1. Introduction
Millions of cells are found in the human body, and each category has its particular role like the light perception, uptake of oxygen, nerve conduction, and protection mechanism. There are present 23 pairs of sub-cellular structures, called chromosomes, within the nucleus of all cells that mark up the human body and are responsible for transforming genetic information from generation to generation. There are two types of chromosomes in humans, i.e., the sex chromosomes (X and Y) and the autosomes. The sex chromosomes determine the gender of a person. A male child receives a Y chromosome from his father and X from the mother, whereas a female child inherits X chromosomes from her parents [
1]. Chromosomes are made up of firmly packed chains of deoxyribonucleic acid (DNA) that store the genetic information for protein synthesis.
A specific regulatory mechanism at the DNA level controls which protein will be formed in what amount and at what time. Proteins are made up of amino acids and are responsible for the structure and function of the different body organs. Not all proteins are required in every cell, and the gene coding for the desired protein is switched off in other cells [
2]. Different forms of the identical gene are termed alleles. All individuals can inherit only one alternate form of a gene. The mutual effect of alleles results in various physical appearances among individuals like eye color, hair color, and the shape of the nose [
3,
4]. Alternation in gene sequence affects protein function that ultimately loses its ability to control the specific phenotype. That dysfunctional gene performance is termed as mutation, which is responsible for diseases. Diseases can occur due to a fault in a single gene or a set of genes. Furthermore, if the gene mutations occur in the egg or sperm cell, the children may inherit the faulty gene from their parents [
5]. According to the grade of gene mutation, the mutations are characterized into the following types:
Chromosomal diseases occur when the complete chromosome or large sections of a chromosome are lost, duplicated, or otherwise changed. For example, Down’s syndrome is an example of chromosomal irregularity.
Single-gene disorders occur when gene alteration affects one gene, like sickle-cell anemia, where a defect in the Hemoglobin gene affects the red blood cells.
Multifactorial disorders occur as a result of mutations in several genes, generally linked with conservational reasons. For example, diabetes is a multifactorial syndrome.
Mitochondrial syndromes: are rare illnesses caused by mutations in non-chromosomal DNA found within the subcellular organelles, i.e., mitochondria. These disorders can be found to affect any part of the body like the brain and the muscles.
Genetic disorders can be managed by the variations in the genome that are usually interconnected with the disease. While considering the ethnic-specific (regional and geographical-based) Mendelian disease, various genetic disorders arise, such as Thalassemia and β- Thalassemia, which has the highest ratio among South Asia [
6,
7]. Other common Mendelian diseases occur due to consanguinity. Research has been conducted on recessive genetic dysfunction with interfamilial marriages in Asian and Arab societies [
8].
The completion of the human genome sequence utilized [
9] the genomic techniques for detecting gene mutation diseases. Different databases represent gene-related information such as gene disorder, protein domains, interspecies DNA sequence comparisons, and associated conditions in literature [
10]. Concurrently, advances in technology have led to the identification of human gene mutation and variations. The knowledge and organization of these alterations in structured knowledge-base will be crucial for diagnosis and clinicians and researchers. Genetic or mutation databases are referred to as online repositories of genomic variants that are described for a single (locus-specific) or multiple (general) genes or specifically for a population or ethnic group (national/ethnic). The following are the primary applications of mutation databases i.e. 1) to facilitate diagnosis at the DNA level and to define an optimal strategy for variant detection, 2) to provide information about variant-specific phenotypic patterns, and 3) to correlate locus-specific variant information with genome-wide features. The available online databases use different sources for collecting the information of other communities are classified in central online databases i.e. Online Mendelian Inheritance in Man (OMIM) [
11] and the Human Gene Mutation Database (HGMD) [
10]. These databases only contain the published mutations. While databases that are made for specific loci are called locus-specific databases (LSDBs) [
12]. LSDBs may not collect the information of a particular nation or ethnic. Other databases are limited to ethnic, country, or geographic region called national mutation databases (NEMDBs). The NEMDBs which have recently emerged, aim to record the varying mutation spectrum observed for any gene (or multiple genes) associated with a genetic disorder across different population groups around the world. The Human Genome Variation Society (HGVS) has a dedicated and ever-growing website, and the last updated by March, 2021. The list of databases in HGVS is not updated for years. The page for national and ethnic mutation databases was visited on March 12th, 2022. It was fund that this page comprises a total of 11 links, but only four of these links are actually working. On the other hand, the page for "Locus-specific mutation database" holds 1,646 links and the total mutations were found to be 145, 964 in number.
The technology of genetic analysis and accomplishment of human genome sequencing has expressively contributed to finding the genomic variations. With the growth of genetic disease identification, there is a need to gather quality control and document all the related information regarding mutation. Such mutation data and their corresponding phenotype are the guarantee for future accessibility of this information to medicinal experts, researchers, and clinicians. Integrating discoveries into existing databases makes them depositors and guardians of our science and critical elements for the progress of scientific research [
13]. The availability of NEMDBs provides beneficial information to the patients and clinicians of specific ethnic groups. To the best of our knowledge, there is no recent paper available that provides a detailed review of ethnic details, methods and materials, and cross-comparison of the available NEMDBs. This paper aims to present state-of-the-art ethnic-specific or national mutation databases, their sources of data collection, Contents availability in these databases, and their ways of querying data. Also, the paper analyzes the comparison between available NEMDBs. In conducting the study, we used "scholar.google.com" as a search tool for gathering the papers published in the 1990s to 2021. We have used the strings "mutation database, "gene disorder," and "genetic disease" for searching the papers.
The rest of the article is organized as follows:
Section 2 presents the detailed background of the available NEMDBs. In section 3, the methodology along with descriptive details of all the NEMDBs is described.
Section 4 is about concluding the review and future recommendations. Finally, in
Section 5, the article is concluded.
