quantitative metrics should not be negligible in that the pixel-wise accurate reconstruction of dermoscopic images is of dermatologists’ great interest especially for clinical purpose. Second, the SR image reconstructed from a clinical skin image was not directly assessed compared to the real dermoscopic image due to the absence of ground truth. The model assessment for clinical skin images is meaningful because dermoscopic images are acquired through microscopic imaging devices and consequently have different optical interpretations from clinical skin images [
27]. However, few public datasets including both dermoscopic images and the corresponding clinical images are currently available to evaluate a super resolution model.
In those reasons, the clinical-to-dermoscopic image reconstruction remains still challenging, nevertheless our study lays an important and pioneering foundation for realizing an AI-assisted low-cost skin disease diagnosis tool which may be practically useful in underdeveloped countries.
Author Contributions
Conceptualization, W.Y.; methodology, W.Y.; software, B.L.; validation, B.L., W.Y.; formal analysis, B.L.; investigation, B.L.; writing—original draft preparation, W.Y.; writing—review and editing, W.Y.; visualization, B.L., W.Y.; supervision, W.Y.; project administration, W.Y.; funding acquisition, W.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the BK21 FOUR (Fostering Outstanding Universities for Research), the Basic Science Research Program (NRF-2022R1F1A1075204), and the Regional Innovation Strategy Project (2021RIS-004), funded by the Ministry of Education(MOE, Korea) and National Research Foundation of Korea(NRF), and was also supported in part by the Start-up Growth Technology Development Project (S3228660) funded by the Ministry of SMEs and Startups.
Data Availability Statement
The source codes presented in this study are available on request from the corresponding author, and will be released publicly in a near future.
Acknowledgments
The authors thank Soo Ye Kim from Adobe Research and Hyunjae Zhang from F&D Partners Corporation, for insightful discussions on image inpainting and its industrial demands, and Youngchan Lee and Gyubin Lee from AIIP Lab, for assisting data processing.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gulati, S.; Bhogal, R.K. Classification of Melanoma from Dermoscopic Images Using Machine Learning. 2020, Vol. 159. [CrossRef]
- Ünver, H.M.; Ayan, E. Skin lesion segmentation in dermoscopic images with combination of yolo and grabcut algorithm. Diagnostics 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.G.; Krohling, R.A. The impact of patient clinical information on automated skin cancer detection. Computers in Biology and Medicine 2020, 116. [Google Scholar] [CrossRef]
- Pacheco, A.G.; Lima, G.R.; Salomão, A.S.; Krohling, B.; Biral, I.P.; de Angelo, G.G.; Alves, F.C.; Esgario, J.G.; Simora, A.C.; Castro, P.B.; et al. PAD-UFES-20: A skin lesion dataset composed of patient data and clinical images collected from smartphones. Data in Brief 2020, 32. [Google Scholar] [CrossRef]
- Li, K.; Yang, S.; Dong, R.; Wang, X.; Huang, J. Survey of single image super-resolution reconstruction. IET Image Processing 2020, 14. [Google Scholar] [CrossRef]
- Dong, C.; Loy, C.C.; He, K.; Tang, X. Image Super-Resolution Using Deep Convolutional Networks 2014.
- Lim, B.; Son, S.; Kim, H.; Nah, S.; Lee, K.M. Enhanced Deep Residual Networks for Single Image Super-Resolution. 2017, Vol. 2017-July. [CrossRef]
- Fan, Y.; Yu, J.; Huang, T.S. Wide-activated deep residual networks based restoration for BPG-compressed images. 2018, Vol. 2018-January.
- Zhang, Y.; Tian, Y.; Kong, Y.; Zhong, B.; Fu, Y. Residual Dense Network for Image Super-Resolution. 2018. [CrossRef]
- Seif, G.; Androutsos, D. Large receptive field networks for high-scale image super-resolution. 2018, Vol. 2018-June. [CrossRef]
- Zhang, Y.; Li, K.; Li, K.; Wang, L.; Zhong, B.; Fu, Y. Image super-resolution using very deep residual channel attention networks. 2018, Vol. 11211 LNCS. [CrossRef]
- Ahn, N.; Kang, B.; Sohn, K.A. Fast, accurate, and lightweight super-resolution with cascading residual network. 2018, Vol. 11214 LNCS. [CrossRef]
- Hui, Z.; Wang, X.; Gao, X. Fast and Accurate Single Image Super-Resolution via Information Distillation Network. 2018. [CrossRef]
- Ledig, C.; Theis, L.; Huszar, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-Realistic Single Image Super-Resolution Using a Generative Adversarial Network. 2016. [Google Scholar]
- Wang, X.; Yu, K.; Wu, S.; Gu, J.; Liu, Y.; Dong, C.; Qiao, Y.; Loy, C.C. ESRGAN: Enhanced super-resolution generative adversarial networks. 2019, Vol. 11133 LNCS. [CrossRef]
- Yuan, Y.; Liu, S.; Zhang, J.; Zhang, Y.; Dong, C.; Lin, L. Unsupervised Image Super-Resolution using Cycle-in-Cycle Generative Adversarial Networks. 2018. [Google Scholar]
- Park, S.J.; Son, H.; Cho, S.; Hong, K.S.; Lee, S. SRFeat: Single Image Super-Resolution with Feature Discrimination. 2018, Vol. 11220 LNCS. [CrossRef]
- Lu, Z.; Li, J.; Liu, H.; Huang, C.; Zhang, L.; Zeng, T. Transformer for Single Image Super-Resolution. 2021. [Google Scholar]
- Ma, C.; Rao, Y.; Cheng, Y.; Chen, C.; Lu, J.; Zhou, J. Structure-preserving super resolution with gradient guidance. 2020. [CrossRef]
- Kim, S.Y.; Aberman, K.; Kanazawa, N.; Garg, R.; Wadhwa, N.; Chang, H.; Karnad, N.; Kim, M.; Liba, O. Zoom-to-Inpaint: Image Inpainting with High-Frequency Details. 2020. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. 2015. [Google Scholar]
- Eigen, D.; Fergus, R. Predicting Depth, Surface Normals and Semantic Labels with a Common Multi-Scale Convolutional Architecture. 2014. [Google Scholar]
- Cassidy, B.; Kendrick, C.; Brodzicki, A.; Jaworek-Korjakowska, J.; Yap, M.H. Analysis of the ISIC image datasets: Usage, benchmarks and recommendations. Medical Image Analysis 2022, 75. [Google Scholar] [CrossRef] [PubMed]
- Kingma, D.P.; Ba, J.L. Adam: A method for stochastic optimization. 2015.
- Goswami, S.; N, R.A. Robust Unpaired Single Image Super-Resolution of Faces. 2022. [Google Scholar]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The Unreasonable Effectiveness of Deep Features as a Perceptual Metric. 2018. [Google Scholar]
- Reiter, O.; Kurtansky, N.; Nanda, J.K.; Busam, K.J.; Scope, A.; Musthaq, S.; Marghoob, A.A. The differences in clinical and dermoscopic features between in situ and invasive nevus-associated melanomas and de novo melanomas. Journal of the European Academy of Dermatology and Venereology 2021, 35. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).