Submitted:
15 February 2023
Posted:
20 February 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
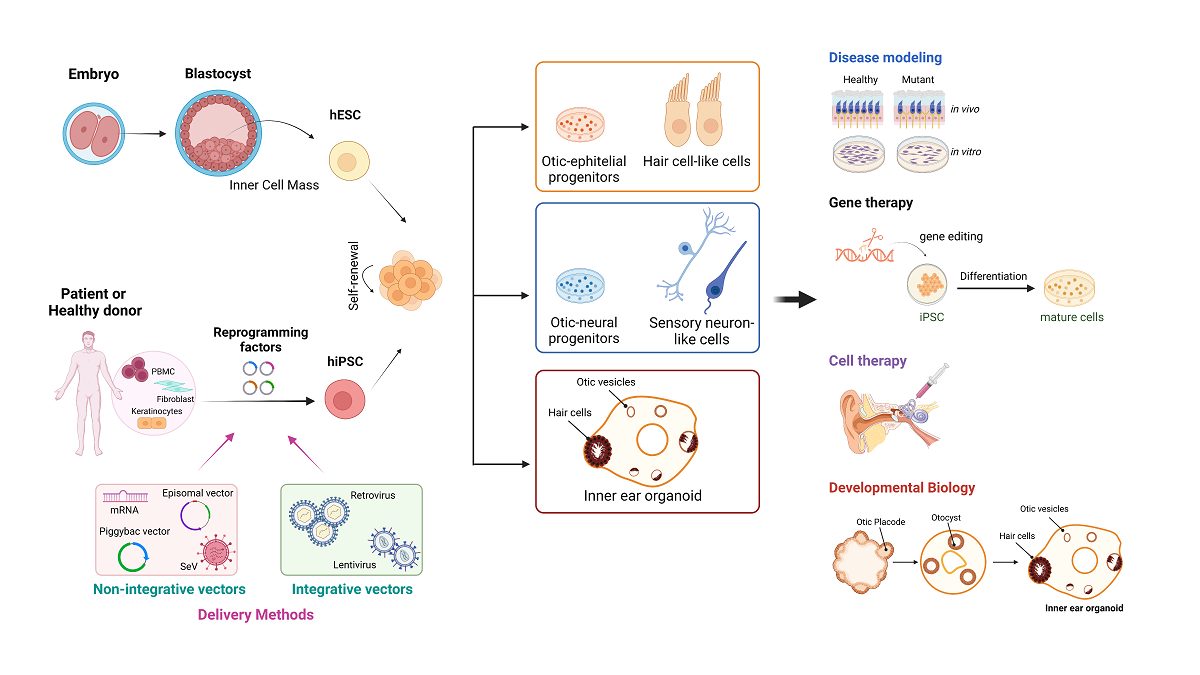
2. Generation of Human Pluripotent Stem Cells
3. Disease modeling by human induced Pluripotent Stem Cells
4. Clinical applications in genetic deafness and vestibular disorders
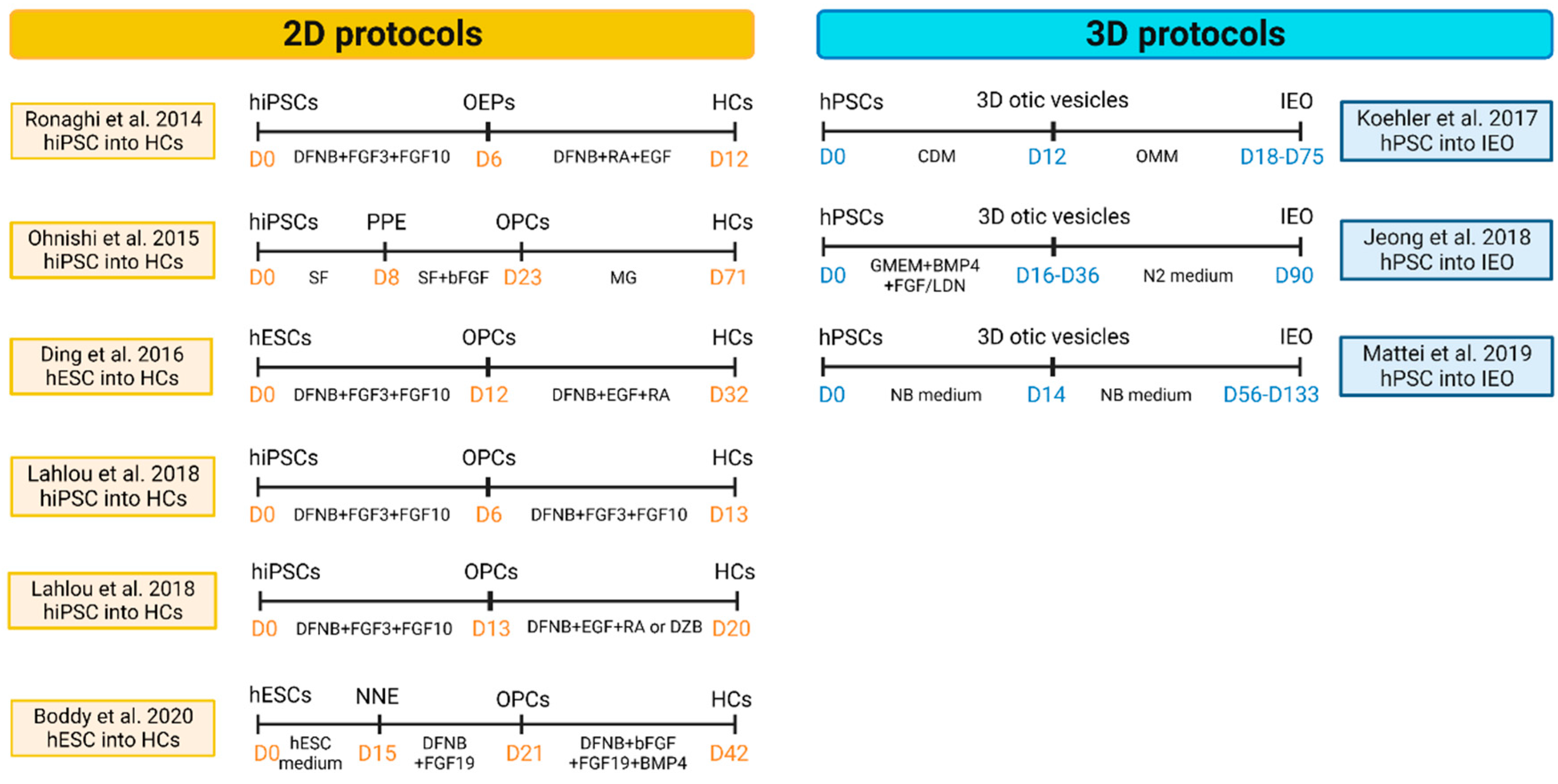
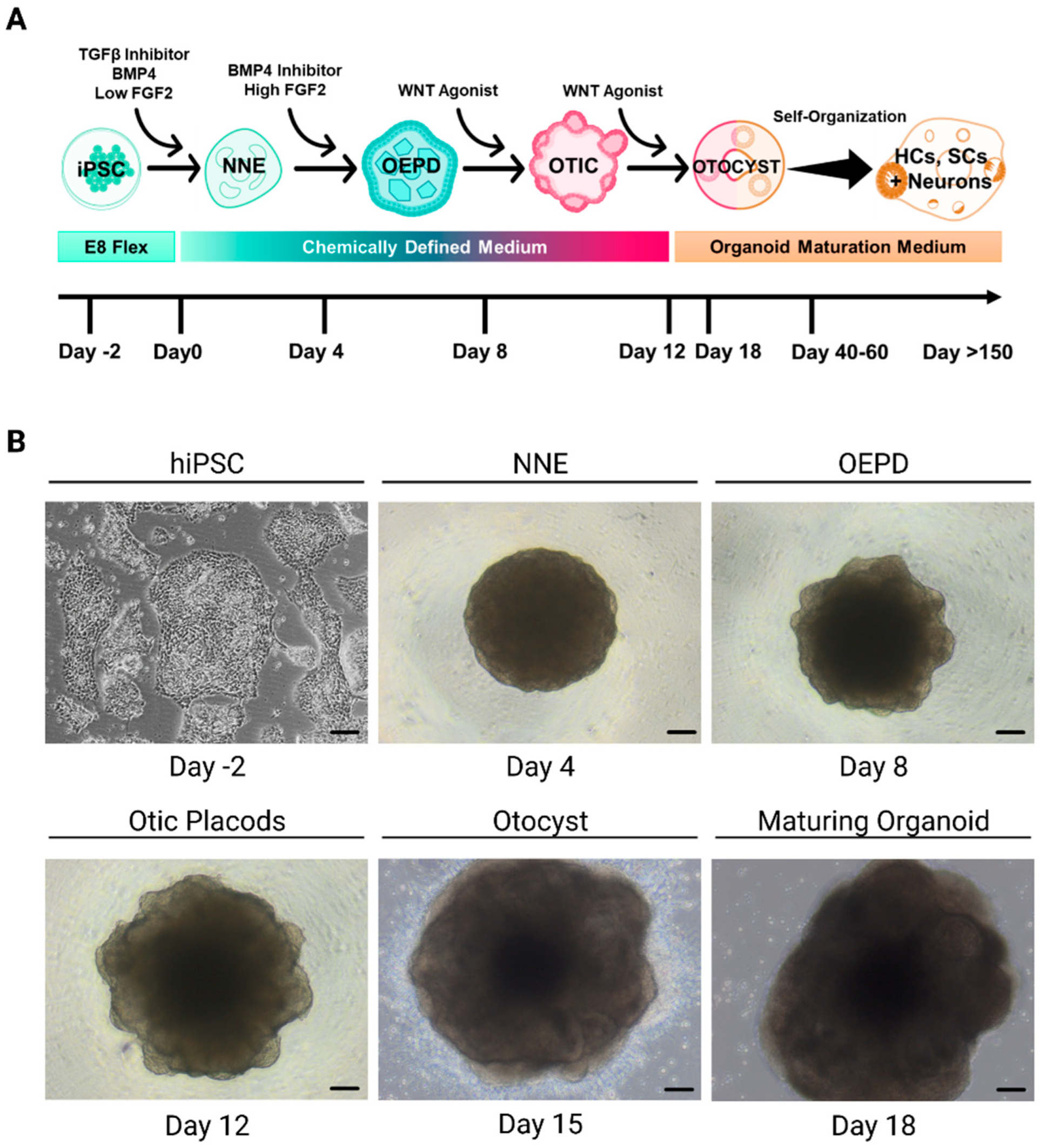
4.1. Modeling inner ear disorders: 2D and 3D cell culture
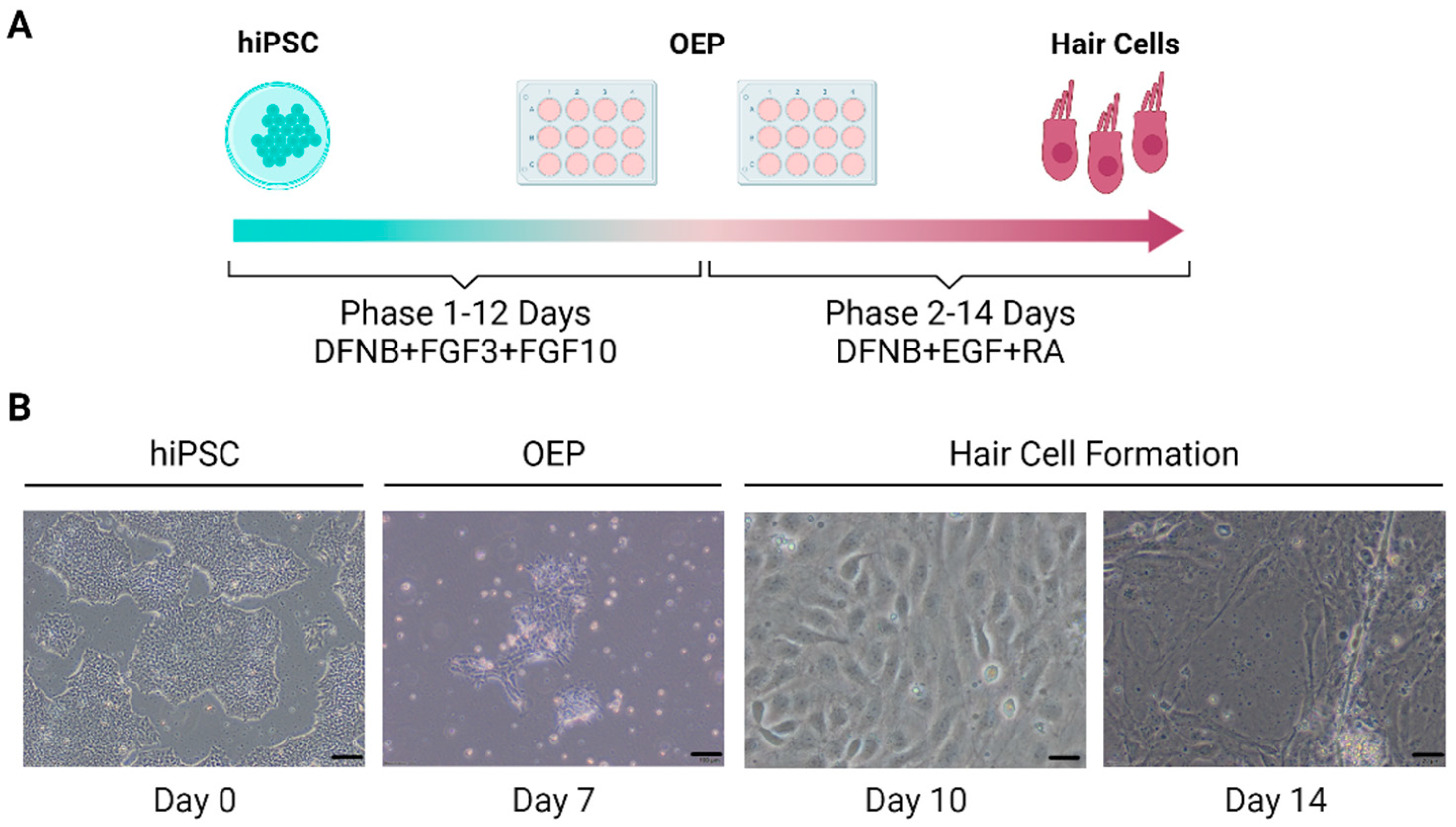
4.1.1. Sensory Epithelia
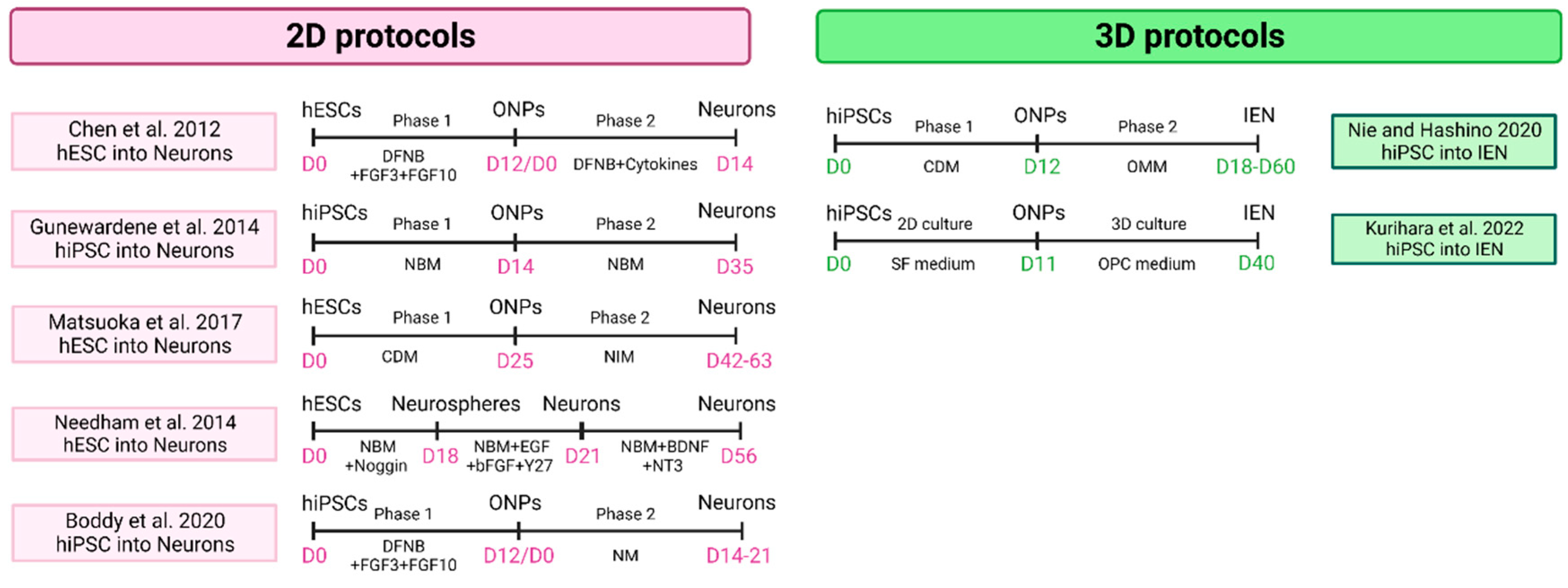
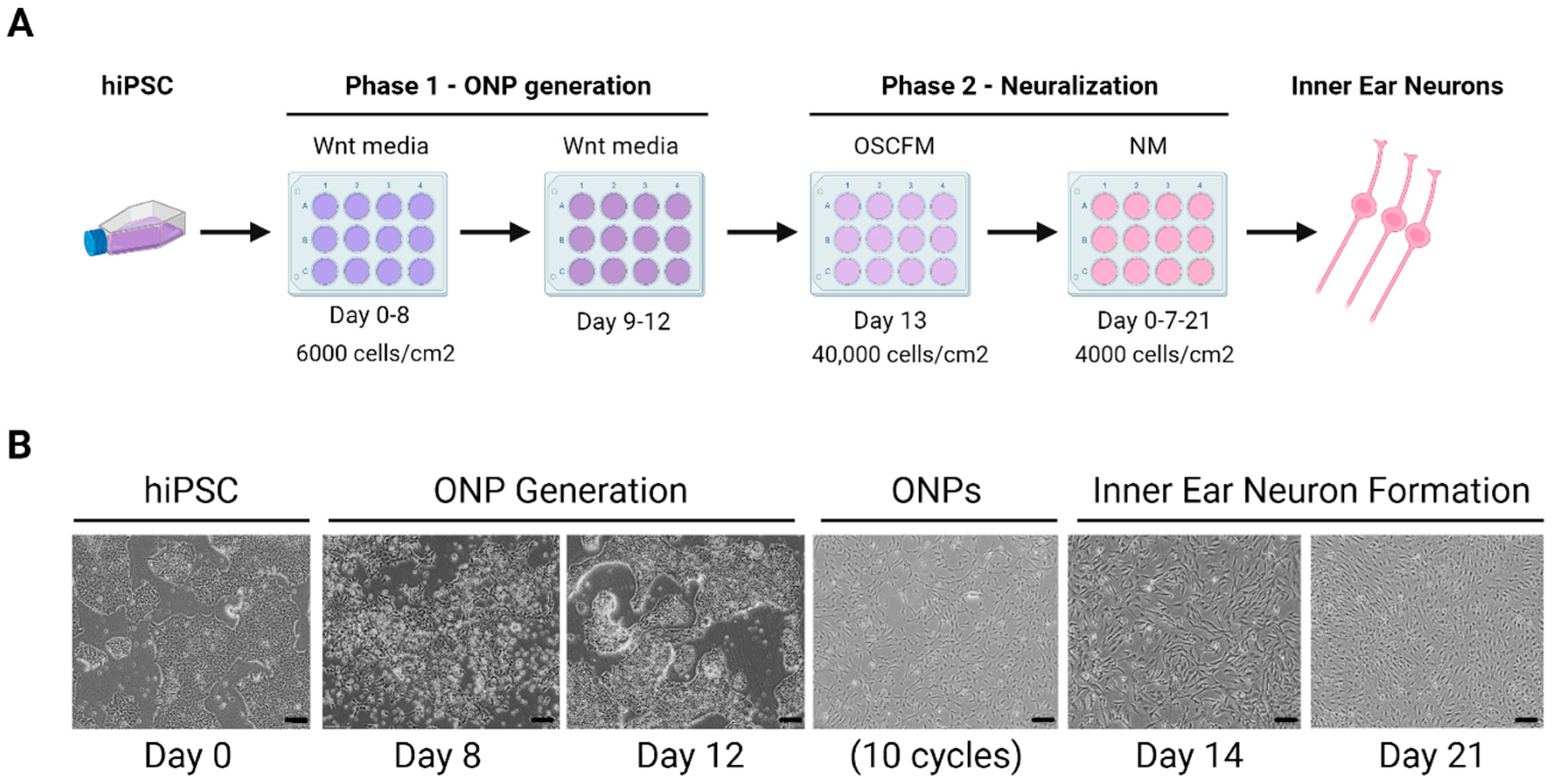
4.1.2. Sensory Neuron
5. Clinical Trials in Inner Ear Disorders
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Avior, Y.; Sagi, I.; Benvenisty, N. Pluripotent Stem Cells in Disease Modelling and Drug Discovery. Nat. Rev. Mol. Cell Biol. 2016, 17, 170–182. [Google Scholar] [CrossRef] [PubMed]
- Bai, X. Stem Cell-Based Disease Modeling and Cell Therapy. Cells 2020, 9, E2193. [Google Scholar] [CrossRef]
- Kim, J.; Koo, B.-K.; Knoblich, J.A. Human Organoids: Model Systems for Human Biology and Medicine. Nat. Rev. Mol. Cell Biol. 2020, 21, 571–584. [Google Scholar] [CrossRef]
- Liu, H.; Zhou, K.; Zhang, X.; Peng, K.A. Fluctuating Sensorineural Hearing Loss. Audiol. Neurootol. 2019, 24, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.-C.; Hashino, E.; Nelson, R.F. Progress in Modeling and Targeting Inner Ear Disorders with Pluripotent Stem Cells. Stem Cell Rep. 2020, 14, 996–1008. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, S.; Shiraki, N.; Yamada, S.; Uwabe, C.; Imai, H.; Matsuda, T.; Yoneyama, A.; Takeda, T.; Takakuwa, T. Morphogenesis of the Inner Ear at Different Stages of Normal Human Development. Anat. Rec. 2015, 298, 2081–2090. [Google Scholar] [CrossRef]
- Kayyali, M.N.; Wright, A.C.; Ramsey, A.J.; Brant, J.A.; Stein, J.M.; O’Malley, B.W.; Li, D. Challenges and Opportunities in Developing Targeted Molecular Imaging to Determine Inner Ear Defects of Sensorineural Hearing Loss. Nanomedicine Nanotechnol. Biol. Med. 2018, 14, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Boddy, S.L.; Chen, W.; Romero-Guevara, R.; Kottam, L.; Bellantuono, I.; Rivolta, M.N. Inner Ear Progenitor Cells Can Be Generated in Vitro from Human Bone Marrow Mesenchymal Stem Cells. Regen. Med. 2012, 7, 757–767. [Google Scholar] [CrossRef] [PubMed]
- Durán Alonso, M.B.; Feijoo-Redondo, A.; Conde de Felipe, M.; Carnicero, E.; García, A.S.; García-Sancho, J.; Rivolta, M.N.; Giráldez, F.; Schimmang, T. Generation of Inner Ear Sensory Cells from Bone Marrow-Derived Human Mesenchymal Stem Cells. Regen. Med. 2012, 7, 769–783. [Google Scholar] [CrossRef]
- Mittal, R.; Ocak, E.; Zhu, A.; Perdomo, M.M.; Pena, S.A.; Mittal, J.; Bohorquez, J.; Eshraghi, A.A. Effect of Bone Marrow-Derived Mesenchymal Stem Cells on Cochlear Function in an Experimental Rat Model. Anat. Rec. Hoboken NJ 2007 2020, 303, 487–493. [Google Scholar] [CrossRef]
- Xu, Y.-P.; Shan, X.-D.; Liu, Y.-Y.; Pu, Y.; Wang, C.-Y.; Tao, Q.-L.; Deng, Y.; Cheng, Y.; Fan, J.-P. Olfactory Epithelium Neural Stem Cell Implantation Restores Noise-Induced Hearing Loss in Rats. Neurosci. Lett. 2016, 616, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Tang, Z.; Chen, J.; Shi, H.; Chen, J.; Wang, C.; Zhang, C.; Li, L.; Chen, P.; Wang, J. Induction of Differentiation of Human Embryonic Stem Cells into Functional Hair-Cell-like Cells in the Absence of Stromal Cells. Int. J. Biochem. Cell Biol. 2016, 81, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Lahlou, H.; Lopez-Juarez, A.; Fontbonne, A.; Nivet, E.; Zine, A. Modeling Human Early Otic Sensory Cell Development with Induced Pluripotent Stem Cells. PloS One 2018, 13, e0198954. [Google Scholar] [CrossRef]