2. Background Study
Bioinformatics recently emerged in science, in which Molecular biology, Information technology, Computer science, and Mathematics merge to form a single discipline. Database designing, categorization, protein structure prediction, RNA folding, mutation map generation are certain areas controlled and organized by bioinformatics. A biological database typically contains structured and persistent data generally linked with computerized software. The database may be utilized for retrieving, updating, storing, and querying the data within the system. Margaret Dayhoff first established the protein sequence database in the 1960s; afterward, DNA sequencing systems in the 1980s developed GenBank as the first nucleotide sequence database [
14]. The genetic material's quality data collection and records are constantly updated in a knowledge base known as mutation databases. The aim of mutation databases was too imminent accessible of such data by medicinal professionals, non-professionals, clinicians, researchers (carried out a study on genetic variations) [
15]. Bioinformatics mainly involves the experimental design of the analysis of biomedical literature. PubMed is one of the most common scientific databases, hosted by the National Center for Biotechnology Information (NCBI) in 1997. It contains several medical-related articles [
16]. PubMed gives access to 38 different databases concerning biomedical research and the analysis of erratic genetic diseases. In addition, there are some other repositories related to gene-disease such as MeSH, ISI web, Medline, which provide comprehensive data about a particular gene and disease [
13]. Access to such databases is publically available for all users. Mainly PubMed contains biologically related subject data, but it also holds a large number of interdisciplinary subjects [
17]. PubMed is one the most influential and updated sites in bioinformatics consisting of the web-based system, i.e., PubMed Assistant [
18], Alibaba [
19], and PubMed-Ex [
20]. PubMed Assistant carries different features like keyword highlighting and direct export to citation managers. Whereas Alibaba and PubMed-EX are semantically enriched over categorizing gene, protein, disease, and other biomedical entities from the text [
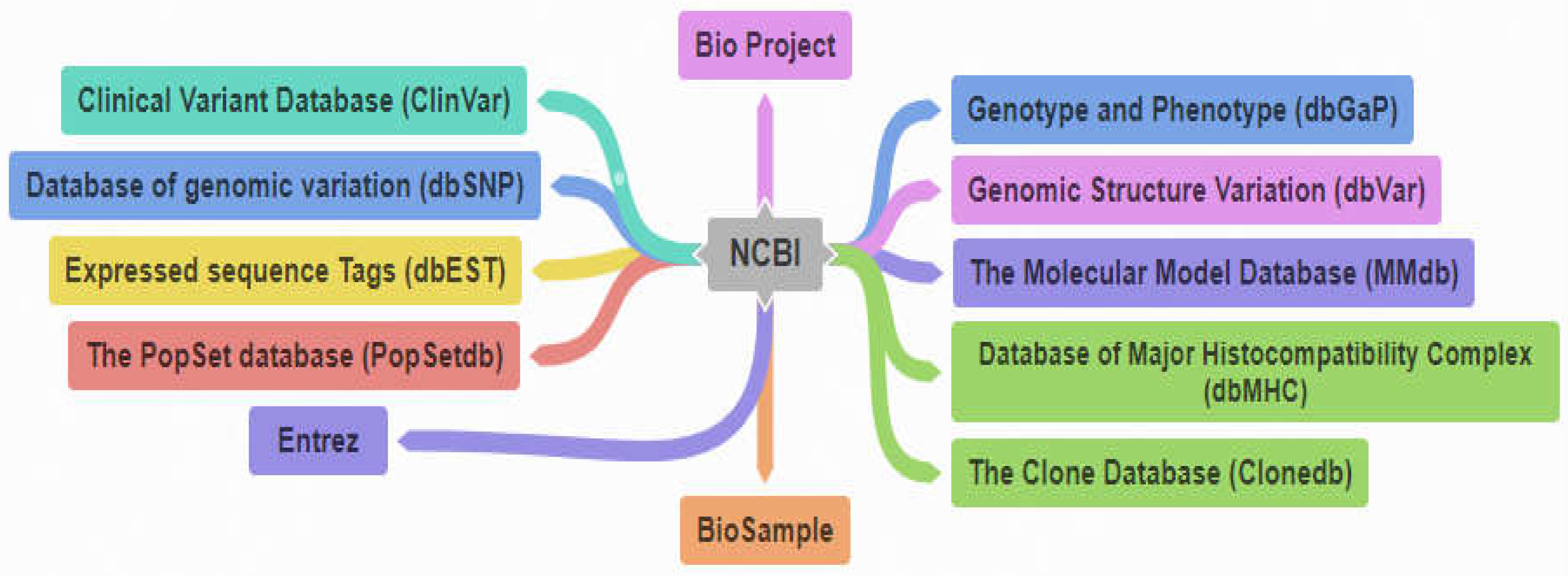
21]. National Institutes of Health, in 1988, established "The National Center for Biotechnology Information (NCBI)," a central system for different resources and databases that can be accessed through the NCBI web page at
www.ncbi.nlm.nih.gov. The primary resources in the NCBI include dbSNP (Database of Short Genetic Variations) database, Database of Genomic Structural Variation (dbVar), Entrez (Entrez is an integrated database retrieval system that gives access to a diverse set of 35 databases), Clone database (CloneDB), BioProject Database [
13,
22], and clinical central variant database (ClinVar) [
23]. A brief description of different NCBI database are shown in
Table 1. The various databases controlled and working under NCBI are graphically represented in
Figure 11.
Table 1 shows the various databases that are controlled under NCBI. In this table, the first column holds the references for the various NCBI controlled databases, second column contains the specific names of these databases while the third column hold the brief description and URLs of these databases.
3. Catalog of Human Variation Databases
Mutation databases are knowledge-base where allelic variations are defined and assigned within an explicit gene. Generally, three types of databases are accessible, i.e. central database and locus databases, and ethnic databases [
31]. The primary mutation database consists of shared info on genome variation and designed different tools to analyze previously collected data.
3.1. Cenetral Databases
The first mutation database, "Online Mendelian Inheritance in Man (OMIM) was begun in the 1970s by Prof. Victor McKusick. OMIM usually collects only the important mutations, containing information about phenotypes, gene function, and allelic variants which is useful for researchers, students, and clinicians [
11]. OMIM has been frequently updated over time and it cn be easily accessed using the link,
https://omim.org/. The OMIM statistics of April 11th, 2022 the updated version of OMIM consists of 26,057 entries. The OMIM entry is specified by a unique six-digit number. The phenotype entries and gene entries are separated by a special symbol. OMIM recorded the allelic variants, clinical synopsis, and gene map locus, and the variants are easily searched by OMIM number, gene disorder, gene name, simple text. The general review of OMIM was presented by [
32] describing the content, advanced features, database organization, and modification of gene architecture. All the contents are peer-reviewed and curated by journals and researchers.
Another well-known database is HGMD (The Human Gene Mutation Database) established in 1996 for the study of mutation disorders in human genetics [
33]. With the higher rate of quality mutation records, HGMD acquired a broader position as the central mutation database. HGMD provides all known gene lesions causing human inherited disease published in the peer-reviewed literature. HGMD data have been widely used by international collaborative research projects, as well as for clinical settings [
34]. These studies help to improve the mutational spectra in human genetics. HGMD includes the mutation causing human inherited disease along with location, frequency, and local DNA frequency environment [
35]. HGMD updates the version of the databases frequently. HGMD is accessible in two versions: The public version of HGMD (
http://www.hgmd.org) is freely accessible by registered users from academic institutions. The Professional version is offered for commercial and educational/non-profit users by subscribing to BIOBASE GmbH (
http://www.biobase-international.com) and under license via QIAGEN Inc [
36]. HGMD professional version facilitates users by providing a feedback function in case of missing or new data, requesting changes, or asking for analysis of listed variants. In addition, HGMD Professional version offers more advanced features than the public version. The latest version of HGMD is released in 2017 and the statistics over April 2021, the database consists of 352731 gene lesions entries in HGMD professional release, where 234987 entries in academic/non-profits curated manually and from journals.
3.2. Locus-Specific Mutation Databases (LSDBs)
In contrast to OMIM, and the HGMD, Locus-Specific Databases (LSDBs), also referred to as Gene variant databases usually curate information on sequence variants (mutations) disorder. LSDBs are considered the first comprehensive database containing the record of a particular gene locus initiated in 1976; hemoglobin mutations were originated for publishing as a Syllabus of Human Hemoglobin Variants. LSDBs are most commonly used in the context of DNA-based diagnosis and to give an up-to-date overview of the genetic variants to clinicians, scientists, and patients. The main purpose of LSDBs includes 1) quality data collection, analyzing, and reporting, 2) validating of all types of data having different variants, 3) results estimation and 4) transparency.
Gene variant databases two have advantages over central databases. 1) Most LSDBs are accessed publically, as they are mainly supported by academic researchers, who aim to share genetic information broadly. 2) An expert on a specific gene mutation or particular gene family is required to assess the biological significance of the variant sequence information deposited in LSDBs [
37]. Mainly Gene variant databases focus on a different variation of a single gene and are generally governed by a group of researchers having scientific expertise and knowledge about a particular gene or phenotype. The experts regularly curate the published and unpublished mutations in locus-specific databases (LSDBs). Generally, LSDBs clearly explained the content and the purpose of the database having active links/cross-references to other mutation databases for clinical information and PubMed/Medline for additional information. Hence, to guarantee that quality data are submitted to the mutation databases, most LSDBs maintain a standard set of data fields i.e. exon number, mutation description, mutation identification technique, and its origin (familial, sporadic, de novo) [
38,
39]. The main source of data for completing LSDBS is direct submission, published literature, and other variant databases (i.e. OMIM, dbSNP, and HGMD). Generally, the data is curated being obtained from publications or the data that a curator finds in their own institute. PubMed is one of the best way to for searching gene related articles using keywords such as “mutation” [
40].
Generally, the genetic databases system has been supported by various “LSDB-in-a-box” over time. This approach was used as a solution intended to achieve the aim of database creation and has encompassed Universal Mutation Database (UMD) [
41], MUTbase [
42], and Mutation Storage and Retrieval (MuStaRt
TM) [
43], and LOVD [
44].
The widely accepted LSDB-in-a-box named LOVD (
http://www.lovd.nl/) is used as the most solution and is freely available software. LOVD has been updated over time and was released in December 2012. The LOVD 3.0 is mainly used as a tool for gene-centric group and for displaying variants of DNA. Also, it also provides space for storing patient-centric data and NGS data, even of variants that lie outside of genes. A particularly attractive feature of LOVD is that its creators have established a database for most human protein-coding genes on their servers (
http://databases.lovd.nl/whole_genome/genes) and have invited interested parties to assume responsibility for maintaining databases for one or more genes of interest.
3.3. The National and Ethnic Mutation Databases (NEMDBs)
Different mutation spectrums could be because of another genetic disorder in particular population groups across the world, which provides a diverse dimension for the researchers. Such range is led to another group of mutation frequency databases i.e., The National and Ethnic Mutation Databases (NEMDBs) [
14], which comprises extensive genetic heterogeneity of the specific ethnic group. The Human Genome Variation Society (HGVS) maintains a dedicated and ever-growing website (
www.hgvs.org), documenting a catalog of central databases, LSDBs, and NEMDBs. Regional or Ethnic Databases offers valuable information for the literature history of population, testing genetics and association of diseases. The National and ethnic specific databases (NEMDBs) are graphically represented in
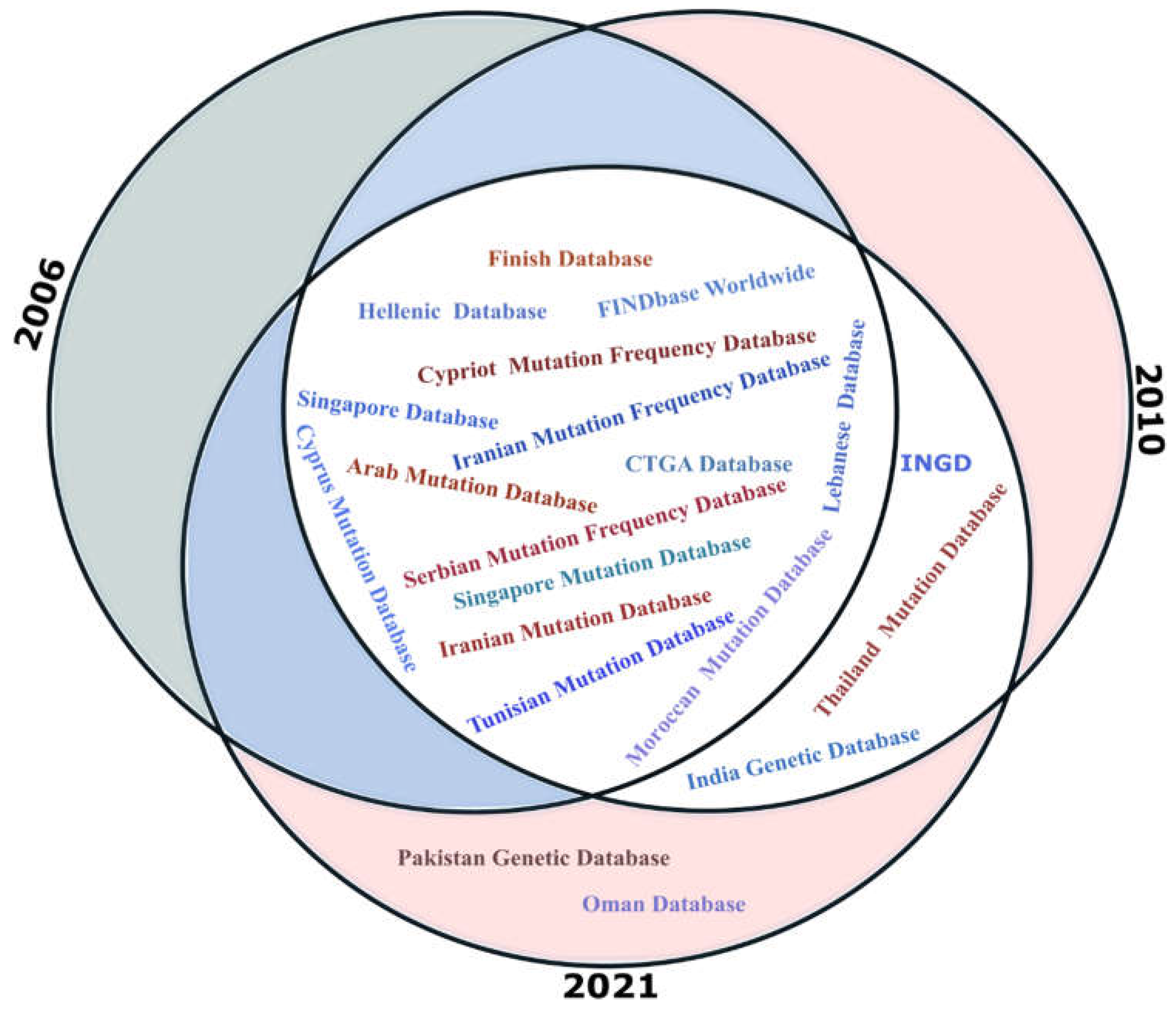
Figure 22.