- Ohnishi, H.; Skerleva, D.; Kitajiri, S.; Sakamoto, T.; Yamamoto, N.; Ito, J.; Nakagawa, T. Limited Hair Cell Induction from Human Induced Pluripotent Stem Cells Using a Simple Stepwise Method. Neurosci. Lett. 2015, 599, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Jeong, M.; O’Reilly, M.; Kirkwood, N.K.; Al-Aama, J.; Lako, M.; Kros, C.J.; Armstrong, L. Generating Inner Ear Organoids Containing Putative Cochlear Hair Cells from Human Pluripotent Stem Cells. Cell Death Dis. 2018, 9, 922. [Google Scholar] [CrossRef] [PubMed]
- Koehler, K.R.; Mikosz, A.M.; Molosh, A.I.; Patel, D.; Hashino, E. Generation of Inner Ear Sensory Epithelia from Pluripotent Stem Cells in 3D Culture. Nature 2013, 500, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Koehler, K.R.; Nie, J.; Longworth-Mills, E.; Liu, X.-P.; Lee, J.; Holt, J.R.; Hashino, E. Generation of Inner Ear Organoids Containing Functional Hair Cells from Human Pluripotent Stem Cells. Nat. Biotechnol. 2017, 35, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Mattei, C.; Lim, R.; Drury, H.; Nasr, B.; Li, Z.; Tadros, M.A.; D’Abaco, G.M.; Stok, K.S.; Nayagam, B.A.; Dottori, M. Generation of Vestibular Tissue-Like Organoids From Human Pluripotent Stem Cells Using the Rotary Cell Culture System. Front. Cell Dev. Biol. 2019, 7, 25. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef]
- Choi, J.; Lee, S.; Mallard, W.; Clement, K.; Tagliazucchi, G.M.; Lim, H.; Choi, I.Y.; Ferrari, F.; Tsankov, A.M.; Pop, R.; et al. A Comparison of Genetically Matched Cell Lines Reveals the Equivalence of Human IPSCs and ESCs. Nat. Biotechnol. 2015, 33, 1173–1181. [Google Scholar] [CrossRef]
- Ortuño-Costela, M.D.C.; Cerrada, V.; García-López, M.; Gallardo, M.E. The Challenge of Bringing IPSCs to the Patient. Int. J. Mol. Sci. 2019, 20, E6305. [Google Scholar] [CrossRef]
- Aasen, T.; Raya, A.; Barrero, M.J.; Garreta, E.; Consiglio, A.; Gonzalez, F.; Vassena, R.; Bilić, J.; Pekarik, V.; Tiscornia, G.; et al. Efficient and Rapid Generation of Induced Pluripotent Stem Cells from Human Keratinocytes. Nat. Biotechnol. 2008, 26, 1276–1284. [Google Scholar] [CrossRef]
- Esteban, M.A.; Wang, T.; Qin, B.; Yang, J.; Qin, D.; Cai, J.; Li, W.; Weng, Z.; Chen, J.; Ni, S.; et al. Vitamin C Enhances the Generation of Mouse and Human Induced Pluripotent Stem Cells. Cell Stem Cell 2010, 6, 71–79. [Google Scholar] [CrossRef]
- Loh, Y.-H.; Agarwal, S.; Park, I.-H.; Urbach, A.; Huo, H.; Heffner, G.C.; Kim, K.; Miller, J.D.; Ng, K.; Daley, G.Q. Generation of Induced Pluripotent Stem Cells from Human Blood. Blood 2009, 113, 5476–5479. [Google Scholar] [CrossRef]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of Induced Pluripotent Stem Cells without Myc from Mouse and Human Fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef]
- Subramanyam, D.; Lamouille, S.; Judson, R.L.; Liu, J.Y.; Bucay, N.; Derynck, R.; Blelloch, R. Multiple Targets of MiR-302 and MiR-372 Promote Reprogramming of Human Fibroblasts to Induced Pluripotent Stem Cells. Nat. Biotechnol. 2011, 29, 443–448. [Google Scholar] [CrossRef]