The range of mutations of a particular ethnic group helps develop strategies for detecting mutations or exploring the related diagnosis. Moreover, it brings consciousness of genetic disorders and provides information about the gene-related history of humans. The range of mutations observed for genes often differs not only between populations around the world but also varies within a geographical region, between ethnic groups of databases. NEMDBs contain information that can be used to stratify national molecular diagnostic services, study human demographic history, admixture patterns, and gene/mutation flow, among others [
45]. Designing such databases aims to find novel mutation screening in ethnic-specific groups via good coordination of genetic testing [
46]. For example, in various Mendelian disorders, the disease is caused due to a single gene mutation. On the other hand, each ethnic group has more chances of certain conditions than other nations, such as cystic fibrosis in Caucasians, hemochromatosis in Jews, sickle cell anemia in people of Negroid, and Thalassemia in the Mediterranean and Southeast Asian population [
47,
48]. Moreover, in hemoglobin, genes reveal different mutation occurrences in the same ethnic and geographical group of people. Mainly NEMDBs are alienated into two categories i.e. National Mutation Genetic Databases (NMDBs) that record the existing gene of ethnic society but a small number of frequency mutations.
Table 2 shows the listed NMDBs reported by [
49].
A list of various national mutation databases (NMDBs)showed in table.
While the other category is Nation Mutation Frequency Databases are (NEMDBs), which observed the frequency of inherited mutation in various populations of an ethnic group [
49,
53]. NEMDBs benefit the community by providing vital information for the design and implementation of mutation disease detection as well as raising awareness among clinicians, bio-scientists, and the general public about the range of most common genetic disorders that affect specific populations and ethnic groups.
Table 3 holds some common National or ethnic mutation databases (NEMDBs).
A brief description of NEMBDs are listed in the
Table 3. Most of genetic databases are frequently updated, but few of them are not accessbile.
4. Materials and Methods
A biological database has a structured form of persistent data that is generally linked with computerized software. These databases contain much information that they are referred to as knowledge-base. For curating published and unpublished mutations, these databases require an increasing number of experts in the mutations and diseases of specific genes. Mostly, mutation databases had web-based access that shows and describes the contents and minimum set of cross-references (active links) to access detailed information. Usually, these databases have ties to central mutation knowledge bases for genetic variation, e.g., NCBI (National Center for Biotechnology Information), OMIM (Online Mendelian Inheritance in Man), HGMD (Human Gene Mutation Database) for clinically related information, Medline/PubMed for access to published references, GenBank/EMBL (European molecular biology laboratory)/ DDBJ(DNA Databank of Japan) for detailed DNA sequence information (Bianco et al., 2013). These databases are designed using different methods and techniques for collecting mutation-related information, database scheme, and querying string/option for retrieving data. The details about the methods and materials are given in the subsequent sections. The NEMDB data can be analyzed by the factors like data quality and consistency, querying the database, system/ designing the database, disease content.
4.1. System Design
The design of most of the mutation databases is user-friendly and free to access the data. However, some databases are password-protected, and the users need to have registration for data access. The registration check ensures that the user must agree to some guidelines that cover data submission, privacy, data authenticity, and acknowledgment. There is a need to have a universal database management system platform that fulfills the basic requirement of a database such as a friendly interface, the searching/querying option, and have some privileges for curators. Despite all these, software was designed according to such preliminaries named ETHNOS software (from ETHnic and National Database Operating Software) used for mutation databases. The ETHNOS-based software is used to satisfy the essential requirement of the NMDBs databases. The administrators of ETHNOS are providing services to all those researchers who wish to implement the software for their database development purposes (detailed information can be found on the database website) [
14,
53]. Ethnos supported the creation of the various databases i.e. Hellenic, Cypriot, Iranian, Lebanese, and Serbian NEMDBs, however, Ethnos could not handle querying capacity and larger dataset [
14,
15,
60].
Frequency of INherited Disorders database (FINDbase), a relational database established on an upgraded version of Ethnos software that is capable of handling larger datasets refers to the frequency of low alleles leading to inherited disorders in various ethnic populations across the world [
61]. FINDbase is an inclusive online resource supporting the occurrence of clinically relevant genomic variation allele frequency information, serving a well-defined scientific discipline. This material is accessible on FINDbase in two modules, Causative Genomic Variants and Pharmacogenomics Biomarkers (PGx). The current updated FINDbase data module focused primarily on the data collection of PGx for growing the existing data sets with information about occurrences of clinically PGx biomarkers in European and other populations but also aims to interlinked PGx data module to DruGeVar [
62]
http://drugevar.genomicmedicinealliance.org/. Moreover, certain databases are based on a three-tier architecture model (user/client, application server/web interface, and RBDMS).
Other mutation databases have used Leiden Open source Variation Database (LOVD) platform [
44]. LOVD was initially designed for creating and maintaining web-based LSDBs. It is platform-independent software that uses PHP and MySQL only. LOVD software has many variations, such as LOVD v.2.0 [
63] and LOVD v.3.0, each following the Human Genome Variation Society (HGVS). The front ends of all databases are based on the hypertext markup language (HTML) with some JavaScript, Php, ASP.Net, and relying on Cascading Style Sheet support. The basic purpose of LOVD is to facilitate the curators by giving flexible tools for gene mutation and a display of DNA variants. LOVD v.3.0 was updated by 15 June 2021. The data can be retrieved by utilizing LOVD API.
4.2. The Quality Data Collection
Data collection is the essential phase of database development. The mutation databases are used to collect data from different disparate sources. The literature reveals that all mutation data have been obtained from various sources such as published materials on PubMed, peer-reviewed and scientific literature, meeting reports, and particularly from experts and genetic services [
64].
Table 3 shows different data collection methods of the mutation databases they use for gathering mutation-related information. Data can also be identified through automated text mining and manual journal screening. In addition, there are some mutation databases where the data have been linked to unpublished mutation data presented in publicly available locus-specific mutation databases (LSDBs); for example, the mutation databases may have a link to the HGMD database that facilitate users with access to LSDBs, for both published and unpublished materials [
10].
Each NEMDB created with Ethnos software has its own data folder in the Golden Helix Server and consists of three distinct functionalities. a) The disease overviews consist of indexed multiple flat-file database technique. The records can cover different lines and contain either plain text or valid HTML code. b) The allele frequency search option can be used in either an unrestricted or secure password-protected environment. A single flat-file database technique also utilized here. The tab-delimited text file contains information such as population, ethnic group, gene, Online Mendelian Inheritance in Man (OMIM) identification (ID), mutation, number (No.) of chromosomes/ families, allele and carrier frequency (percent), every time depending on the NEMDB in question. c) Genetic research laboratories: The indexed multiple flat-file database technique, as with the disease summaries option, is used here as well, though the files are in a different format.
4.3. Querying the Database
The gene mutation databases can be accessed using different search strings and query options. Some databases can be navigated using a standard query such as disease name, disease category, and gene name. Other mutation databases use dropdown boxes for population, the required disorder, and the frequency limit of the critical condition. Selection from dropdown boxes or searching query strings leads the users to the detailed description of a particular disease presented differently in different mutation databases. The detailed report may contain gene name, phenotype, chromosomal information, inheritance model, allele, protein variant, and their link/references to PubMed.
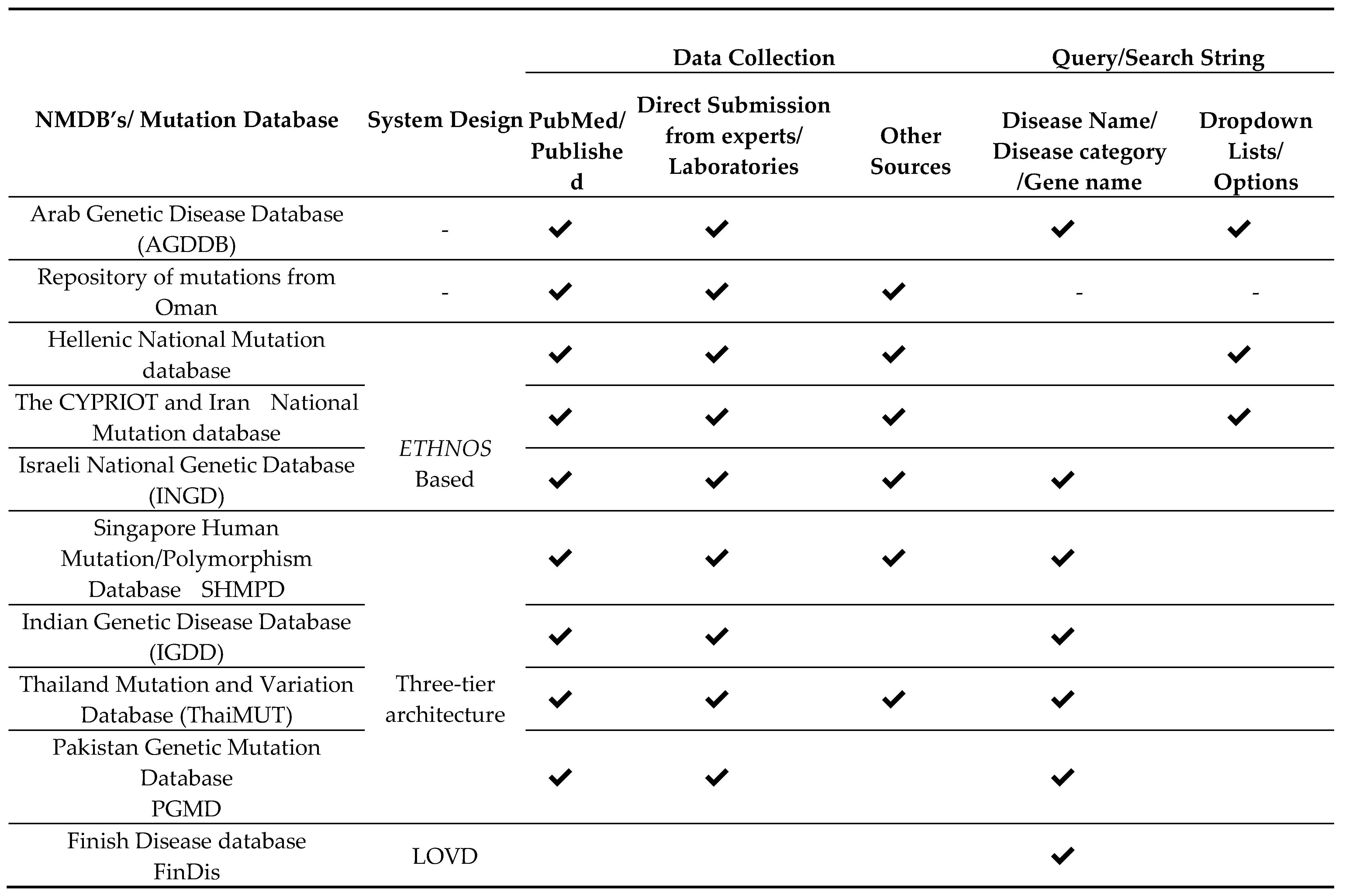
Table 4 shows the system design, data collection and data quality in the available NEMDBs. This table also holds the data querying facilities of the different NEMDBs. In this table, first column contains the various fully functional and accessible NEMDBs. Second column is reserved to the system/database design of each of these databases. In third column, the data collection ways of these databases have been reported. Finally, in fourth column, the data querying facilities of these databases are recorded. Note that this table only contains details about all NEMDBs that are either published on online accessible.
4.4. Disease-Related Content
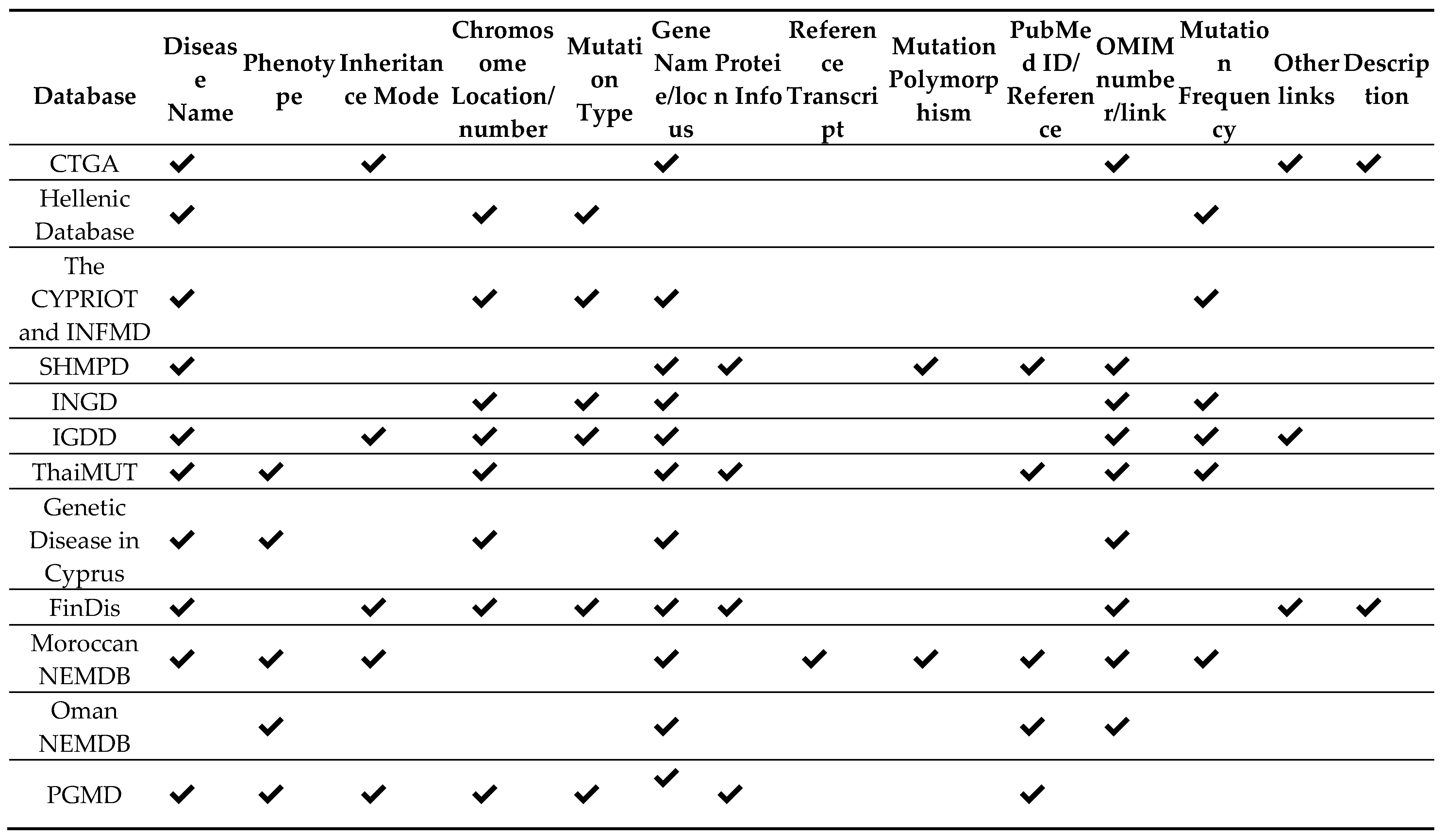
The available studied NEMDBs contains information about a particular disorder of a specific ethnic group or population. The information in most of the NEMDBs is presented in tabular form, while some databases have included the details in textual form. The information about the disorder may hold gene name, phenotype, a disease associated, OMIM number, inheritance model, polymorphism, ethnic group, mutation frequency, references, and other essential links; however, not all of the NEMDBs are enriched in content. The disease-related contents of different NEMDBs can be seen in
Table 5. Some NEMDBs contain extra information such as HGVS nomenclature and population group is found in the Cypriot database, the ethnic group in Israeli mutation database and Nucleotide change in Oman database and the database for the genetic disease of Cyprus contains additional information band, transcript and the tissues associated with a specific disease.