- Li, W.; Zhou, H.; Abujarour, R.; Zhu, S.; Young Joo, J.; Lin, T.; Hao, E.; Schöler, H.R.; Hayek, A.; Ding, S. Generation of Human-Induced Pluripotent Stem Cells in the Absence of Exogenous Sox2. Stem Cells Dayt. Ohio 2009, 27, 2992–3000. [Google Scholar] [CrossRef]
- Lin, S.-L.; Chang, D.C.; Chang-Lin, S.; Lin, C.-H.; Wu, D.T.S.; Chen, D.T.; Ying, S.-Y. Mir-302 Reprograms Human Skin Cancer Cells into a Pluripotent ES-Cell-like State. RNA N. Y. N 2008, 14, 2115–2124. [Google Scholar] [CrossRef]
- Zhao, Y.; Yin, X.; Qin, H.; Zhu, F.; Liu, H.; Yang, W.; Zhang, Q.; Xiang, C.; Hou, P.; Song, Z.; et al. Two Supporting Factors Greatly Improve the Efficiency of Human IPSC Generation. Cell Stem Cell 2008, 3, 475–479. [Google Scholar] [CrossRef]
- Mali, P.; Ye, Z.; Hommond, H.H.; Yu, X.; Lin, J.; Chen, G.; Zou, J.; Cheng, L. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells Dayt. Ohio 2008, 26, 1998–2005. [Google Scholar] [CrossRef]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef]
- Liao, J.; Wu, Z.; Wang, Y.; Cheng, L.; Cui, C.; Gao, Y.; Chen, T.; Rao, L.; Chen, S.; Jia, N.; et al. Enhanced Efficiency of Generating Induced Pluripotent Stem (IPS) Cells from Human Somatic Cells by a Combination of Six Transcription Factors. Cell Res. 2008, 18, 600–603. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Y.; Jin, H.; Xie, L.; Liu, C.; Jiang, F.; Luo, Y.; Yin, G.; Li, Y.; Wang, J.; et al. Rapid and Efficient Reprogramming of Human Amnion-Derived Cells into Pluripotency by Three Factors OCT4/SOX2/NANOG. Differ. Res. Biol. Divers. 2010, 80, 123–129. [Google Scholar] [CrossRef]
- Zhu, S.; Li, W.; Zhou, H.; Wei, W.; Ambasudhan, R.; Lin, T.; Kim, J.; Zhang, K.; Ding, S. Reprogramming of Human Primary Somatic Cells by OCT4 and Chemical Compounds. Cell Stem Cell 2010, 7, 651–655. [Google Scholar] [CrossRef]
- Ye, L.; Chang, J.C.; Lin, C.; Qi, Z.; Yu, J.; Kan, Y.W. Generation of Induced Pluripotent Stem Cells Using Site-Specific Integration with Phage Integrase. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 19467–19472. [Google Scholar] [CrossRef]
- Zhou, W.; Freed, C.R. Adenoviral Gene Delivery Can Reprogram Human Fibroblasts to Induced Pluripotent Stem Cells. Stem Cells Dayt. Ohio 2009, 27, 2667–2674. [Google Scholar] [CrossRef]
- Ban, H.; Nishishita, N.; Fusaki, N.; Tabata, T.; Saeki, K.; Shikamura, M.; Takada, N.; Inoue, M.; Hasegawa, M.; Kawamata, S.; et al. Efficient Generation of Transgene-Free Human Induced Pluripotent Stem Cells (IPSCs) by Temperature-Sensitive Sendai Virus Vectors. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 14234–14239. [Google Scholar] [CrossRef]
- Fusaki, N.; Ban, H.; Nishiyama, A.; Saeki, K.; Hasegawa, M. Efficient Induction of Transgene-Free Human Pluripotent Stem Cells Using a Vector Based on Sendai Virus, an RNA Virus That Does Not Integrate into the Host Genome. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 348–362. [Google Scholar] [CrossRef]
- Kaji, K.; Norrby, K.; Paca, A.; Mileikovsky, M.; Mohseni, P.; Woltjen, K. Virus-Free Induction of Pluripotency and Subsequent Excision of Reprogramming Factors. Nature 2009, 458, 771–775. [Google Scholar] [CrossRef]
- Woltjen, K.; Michael, I.P.; Mohseni, P.; Desai, R.; Mileikovsky, M.; Hämäläinen, R.; Cowling, R.; Wang, W.; Liu, P.; Gertsenstein, M.; et al. PiggyBac Transposition Reprograms Fibroblasts to Induced Pluripotent Stem Cells. Nature 2009, 458, 766–770. [Google Scholar] [CrossRef]