Table 5 shows information about different disease in the available NEMDBs. Note that we have taken the 14 features, each of which are available in more than single NEMDB, however, there are some NEMDBs that contains more information beyond the ones mentioned in table.
5. Discussion
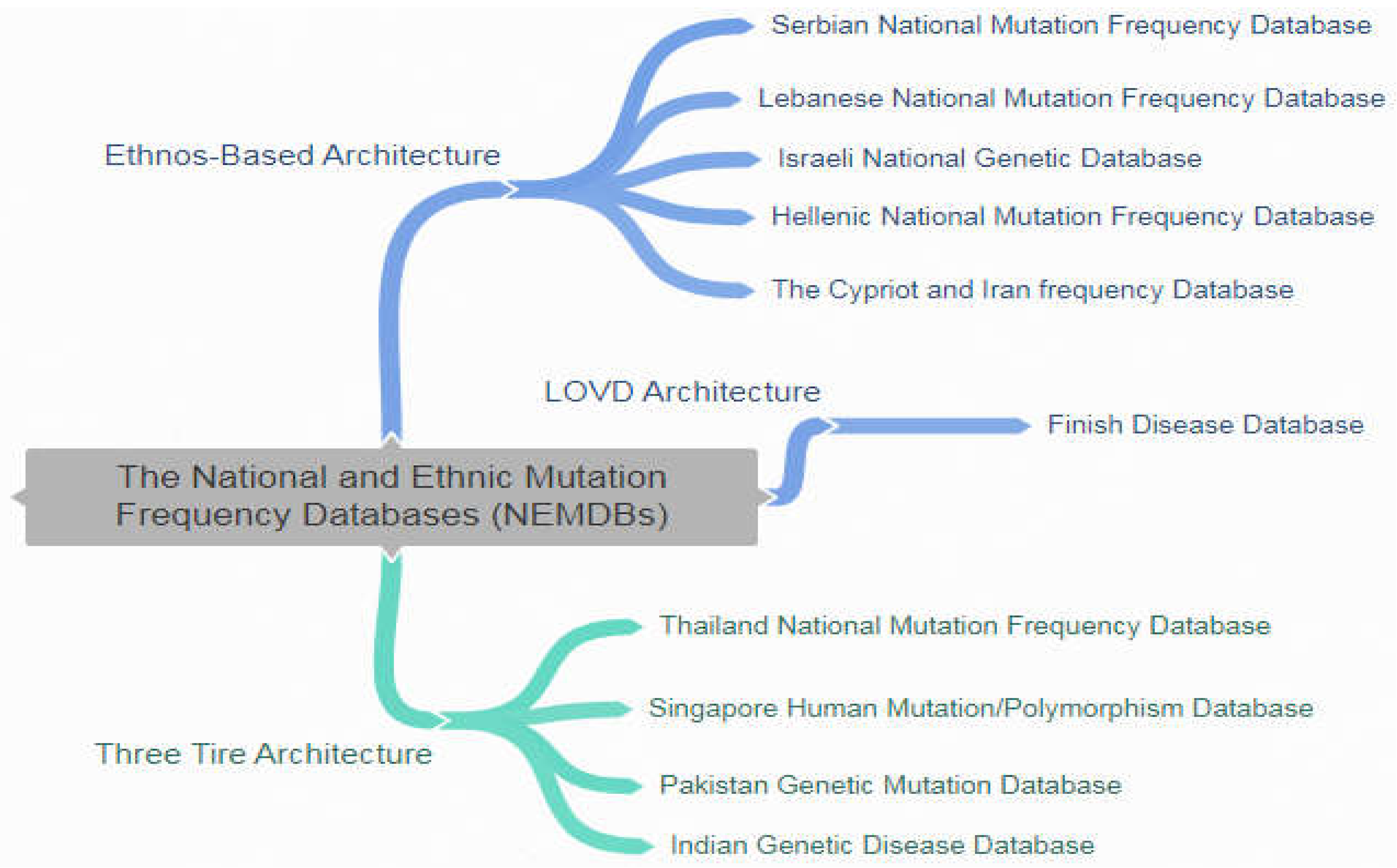
NEMDBs are online repositories containing information about the genetic disorder of a particular population or ethnic group. The available NEMDBs cover a variety of disorders and the corresponding gene mutations in different ethnic groups or geographic regions. In this paper, we discussed NEMDBs developed for specific ethnic groups, mostly for different nations/countries. The database have been developed over different time period and are shown in figure 3. All these databases are using their developed platform and most of the databases are linked with central databases. Unfortunately, some of the mutation disorders may appear in different ethnic groups as these ethnic groups share the same environmental change and hence similar or same mutation disorders. The mutation databases can be accessed on their own specified URLs however, some of the databases are kept on a mutual server (i.e. Golden Helix) that is not accessible everywhere. Moreover, these databases have some shortcomings that are:
The mutation databases (NEMDBs) have used different platforms for the development and hence the data querying, data representation, adding new records, and all the other features of these databases are different from each other.
Most of these databases are linked to the central databases but not with each other and due to this reason, these databases may have duplicate data. The data duplication may occur due to the same/similar ethnic boundaries of different nations.
Some of the databases are not been updated over time, hence not covering the new mutation disorders if occurred.
Some databases provide details about a very limited number of mutation disorders. These databases may inherit records from other databases as the same population can have similar mutation disorders as in other databases, though not included.
Figure 3.
A catalog of National Ethnic Frequency and mutation Database.
Figure 3.
A catalog of National Ethnic Frequency and mutation Database.