- Si-Tayeb, K.; Noto, F.K.; Sepac, A.; Sedlic, F.; Bosnjak, Z.J.; Lough, J.W.; Duncan, S.A. Generation of Human Induced Pluripotent Stem Cells by Simple Transient Transfection of Plasmid DNA Encoding Reprogramming Factors. BMC Dev. Biol. 2010, 10, 81. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human Induced Pluripotent Stem Cells Free of Vector and Transgene Sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef]
- Yu, J.; Chau, K.F.; Vodyanik, M.A.; Jiang, J.; Jiang, Y. Efficient Feeder-Free Episomal Reprogramming with Small Molecules. PLOS ONE 2011, 6, e17557. [Google Scholar] [CrossRef]
- Okita, K.; Matsumura, Y.; Sato, Y.; Okada, A.; Morizane, A.; Okamoto, S.; Hong, H.; Nakagawa, M.; Tanabe, K.; Tezuka, K.; et al. A More Efficient Method to Generate Integration-Free Human IPS Cells. Nat. Methods 2011, 8, 409–412. [Google Scholar] [CrossRef]
- Jia, F.; Wilson, K.D.; Sun, N.; Gupta, D.M.; Huang, M.; Li, Z.; Panetta, N.J.; Chen, Z.Y.; Robbins, R.C.; Kay, M.A.; et al. A Nonviral Minicircle Vector for Deriving Human IPS Cells. Nat. Methods 2010, 7, 197–199. [Google Scholar] [CrossRef]
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of Human Induced Pluripotent Stem Cells by Direct Delivery of Reprogramming Proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of Human Fibroblasts to Pluripotent Stem Cells Using MRNA of Four Transcription Factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193. [Google Scholar] [CrossRef]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified MRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef]
- Miyoshi, N.; Ishii, H.; Nagano, H.; Haraguchi, N.; Dewi, D.L.; Kano, Y.; Nishikawa, S.; Tanemura, M.; Mimori, K.; Tanaka, F.; et al. Reprogramming of Mouse and Human Cells to Pluripotency Using Mature MicroRNAs. Cell Stem Cell 2011, 8, 633–638. [Google Scholar] [CrossRef] [PubMed]
- Trokovic, R.; Weltner, J.; Nishimura, K.; Ohtaka, M.; Nakanishi, M.; Salomaa, V.; Jalanko, A.; Otonkoski, T.; Kyttälä, A. Advanced Feeder-Free Generation of Induced Pluripotent Stem Cells Directly From Blood Cells. Stem Cells Transl. Med. 2014, 3, 1402–1409. [Google Scholar] [CrossRef]
- Schlaeger, T.M.; Daheron, L.; Brickler, T.R.; Entwisle, S.; Chan, K.; Cianci, A.; DeVine, A.; Ettenger, A.; Fitzgerald, K.; Godfrey, M.; et al. A Comparison of Non-Integrating Reprogramming Methods. Nat. Biotechnol. 2015, 33, 58–63. [Google Scholar] [CrossRef]
- Kogut, I.; McCarthy, S.M.; Pavlova, M.; Astling, D.P.; Chen, X.; Jakimenko, A.; Jones, K.L.; Getahun, A.; Cambier, J.C.; Pasmooij, A.M.G.; et al. High-Efficiency RNA-Based Reprogramming of Human Primary Fibroblasts. Nat. Commun. 2018, 9. [Google Scholar] [CrossRef]
- Wang, A.Y.L. Application of Modified MRNA in Somatic Reprogramming to Pluripotency and Directed Conversion of Cell Fate. Int. J. Mol. Sci. 2021, 22, 8148. [Google Scholar] [CrossRef]
- Cabrera, S.; Ji, A.-R.; Frejo, L.; Ramos-Mejia, V.; Romero, T.; Real, P.; Lopez-Escamez, J.A. Generation of Human IPSC Line GRX-MCiPS4F-A2 from Adult Peripheral Blood Mononuclear Cells (PBMCs) with Spanish Genetic Background. Stem Cell Res. 2015, 15, 337–340. [Google Scholar] [CrossRef]
- Lamolda, M.; Montes, R.; Simón, I.; Perales, S.; Martínez-Navajas, G.; Lopez-Onieva, L.; Ríos-Pelegrina, R.; del Moral, R.G.; Griñan-Lison, C.; Marchal, J.A.; et al. GENYOi005-A: An Induced Pluripotent Stem Cells (IPSCs) Line Generated from a Patient with Familial Platelet Disorder with Associated Myeloid Malignancy (FPDMM) Carrying a p.Thr196Ala Variant. Stem Cell Res. 2019, 41, 101603. [Google Scholar] [CrossRef]
- Cimmino, L.; Neel, B.G.; Aifantis, I. Vitamin C in Stem Cell Reprogramming and Cancer. Trends Cell Biol. 2018, 28, 698–708. [Google Scholar] [CrossRef]
- Steinle, H.; Weber, M.; Behring, A.; Mau-Holzmann, U.; Schlensak, C.; Wendel, H.P.; Avci-Adali, M. Generation of IPSCs by Nonintegrative RNA-Based Reprogramming Techniques: Benefits of Self-Replicating RNA versus Synthetic MRNA. Stem Cells Int. 2019, 2019. [Google Scholar] [CrossRef]
- Park, I.-H.; Arora, N.; Huo, H.; Maherali, N.; Ahfeldt, T.; Shimamura, A.; Lensch, M.W.; Cowan, C.; Hochedlinger, K.; Daley, G.Q. Disease-Specific Induced Pluripotent Stem Cells. Cell 2008, 134, 877–886. [Google Scholar] [CrossRef]
- Francis, K.R.; Wei, L. Human Embryonic Stem Cell Neural Differentiation and Enhanced Cell Survival Promoted by Hypoxic Preconditioning. Cell Death Dis. 2010, 1, e22–e22. [Google Scholar] [CrossRef]
- Hu Bao-Yang; Weick Jason P.; Yu Junying; Ma Li-Xiang; Zhang Xiao-Qing; Thomson James A.; Zhang Su-Chun Neural Differentiation of Human Induced Pluripotent Stem Cells Follows Developmental Principles but with Variable Potency. Proc. Natl. Acad. Sci. 2010, 107, 4335–4340. [CrossRef]
- Kuruş, M.; Akbari, S.; Eskier, D.; Bursalı, A.; Ergin, K.; Erdal, E.; Karakülah, G. Transcriptome Dynamics of Human Neuronal Differentiation From IPSC. Front. Cell Dev. Biol. 2021, 0. [Google Scholar] [CrossRef] [PubMed]
- Israel, M.A.; Yuan, S.H.; Bardy, C.; Reyna, S.M.; Mu, Y.; Herrera, C.; Hefferan, M.P.; Van Gorp, S.; Nazor, K.L.; Boscolo, F.S.; et al. Probing Sporadic and Familial Alzheimer’s Disease Using Induced Pluripotent Stem Cells. Nature 2012, 482, 216–220. [Google Scholar] [CrossRef]
- Juopperi, T.A.; Kim, W.R.; Chiang, C.-H.; Yu, H.; Margolis, R.L.; Ross, C.A.; Ming, G.; Song, H. Astrocytes Generated from Patient Induced Pluripotent Stem Cells Recapitulate Features of Huntington’s Disease Patient Cells. Mol. Brain 2012, 5, 17. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Song, J.; Huang, H.; Chen, H.; Qian, K. Modeling Hallmark Pathology Using Motor Neurons Derived from the Family and Sporadic Amyotrophic Lateral Sclerosis Patient-Specific IPS Cells. Stem Cell Res. Ther. 2018, 9, 315. [Google Scholar] [CrossRef]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue in Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Nakano, T.; Ando, S.; Takata, N.; Kawada, M.; Muguruma, K.; Sekiguchi, K.; Saito, K.; Yonemura, S.; Eiraku, M.; Sasai, Y. Self-Formation of Optic Cups and Storable Stratified Neural Retina from Human ESCs. Cell Stem Cell 2012, 10, 771–785. [Google Scholar] [CrossRef]
- Fligor, C.M.; Huang, K.-C.; Lavekar, S.S.; VanderWall, K.B.; Meyer, J.S. Differentiation of Retinal Organoids from Human Pluripotent Stem Cells. Methods Cell Biol. 2020, 159, 279–302. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef] [PubMed]
- Chari, D.A.; Chan, D.K. Diagnosis and Treatment of Congenital Sensorineural Hearing Loss. Curr. Otorhinolaryngol. Rep. 2017, 5, 251–258. [Google Scholar] [CrossRef]
- Renauld, J.M.; Basch, M.L. Congenital Deafness and Recent Advances Towards Restoring Hearing Loss. Curr. Protoc. 2021, 1, e76. [Google Scholar] [CrossRef]
- Sheikh, A.; Bint-e-Zainab; Shabbir, K.; Imtiaz, A. Structure and Physiology of Human Ear Involved in Hearing; IntechOpen, 2022; ISBN 978-1-80355-190-6.