One solution to make these databases more effective and convenient is to utilize the concept of Linked Open Data (LOD). The use of Linked open data will break the barriers among different formats of these databases and the databases will be able to be connected and queried. LOD not only makes the human readers serve web pages but it generates the data in such a form that it can be directly read and understood by computers. We suggest LOD be used for linking all the mutation databases so that information from different formats can be queried easily. Using LOD, one can get the desired results from different data sources.
Another way of enhancing the effectiveness of the NEMDBs is to use Local as View (LAV) and Global as View (GAV) approaches. These approaches are used as data integration, where the data from different disparate sources are integrated and a single view over these sources is presented. The GAV approach uses the concept of mediation, where a virtual view of the databases (Global View) is presented and the actual data is stored at different separate data sources. The presentation of virtual view is simply called the mediator which is made from the combination of different data sources. Using a GAV approach, whenever a query is submitted for data extraction, the query is mapped with other queries and is sent to the data sources. The sources then check for the required results and send the results back to the mediator. The query mapping process in the GAV approach is done using wrappers (Used to present the local views of data sources and map it with the global views). The GAV approach has a major limitation that is the addition or removal of sources needs to make changes in the global schema and their mapping with local schema. On the other hand, the local as view (LAV) approach does not need the database administrator to completely change the schemas when sources are to be added or removed. In the LAV approach, the relations are displayed over the views of Global Schemas. In these systems, the local schemas describe Global Schemas in such a way that adding or deleting sources does not alter and it's due to the reason that Global schemas are designed independently of source schemas. The LAV approach is better in terms of deletion of sources from the system or adding new sources to the system however, it has very high time complexity. To overcome the weaknesses of both LAV and GAV approaches, we suggest of combine these approaches so that the strengths of both of the approaches can be used for effective data integration of Gene mutation Databases.
6. Conclusion
The rapid growth of bioinformatics projects has been changed with huge amounts of data being generated in a medical organization. The mutation databases play a vital role in genetic healthcare or research works related to mutation disorders. In this paper, we investigated the available genetic mutation databases (NEMDBs) that have been developed for different ethnic groups. The databases are evaluated and compared in terms of their data collection methods, their used system design, the ways of querying these databases, the various links/URLs used for these databases, and the disease-related contents available in each database. This paper also examines the weaknesses found in the developed databases and suggestions that can be brought more effectiveness for accessing mutation-related data. We aim that this study will open new ways for researchers in the area of genetic disorders.
Author Contributions
The following statements should be used “Conceptualization, I.Q. and S.K.; methodology, S.K; formal analysis, S.K, M.A and S.K (Shahnaz khan).; investigation, S.K.; resources, S.K.; data curation, S.K, M.A.; writing—original draft preparation, S.K.; writing—review and editing, W.K, I.Q. and M.A; visualization, S.K and S.K(Shahnaz khan.).; supervision, I.R, I.Q.; project administration, I.Q.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data and web links are available in the article.
Acknowledgments
The authors are grateful to Pakistan Science Foundation for providing partial financial support for project of mutation database “Pakistan Genetic Mutation Database (PGMD)” design and developed by Dr. Iqbal Qasim.
Conflicts of Interest
The authors declare no conflict of interest.
Notes
| 1 |
https://coggle.it/diagram/YUgtHQ9uj-ii0cse/t/ncbi. |
| 2 |
https://coggle.it/diagram/YTSDZgEJq_PwvBo1/t/the-national-and-mutation-frequency-databases-nemdbs. |
References
- Agris, P.F., The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function, in Progress in nucleic acid research and molecular biology. 1996, Elsevier. p. 79-129.
- Baltimore, D., Our genome unveiled. Nature, 2001. 409(6822): p. 814.
- Venter, J.C., et al., The sequence of the human genome. science, 2001. 291(5507): p. 1304-1351.
- Adams, P.C., et al., Hemochromatosis and iron-overload screening in a racially diverse population. New England Journal of Medicine, 2005. 352(17): p. 1769-1778. [CrossRef]
- Bhardwaj, U., Y.-H. Zhang, and E.R. McCabe, Neonatal hemoglobinopathy screening: molecular genetic technologies. Molecular genetics and metabolism, 2003. 1(80): p. 129-137. [CrossRef]
- Pradhan, S., et al., Indian genetic disease database. Nucleic acids research, 2010. 39(suppl_1): p. D933-D938. [CrossRef]
- Bhardwaj, U., et al., Molecular genetic confirmatory testing from newborn screening samples for the common African-American, Asian Indian, Southeast Asian, and Chinese β-thalassemia mutations. American journal of hematology, 2005. 78(4): p. 249-255.
- Hoodfar, E. and A.S. Teebi, Genetic referrals of Middle Eastern origin in a western city: inbreeding and disease profile. Journal of medical genetics, 1996. 33(3): p. 212-215. [CrossRef]
- Consortium, I.H.G.S., Correction: initial sequencing and analysis of the human genome. Nature, 2001. 412(6846): p. 565.
- Stenson, P.D., et al., Human gene mutation database (HGMD®): 2003 update. Human mutation, 2003. 21(6): p. 577-581. [CrossRef]
- Hamosh, A., et al., Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic acids research, 2005. 33(suppl_1): p. D514-D517. [CrossRef]
- Claustres, M., et al., Time for a unified system of mutation description and reporting: a review of locus-specific mutation databases. Genome research, 2002. 12(5): p. 680-688. [CrossRef]
- Bianco, A.M., et al., Database tools in genetic diseases research. Genomics, 2013. 101(2): p. 75-85. [CrossRef]
- Patrinos, G.P., et al., Hellenic National Mutation database: a prototype database for mutations leading to inherited disorders in the Hellenic population. Human mutation, 2005. 25(4): p. 327-333. [CrossRef]
- Kleanthous, M., et al., The cypriot and Iranian national mutation frequency databases. Human mutation, 2006. 27(6): p. 598-599. [CrossRef]
- Hunter, L. and K.B. Cohen, Biomedical language processing: what's beyond PubMed? Molecular cell, 2006. 21(5): p. 589-594.
- Lu, Z., PubMed and beyond: a survey of web tools for searching biomedical literature. Database, 2011. 2011. [CrossRef]
- Ding, J., et al., PubMed Assistant: a biologist-friendly interface for enhanced PubMed search. Bioinformatics, 2006. 22(3): p. 378-380. [CrossRef]
- Plake, C., et al., AliBaba: PubMed as a graph. Bioinformatics, 2006. 22(19): p. 2444-2445. [CrossRef]
- Tsai, R.T.-H., et al., PubMed-EX: a web browser extension to enhance PubMed search with text mining features. Bioinformatics, 2009. 25(22): p. 3031-3032. [CrossRef]
- Galperin, M.Y. and G.R. Cochrane, Nucleic acids research annual database issue and the NAR online molecular biology database collection in 2009. Nucleic Acids Research, 2009. 37(suppl_1): p. D1-D4. [CrossRef]
- Scriver, C.R., et al., PAHdb: a locus-specific knowledgebase. Human mutation, 2000. 15(1): p. 99-104. [CrossRef]
- Landrum, M.J., et al., ClinVar: improvements to accessing data. Nucleic acids research, 2020. 48(D1): p. D835-D844. [CrossRef]
- Sayers, E.W., et al., Database resources of the national center for biotechnology information. Nucleic acids research, 2012. 40(D1): p. D13-D25.