- Zhang, W.; Kim, S.M.; Wang, W.; Cai, C.; Feng, Y.; Kong, W.; Lin, X. Cochlear Gene Therapy for Sensorineural Hearing Loss: Current Status and Major Remaining Hurdles for Translational Success. Front. Mol. Neurosci. 2018, 11, 221. [Google Scholar] [CrossRef]
- Lopez-Escamez, J.A.; Carey, J.; Chung, W.-H.; Goebel, J.A.; Magnusson, M.; Mandalà, M.; Newman-Toker, D.E.; Strupp, M.; Suzuki, M.; Trabalzini, F.; et al. Diagnostic Criteria for Menière’s Disease. J. Vestib. Res. Equilib. Orientat. 2015, 25, 1–7. [Google Scholar] [CrossRef]
- Lopez-Escamez, J.A.; Batuecas-Caletrio, A.; Bisdorff, A. Towards Personalized Medicine in Ménière’s Disease. F1000Research 2018, 7, F1000 Faculty Rev-1295. [Google Scholar] [CrossRef]
- Gibson, W.P.R. Meniere’s Disease. Adv. Otorhinolaryngol. 2019, 82, 77–86. [Google Scholar] [CrossRef]
- Perez-Carpena, P.; Lopez-Escamez, J.A. Current Understanding and Clinical Management of Meniere’s Disease: A Systematic Review. Semin. Neurol. 2020, 40, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Frejo, L.; Martin-Sanz, E.; Teggi, R.; Trinidad, G.; Soto-Varela, A.; Santos-Perez, S.; Manrique, R.; Perez, N.; Aran, I.; Almeida-Branco, M.S.; et al. Extended Phenotype and Clinical Subgroups in Unilateral Meniere Disease: A Cross-Sectional Study with Cluster Analysis. Clin. Otolaryngol. Off. J. ENT-UK Off. J. Neth. Soc. Oto-Rhino-Laryngol. Cervico-Facial Surg. 2017, 42, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Martín-Sierra, C.; Gallego-Martinez, A.; Requena, T.; Frejo, L.; Batuecas-Caletrío, A.; Lopez-Escamez, J.A. Variable Expressivity and Genetic Heterogeneity Involving DPT and SEMA3D Genes in Autosomal Dominant Familial Meniere’s Disease. Eur. J. Hum. Genet. EJHG 2017, 25, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Roman-Naranjo, P.; Gallego-Martinez, A.; Soto-Varela, A.; Aran, I.; Moleon, M.D.C.; Espinosa-Sanchez, J.M.; Amor-Dorado, J.C.; Batuecas-Caletrio, A.; Perez-Vazquez, P.; Lopez-Escamez, J.A. Burden of Rare Variants in the OTOG Gene in Familial Meniere’s Disease. Ear Hear. 2020, 41, 1598–1605. [Google Scholar] [CrossRef]
- Roman-Naranjo, P.; Moleon, M.D.C.; Aran, I.; Escalera-Balsera, A.; Soto-Varela, A.; Bächinger, D.; Gomez-Fiñana, M.; Eckhard, A.H.; Lopez-Escamez, J.A. Rare Coding Variants Involving MYO7A and Other Genes Encoding Stereocilia Link Proteins in Familial Meniere Disease. Hear. Res. 2021, 409, 108329. [Google Scholar] [CrossRef]
- Roman-Naranjo, P.; Parra-Perez, A.M.; Escalera-Balsera, A.; Soto-Varela, A.; Gallego-Martinez, A.; Aran, I.; Perez-Fernandez, N.; Bächinger, D.; Eckhard, A.H.; Gonzalez-Aguado, R.; et al. Defective α-Tectorin May Involve Tectorial Membrane in Familial Meniere Disease. Clin. Transl. Med. 2022, 12, e829. [Google Scholar] [CrossRef]
- T, R.; S, C.; C, M.-S.; Sd, P.; A, L.; Ja, L.-E. Identification of Two Novel Mutations in FAM136A and DTNA Genes in Autosomal-Dominant Familial Meniere’s Disease. Hum. Mol. Genet. 2015, 24. [Google Scholar] [CrossRef]
- Hoffman, H.J.; Dobie, R.A.; Losonczy, K.G.; Themann, C.L.; Flamme, G.A. Declining Prevalence of Hearing Loss in US Adults Aged 20 to 69 Years. JAMA Otolaryngol. Neck Surg. 2017, 143, 274. [Google Scholar] [CrossRef]
- Zheng, G.; Liu, Y.; He, J.; Li, S.; Zhang, Q.; Duan, M.; Yang, J.; Jin, Y. A Comparison of Local Endolymphatic Sac Decompression, Endolymphatic Mastoid Shunt, and Wide Endolymphatic Sac Decompression in the Treatment of Intractable Meniere’s Disease: A Short-Term Follow-Up Investigation. Front. Neurol. 2022, 13, 810352. [Google Scholar] [CrossRef]
- De Luca, P.; Cassandro, C.; Ralli, M.; Gioacchini, F.M.; Turchetta, R.; Orlando, M.P.; Iaccarino, I.; Cavaliere, M.; Cassandro, E.; Scarpa, A. Dietary Restriction for The Treatment of Meniere’s Disease. Transl. Med. UniSa 2020, 22, 5–9. [Google Scholar]
- Castiglione, A.; Benatti, A.; Velardita, C.; Favaro, D.; Padoan, E.; Severi, D.; Pagliaro, M.; Bovo, R.; Vallesi, A.; Gabelli, C.; et al. Aging, Cognitive Decline and Hearing Loss: Effects of Auditory Rehabilitation and Training with Hearing Aids and Cochlear Implants on Cognitive Function and Depression among Older Adults. Audiol. Neurotol. 2016, 21, 21–28. [Google Scholar] [CrossRef]
- Gao, X.; Grayden, D.; McDonnell, M. Unifying Information Theory and Machine Learning in a Model of Electrode Discrimination in Cochlear Implants. PLOS ONE 2021, 16, e0257568. [Google Scholar] [CrossRef]
- de Cates, C.; Winters, R. Intratympanic Steroid Injection. In StatPearls; StatPearls Publishing: Treasure Island (FL), 2022. [Google Scholar]
- Shin, S.-H.; Byun, S.W.; Park, S.; Kim, E.H.; Kim, M.W.; Lee, H.Y. Optimal First-Line Therapy for Acute Low-Tone Sensorineural Hearing Loss. J. Audiol. Otol. 2021, 25, 209–216. [Google Scholar] [CrossRef]
- Goudey, B.; Plant, K.; Kiral, I.; Jimeno-Yepes, A.; Swan, A.; Gambhir, M.; Büchner, A.; Kludt, E.; Eikelboom, R.H.; Sucher, C.; et al. A MultiCenter Analysis of Factors Associated with Hearing Outcome for 2,735 Adults with Cochlear Implants. Trends Hear. 2021, 25, 233121652110375. [Google Scholar] [CrossRef]
- Roccio, M.; Senn, P.; Heller, S. Novel Insights into Inner Ear Development and Regeneration for Targeted Hearing Loss Therapies. Hear. Res. 2020, 397, 107859. [Google Scholar] [CrossRef]
- He, Z.; Ding, Y.; Mu, Y.; Xu, X.; Kong, W.; Chai, R.; Chen, X. Stem Cell-Based Therapies in Hearing Loss. Front. Cell Dev. Biol. 2021, 9, 730042. [Google Scholar] [CrossRef]
- Nacher-Soler, G.; Garrido, J.M.; Rodríguez-Serrano, F. Hearing Regeneration and Regenerative Medicine: Present and Future Approaches. Arch. Med. Sci. AMS 2019, 15, 957–967. [Google Scholar] [CrossRef]
- Johnson Chacko, L.; Wertjanz, D.; Sergi, C.; Dudas, J.; Fischer, N.; Eberharter, T.; Hoermann, R.; Glueckert, R.; Fritsch, H.; Rask-Andersen, H.; et al. Growth and Cellular Patterning during Fetal Human Inner Ear Development Studied by a Correlative Imaging Approach. BMC Dev. Biol. 2019, 19, 11. [Google Scholar] [CrossRef]
- Filova, I.; Bohuslavova, R.; Tavakoli, M.; Yamoah, E.N.; Fritzsch, B.; Pavlinkova, G. Early Deletion of Neurod1 Alters Neuronal Lineage Potential and Diminishes Neurogenesis in the Inner Ear. Front. Cell Dev. Biol. 2022, 10, 845461. [Google Scholar] [CrossRef]
- Song, Z.; Jadali, A.; Fritzsch, B.; Kwan, K.Y. NEUROG1 Regulates CDK2 to Promote Proliferation in Otic Progenitors. Stem Cell Rep. 2017, 9, 1516–1529. [Google Scholar] [CrossRef]
- Filova, I.; Dvorakova, M.; Bohuslavova, R.; Pavlinek, A.; Elliott, K.L.; Vochyanova, S.; Fritzsch, B.; Pavlinkova, G. Combined Atoh1 and Neurod1 Deletion Reveals Autonomous Growth of Auditory Nerve Fibers. Mol. Neurobiol. 2020, 57, 5307–5323. [Google Scholar] [CrossRef]
- Sherrill, H.E.; Jean, P.; Driver, E.C.; Sanders, T.R.; Fitzgerald, T.S.; Moser, T.; Kelley, M.W. Pou4f1 Defines a Subgroup of Type I Spiral Ganglion Neurons and Is Necessary for Normal Inner Hair Cell Presynaptic Ca2+ Signaling. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 5284–5298. [Google Scholar] [CrossRef]
- Lahlou, H.; Nivet, E.; Lopez-Juarez, A.; Fontbonne, A.; Assou, S.; Zine, A. Enriched Differentiation of Human Otic Sensory Progenitor Cells Derived From Induced Pluripotent Stem Cells. Front. Mol. Neurosci. 2018, 11, 452. [Google Scholar] [CrossRef]
- Kurihara, S.; Fujioka, M.; Hirabayashi, M.; Yoshida, T.; Hosoya, M.; Nagase, M.; Kato, F.; Ogawa, K.; Okano, H.; Kojima, H.; et al. Otic Organoids Containing Spiral Ganglion Neuron-like Cells Derived from Human-Induced Pluripotent Stem Cells as a Model of Drug-Induced Neuropathy. Stem Cells Transl. Med. 2022, 11, 282–296. [Google Scholar] [CrossRef]
- Boddy, S.L.; Romero-Guevara, R.; Ji, A.-R.; Unger, C.; Corns, L.; Marcotti, W.; Rivolta, M.N. Generation of Otic Lineages from Integration-Free Human-Induced Pluripotent Stem Cells Reprogrammed by MRNAs. Stem Cells Int. 2020, 2020, 3692937. [Google Scholar] [CrossRef]
- Chen, J.-R.; Tang, Z.-H.; Zheng, J.; Shi, H.-S.; Ding, J.; Qian, X.-D.; Zhang, C.; Chen, J.-L.; Wang, C.-C.; Li, L.; et al. Effects of Genetic Correction on the Differentiation of Hair Cell-like Cells from IPSCs with MYO15A Mutation. Cell Death Differ. 2016, 23, 1347–1357. [Google Scholar] [CrossRef]
- Chen, W.; Jongkamonwiwat, N.; Abbas, L.; Eshtan, S.J.; Johnson, S.L.; Kuhn, S.; Milo, M.; Thurlow, J.K.; Andrews, P.W.; Marcotti, W.; et al. Restoration of Auditory Evoked Responses by Human ES-Cell-Derived Otic Progenitors. Nature 2012, 490, 278–282. [Google Scholar] [CrossRef]
- Ronaghi, M.; Nasr, M.; Ealy, M.; Durruthy-Durruthy, R.; Waldhaus, J.; Diaz, G.H.; Joubert, L.-M.; Oshima, K.; Heller, S. Inner Ear Hair Cell-like Cells from Human Embryonic Stem Cells. Stem Cells Dev. 2014, 23, 1275–1284. [Google Scholar] [CrossRef]
- Kenna, M.A.; Feldman, H.A.; Neault, M.W.; Frangulov, A.; Wu, B.-L.; Fligor, B.; Rehm, H.L. Audiologic Phenotype and Progression in GJB2 (Connexin 26) Hearing Loss. Arch. Otolaryngol. Head Neck Surg. 2010, 136, 81–87. [Google Scholar] [CrossRef]
- Fukunaga, I.; Oe, Y.; Danzaki, K.; Ohta, S.; Chen, C.; Shirai, K.; Kawano, A.; Ikeda, K.; Kamiya, K. Modeling Gap Junction Beta 2 Gene-Related Deafness with Human IPSC. Hum. Mol. Genet. 2021, 30, 1429–1442. [Google Scholar] [CrossRef]
- Frejo, L.; Cara, Francisca E; Gallego-Martinez; Lopez-Escamez An Inner Ear Organoid Model of Meniere Disease.; Trieste, Italy, September 2022.
- Frejo, L.; Cara, Francisca E; Gallego-Martinez; Lopez-Escamez DTNA and FAM136A Expression in a 3D Inner Ear Organoid Model of Meniere Disease.; Granada, Spain, May 2022.
- Nie, J.; Hashino, E. Generation of Inner Ear Organoids from Human Pluripotent Stem Cells. Methods Cell Biol. 2020, 159, 303–321. [Google Scholar] [CrossRef]
- Kawasaki, H.; Mizuseki, K.; Nishikawa, S.; Kaneko, S.; Kuwana, Y.; Nakanishi, S.; Nishikawa, S.I.; Sasai, Y. Induction of Midbrain Dopaminergic Neurons from ES Cells by Stromal Cell-Derived Inducing Activity. Neuron 2000, 28, 31–40. [Google Scholar] [CrossRef]
- Ying, Q.-L.; Stavridis, M.; Griffiths, D.; Li, M.; Smith, A. Conversion of Embryonic Stem Cells into Neuroectodermal Precursors in Adherent Monoculture. Nat. Biotechnol. 2003, 21, 183–186. [Google Scholar] [CrossRef]
- Chen, Y.; Xiong, M.; Dong, Y.; Haberman, A.; Cao, J.; Liu, H.; Zhou, W.; Zhang, S.-C. Chemical Control of Grafted Human PSC-Derived Neurons in a Mouse Model of Parkinson’s Disease. Cell Stem Cell 2016, 18, 817–826. [Google Scholar] [CrossRef]
- Oestreicher, E.; Wolfgang, A.; Felix, D. Neurotransmission of the Cochlear Inner Hair Cell Synapse--Implications for Inner Ear Therapy. Adv. Otorhinolaryngol. 2002, 59, 131–139. [Google Scholar] [CrossRef]
- Gunewardene, N.; Bergen, N.V.; Crombie, D.; Needham, K.; Dottori, M.; Nayagam, B.A. Directing Human Induced Pluripotent Stem Cells into a Neurosensory Lineage for Auditory Neuron Replacement. BioResearch Open Access 2014, 3, 162–175. [Google Scholar] [CrossRef]
- Matsuoka, A.J.; Morrissey, Z.D.; Zhang, C.; Homma, K.; Belmadani, A.; Miller, C.A.; Chadly, D.M.; Kobayashi, S.; Edelbrock, A.N.; Tanaka-Matakatsu, M.; et al. Directed Differentiation of Human Embryonic Stem Cells Toward Placode-Derived Spiral Ganglion-Like Sensory Neurons. Stem Cells Transl. Med. 2017, 6, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Needham, K.; Hyakumura, T.; Gunewardene, N.; Dottori, M.; Nayagam, B.A. Electrophysiological Properties of Neurosensory Progenitors Derived from Human Embryonic Stem Cells. Stem Cell Res. 2014, 12, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Hong, F.; Zhang, C.; Li, L.; Wang, C.; Shi, H.; Fu, Y.; Wang, J. Differentiation and Transplantation of Human Induced Pluripotent Stem Cell-Derived Otic Epithelial Progenitors in Mouse Cochlea. Stem Cell Res. Ther. 2018, 9, 230. [Google Scholar] [CrossRef] [PubMed]
- Tang, P.-C.; Alex, A.L.; Nie, J.; Lee, J.; Roth, A.A.; Booth, K.T.; Koehler, K.R.; Hashino, E.; Nelson, R.F. Defective Tmprss3-Associated Hair Cell Degeneration in Inner Ear Organoids. Stem Cell Rep. 2019, 13, 147–162. [Google Scholar] [CrossRef] [PubMed]
- MD, J.B. Safety of Autologous Stem Cell Infusion for Children With Acquired Hearing Loss; clinicaltrials.gov, 2018.