- Boguski, M.S., T.M. Lowe, and C.M. Tolstoshev, dbEST—database for “expressed sequence tags”. Nature genetics, 1993. 4(4): p. 332-333.
- Church, D.M., et al., Public data archives for genomic structural variation. Nature genetics, 2010. 42(10): p. 813-814. [CrossRef]
- Louhichi, A., A. Fourati, and A. Rebaï, IGD: a resource for intronless genes in the human genome. Gene, 2011. 488(1-2): p. 35-40. [CrossRef]
- Mailman, M., et al., Bagoutdinov r, hao l. Kiang a, Paschall J, Phan l, Popova n, Pretel s, Ziyabari l, lee M, shao Y, Wang ZY, sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, shevelev s, Preuss D, Yaschenko e, graeff a, Ostell J, sherry sT. The ncBi dbgaP database of genotypes and phenotypes. Nat Genet, 2007. 39: p. 1181-6. [CrossRef]
- Manolio, T.A., et al., New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nature genetics, 2007. 39(9).
- Sherry, S.T., et al., dbSNP: the NCBI database of genetic variation. Nucleic acids research, 2001. 29(1): p. 308-311. [CrossRef]
- Horaitis, O. and R.G. Cotton, Human mutation databases. Current protocols in bioinformatics, 2005. 9(1): p. 1.10. 1-1.10. 13.
- Hamosh, A., et al., Online Mendelian inheritance in man (OMIM). Human mutation, 2000. 15(1): p. 57-61. [CrossRef]
- Cooper, D.N., et al., Genes, mutations, and human inherited disease at the dawn of the age of personalized genomics. Human mutation, 2010. 31(6): p. 631-655. [CrossRef]
- Stenson, P.D., et al., The Human Gene Mutation Database: towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Human genetics, 2017. 136(6): p. 665-677. [CrossRef]
- Cooper, D.N., et al., On the sequence-directed nature of human gene mutation: the role of genomic architecture and the local DNA sequence environment in mediating gene mutations underlying human inherited disease. Human mutation, 2011. 32(10): p. 1075-1099. [CrossRef]
- Stenson, P.D., et al., The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Human genetics, 2014. 133(1): p. 1-9. [CrossRef]
- Samuels, M.E. and G.A. Rouleau, The case for locus-specific databases. Nature Reviews Genetics, 2011. 12(6): p. 378-379. [CrossRef]
- Celli, J., et al., Curating gene variant databases (LSDBs): toward a universal standard. Human mutation, 2012. 33(2): p. 291-297. [CrossRef]
- Vihinen, M., et al., Guidelines for establishing locus specific databases. Human mutation, 2012. 33(2): p. 298-305. [CrossRef]
- Dalgleish, R., LSDBs and how they have evolved. Human Mutation, 2016. 37(6): p. 532-539. [CrossRef]
- Béroud, C., et al., UMD (Universal mutation database): a generic software to build and analyze locus-specific databases. Human mutation, 2000. 15(1): p. 86-94. [CrossRef]
- Riikonen, P. and M. Vihinen, MUTbase: maintenance and analysis of distributed mutation databases. Bioinformatics (Oxford, England), 1999. 15(10): p. 852-859. [CrossRef]
- Brown, A.F. and M.A. McKie, MuStaR™ and other software for locus-specific mutation databases. Human mutation, 2000. 15(1): p. 76-85. [CrossRef]
- Fokkema, I.F., J.T. den Dunnen, and P.E. Taschner, LOVD: easy creation of a locus-specific sequence variation database using an “LSDB-in-a-box” approach. Human mutation, 2005. 26(2): p. 63-68.
- Scriver, C.R., Human genetics: lessons from Quebec populations. Annual review of genomics and human genetics, 2001. 2(1): p. 69-101. [CrossRef]
- Qasim, I., et al., Pakistan genetic mutation database (PGMD); a centralized Pakistani mutome data source. European journal of medical genetics, 2018. 61(4): p. 204-208. [CrossRef]
- Clark, B. and S. Thein, Molecular diagnosis of haemoglobin disorders. Clinical & Laboratory Haematology, 2004. 26(3): p. 159-176. [CrossRef]
- Tan, E.c., et al., Singapore Human Mutation/Polymorphism Database: a country-specific database for mutations and polymorphisms in inherited disorders and candidate gene association studies. Human mutation, 2006. 27(3): p. 232-235. [CrossRef]
- Patrinos, G.P., National and ethnic mutation databases: recording populations' genography. Human mutation, 2006. 27(9): p. 879-887. [CrossRef]
- Teebi, A.S., et al., Arab genetic disease database (AGDDB): A population-specific clinical and mutation database. Human mutation, 2002. 19(6): p. 615-621. [CrossRef]
- Tadmouri, G.O., et al., CTGA: the database for genetic disorders in Arab populations. Nucleic acids research, 2006. 34(suppl_1): p. D602-D606. [CrossRef]
- Peltonen, L., A. Jalanko, and T. Varilo, Molecular genetics the Finnish disease heritage. Human molecular genetics, 1999. 8(10): p. 1913-1923.
- van Baal, S., et al., ETHNOS: a versatile electronic tool for the development and curation of National Genetic databases. Human genomics, 2010. 4(5): p. 1-8.
- Zlotogora, J. and G.P. Patrinos, The Israeli National Genetic database: a 10-year experience. Human genomics, 2017. 11(1): p. 1-5. [CrossRef]
- Nakouzi, G., K. Kreidieh, and S. Yazbek, A review of the diverse genetic disorders in the Lebanese population: highlighting the urgency for community genetic services. Journal of community genetics, 2015. 6(1): p. 83-105. [CrossRef]
- Sefiani, A., Genetic disorders in Morocco, in Genetic disorders Among Arab populations. 2010, Springer. p. 455-472.
- Ruangrit, U., et al., Thailand mutation and variation database (ThaiMUT). Human mutation, 2008. 29(8): p. E68-E75. [CrossRef]
- Romdhane, L., et al., Genetic diseases in the Tunisian population. American Journal of Medical Genetics Part A, 2011. 155(1): p. 238-267. [CrossRef]
- Rajab, A., et al., Repository of mutations from Oman: The entry point to a national mutation database. F1000Research, 2015. 4. [CrossRef]
- Megarbane, A., et al., The Lebanese National Mutation Frequency database. Eur. J. Hum. Genet, 2006. 14(Suppl 1): p. 365.
- van Baal, S., et al., FINDbase: a relational database recording frequencies of genetic defects leading to inherited disorders worldwide. Nucleic acids research, 2007. 35(suppl_1): p. D690-D695.
- Dalabira, E., et al., DruGeVar: an online resource triangulating drugs with genes and genomic biomarkers for clinical pharmacogenomics. Public Health Genomics, 2014. 17(5-6): p. 265-271. [CrossRef]
- Fokkema, I.F., et al., LOVD v. 2.0: the next generation in gene variant databases. Human mutation, 2011. 32(5): p. 557-563. [CrossRef]
- Coordinators, N.R., Database resources of the national center for biotechnology information. Nucleic acids research, 2016. 44(Database issue): p. D7. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).