- Kanzaki, S.; Toyoda, M.; Umezawa, A.; Ogawa, K. Application of Mesenchymal Stem Cell Therapy and Inner Ear Regeneration for Hearing Loss: A Review. Int. J. Mol. Sci. 2020, 21, E5764. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.-H.; Chen, J.-R.; Zheng, J.; Shi, H.-S.; Ding, J.; Qian, X.-D.; Zhang, C.; Chen, J.-L.; Wang, C.-C.; Li, L.; et al. Genetic Correction of Induced Pluripotent Stem Cells From a Deaf Patient With MYO7A Mutation Results in Morphologic and Functional Recovery of the Derived Hair Cell-Like Cells. Stem Cells Transl. Med. 2016, 5, 561–571. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Tsai, C.-L.; Wei, Y.-H.; Wu, Y.-T.; Hsu, W.-T.; Lin, H.-C.; Hsu, Y.-C. ATOH1/RFX1/RFX3 Transcription Factors Facilitate the Differentiation and Characterisation of Inner Ear Hair Cell-like Cells from Patient-Specific Induced Pluripotent Stem Cells Harbouring A8344G Mutation of Mitochondrial DNA. Cell Death Dis. 2018, 9, 437. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, I.; Fujimoto, A.; Hatakeyama, K.; Aoki, T.; Nishikawa, A.; Noda, T.; Minowa, O.; Kurebayashi, N.; Ikeda, K.; Kamiya, K. In Vitro Models of GJB2-Related Hearing Loss Recapitulate Ca2+ Transients via a Gap Junction Characteristic of Developing Cochlea. Stem Cell Rep. 2016, 7, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Hosoya, M.; Fujioka, M.; Sone, T.; Okamoto, S.; Akamatsu, W.; Ukai, H.; Ueda, H.R.; Ogawa, K.; Matsunaga, T.; Okano, H. Cochlear Cell Modeling Using Disease-Specific IPSCs Unveils a Degenerative Phenotype and Suggests Treatments for Congenital Progressive Hearing Loss. Cell Rep. 2017, 18, 68–81. [Google Scholar] [CrossRef]
- Hosoya, M.; Saeki, T.; Saegusa, C.; Matsunaga, T.; Okano, H.; Fujioka, M.; Ogawa, K. Estimating the Concentration of Therapeutic Range Using Disease-Specific IPS Cells: Low-Dose Rapamycin Therapy for Pendred Syndrome. Regen. Ther. 2019, 10, 54–63. [Google Scholar] [CrossRef]
- Fujioka, M.; Akiyama, T.; Hosoya, M.; Kikuchi, K.; Fujiki, Y.; Saito, Y.; Yoshihama, K.; Ozawa, H.; Tsukada, K.; Nishio, S.-Y.; et al. A Phase I/IIa Double Blind Single Institute Trial of Low Dose Sirolimus for Pendred Syndrome/DFNB4. Medicine (Baltimore) 2020, 99, e19763. [Google Scholar] [CrossRef] [PubMed]
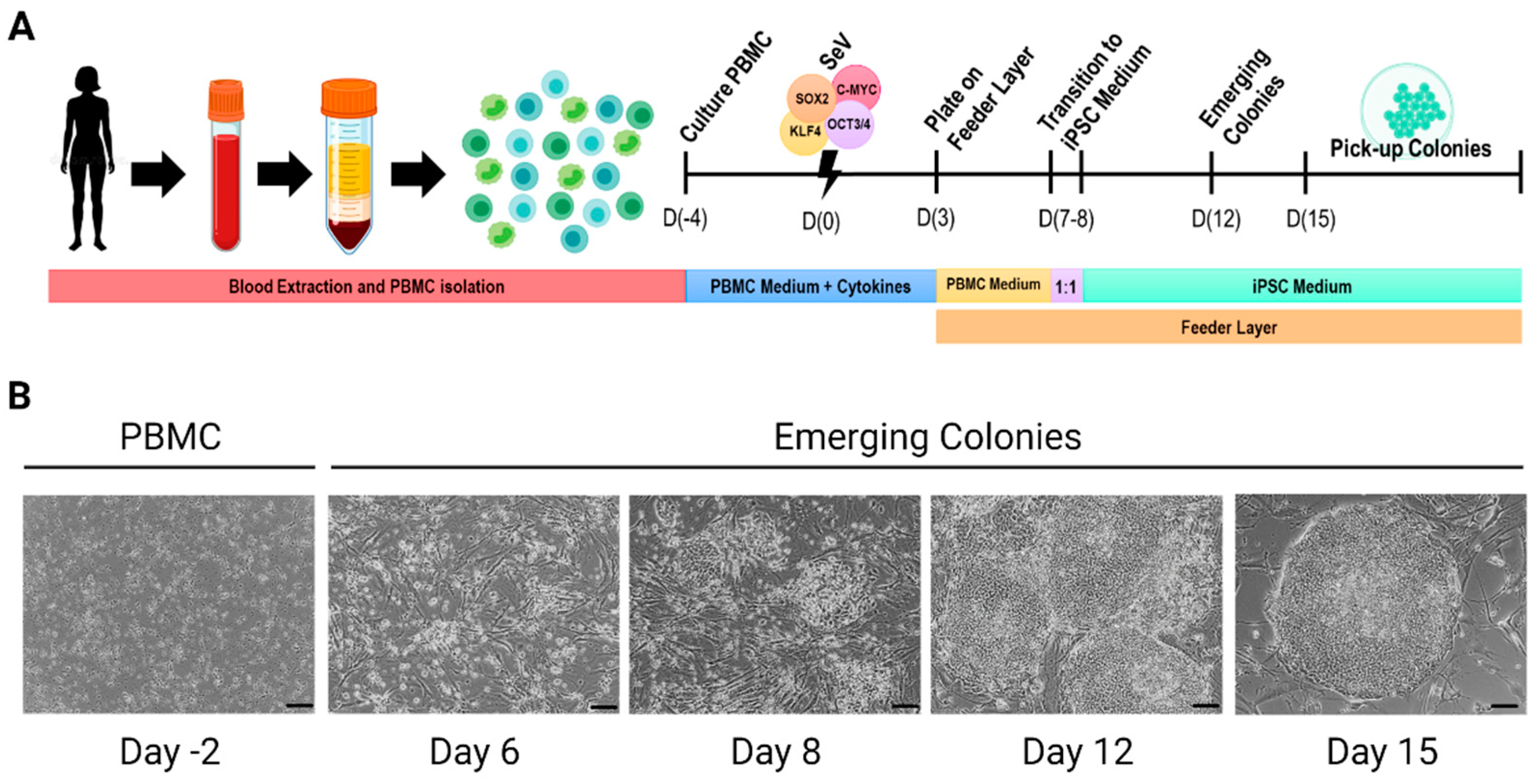
| Type of Vector | Integration | Factors | References | |
|---|---|---|---|---|
| Viral | Retrovirus | Yes | OSKM | [19,22,23,24] |
| OSK | [25,26] | |||
| OK | [27] | |||
| Hsa-miR-302ª/b/c/d | [28] | |||
| Lentivirus | Yes | OSKM | [29] | |
| OSNL | [30,31] | |||
| OSKMNL | [32] | |||
| OSN | [33,33] | |||
| O | [34] | |||
| Bacteriophage | Yes | OSKM | [35] | |
| Adenovirus | No | OSKM | [36] | |
| Sendai Virus | No | OSKM | [37,38] | |
| Non-viral | piggyBac | No | OSKM | [39,40] |
| Plasmid | No | OSNL | [41] | |
| Episomal Vector | No | OSKMNL | [42,43] | |
| OSKM*L | [44] | |||
| Minicircle vector | No | OSNL | [45] | |
| Protein | No | OSKM | [46] | |
| mRNA | No | OSNL | [47] | |
| OSKM (L) | [48] | |||
| microRNA | No | miR-200c, 302ª/b/c/d,369-3p/5p | [49] |
| Gene | RefSeq | Variant | Type of inheritance | Model | References |
| MYO7A | NM_000260.4 | c.1184G>Ayufenc.4118C>T | Compound heterozygous | Hair Cells | [121] |
| MYO15A | NM_016239.4 | c.4642G>Ayufenc.8374G>A | Compound heterozygous | Hair Cells | [102] |
| MERRF/MT-TK | NC_012920.1 | c.8344A>G | Mitochondrial | Hair Cells | [122] |
| SLC26A4 | NM_000441.2 | c.439A>Gyufenc.1229C>Tyufenc.2168A>G | Autosomal recessive | Supporting Cells | [124,125] |
| GJB2 | NM_004004.6 | c.235DelC | Autosomal recessive / Digenic | Supporting Cells | [106,123] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





