1. Introduction
Plants cope with herbivory through constitutive and inducible defences (Agrawal & Karban 1999). While constitutive defences consist of premade traits, like physical barriers or toxins, inducible defences are only established after herbivore attack (reviewed in Vega-Muñoz et al. 2020) and can include reinforcement of constitutive defences. This inducible response comprises a collection of defence programmes, regulated primarily by jasmonic acid (JA) (Wasternack and House, 2013) and salicylic acid (SA) (Vlot et al., 2009) dependent pathways, and aims at reducing the quality of the plant. This can include the synthesis of detrimental proteins, e.g. by the induction of proteinase inhibitors that reduce herbivore protein digestion (Walling 2000), which thereby decrease herbivore survival and reproduction (Karban & Myers, 1989), and thus population growth, promoting plant fitness (Kant et al., 2015). In response to these defensive mechanisms, some herbivores (e.g., arthropods and nematodes) have evolved strategies to overcome such defences either directly by metabolic resistance (Depres et al., 2007; Heckel 2014; reviewed in Groen & Whiteman 2022) or indirectly by suppressing defences in planta (e.g. reviewed in Kant et al., 2015).
Metabolic resistance can be established through detoxification (i.e., metabolic modification, Cianfrogna et al., 2002; degradation, Heckel 2014), sequestration (e.g. Bowers 2022) or the evolution of target-site insensitivity (Berenbaum 1986). Metabolic resistance can be efficient for herbivores living on hosts with a narrow range of secondary chemistry. However, for generalists, stacking different modes of metabolic resistance to cope with a wider variety of defensive substances may be costly and thus countered by natural selection (Ali & Agrawal 2012; Blaazer et al., 2018). As such, it has been suggested that generalists may also cope with their variable hosts by interfering with conserved elements in plant defence pathways (Kant et al., 2015). Such suppression (reviewed in Blaazer et al., 2018) is characterized by a reduction in toxin accumulation (e.g. reduced nicotine accumulation by feeding caterpillars in the leaves of tobacco, Musser et al., 2002) and defence gene expression (Kant et al., 2004) either to intermediate levels (Alba et al., 2014) or even to levels below the levels of unattacked control plants (Sarmento et al., 2011; Schimmel et al., 2018). This suppression boosts the performance of the suppressor (Sarmento et al., 2011) but also can boost the performance of nearby non-suppressors residing on the same plant (Kant et al., 2004) and can act across a diverse host range (Paulo et al., 2018). Suppression of plant defences is well known from phytopathogens and established via molecules, often referred to as “effectors”, secreted into the host during the early and later stages of the infection (reviewed in Jones et al., 2022). Data on the functionality of effectors across multiple hosts is still scarce, but some host plants have evolved mechanisms to detect the activity of effectors and by-pass their suppression effectively, turning the effector into a molecule that can induce defences (i.e., an elicitor, reviewed in Kallure et al., 2022). Although effectors are associated with defence suppression, herbivores that induce defences may also possess them (Villarroel et al., 2016) and thus induction/suppression depends on the host's ability to detect and respond to the collective of secreted substances (Cui et al., 2022). To date, no studies have addressed intraspecific genetic variation in herbivore effectors.
Tetranychus urticae is a generalist spider mite species (Helle & Sabelis 1985) and a model herbivore for studying plant-herbivore interactions (Blaazer et al. 2018). When feeding on tomato plants (Solanum lycopersicum), most of the T. urticae strains studied induce plant defences in the JA and SA pathway, with some being susceptible, leading to decreased herbivore performance and survival (e.g., Kant et al., 2008; Sarmento et al., 2011a, b; Alba et al., 2015; Godinho et al., 2016). As such, most of the populations of this species are believed to be susceptible inducers of tomato JA and SA defences. However, some strains can be resistant to plant defences, despite strong induction upon herbivore feeding (Kant et al., 2008). It was shown that the T. urticae genome has multiple gene families implied in xenobiotic metabolism, which are plastically expressed when adapting to new environments (Zhurov et al., 2014; Wybouw et al., 2015). This suggests that metabolic resistance may be a prominent trait underlying host adaptability in this species (Van Leeuwen and Dermauw, 2016; Blaazer et al., 2018). Nevertheless, adaptation to novel challenging environments was also associated with defence suppression (Wybouw et al., 2015). Indeed, some populations can suppress JA- and SA-related defences, leading to an increased performance of this herbivore (Kant et al., 2008; Sarmento et al., 2011a, b; Alba et al. 2015). Defence suppression by spider mites seems to be associated with the production of salivary effectors of family 84 (effector 84 for short, Villaroel et al., 2016; Schimmel et al., 2017; Liu et al., 2020a). However, the expression of such effectors was only studied in one inducer T. urticae strain (Villaroel et al., 2016; Schimmel et al., 2017; Liu et al., 2020a, b) and one suppressor strain from the sister species T. evansi (Villaroel et al., 2016; Schimmel et al., 2017; Liu et al., 2020a, b). The expression of effector 84 is plastic and changes in response to the presence of con- and heterospecific competitors (Schimmel et al., 2017), to light/dark conditions (Liu et al., 2020a) and differs across mite life stages and sexes (Liu et al., 2020b). Also, it is expected that several effectors in the same individual are associated with defence induction and suppression and host adaptation (Villarroel et al., 2016; Jonckheere et al., 2018; Iida et al., 2019; Cui et al., 2022).
Previous studies focused on the occurrence of traits that allow mites to suppress tomato JA defences in populations living on non-solanaceous host plants e.g. European spindle tree (Euonymus europea), deadnettle (Lamium album) and castor oil (Ricinus communis), in comparison to a benchmark mite strain collected from cultivated tomato and resistant to its JA-defences (strain Houten-1, Kant et al., 2008; Alba et al., 2015). In this study, we focussed on natural populations living on biologically grown field tomatoes, as it was shown that non-suppressor mite strains from bean can evolve defence suppression on tomato after 30 generations (Wybouw et al., 2015). Hence, we aimed to understand if mites naturally occurring on field-grown tomatoes display this trait as well. Here, we compared three T. urticae populations sampled from tomato at three field sites and also an outbred population created from these via controlled crosses. We then assessed their fecundity on tomato plants with (wildtype, WT) and without (def-1) inducible JA-defences, as well as the magnitude of JA defences they induce in WT plants, to discriminate between resistant types (high induction and high performance) and those that can suppress defences (low induction, high performance). We then aligned these data with data on variation in mitochondrial DNA cytochrome oxidase I (COI) and effector 84 (which can suppress JA-defences), in order to compare genetic diversity patterns among mite lines that induce or suppress defences. We found that suppression is the dominant phenotype in mite populations collected from field-grown tomatoes, and that inducers and suppressors predominantly cluster in distinct effector 84 clades.
2. Material and Methods
2.1. Plants
Tomato plants (Solanum lycopersicum L.), of the varieties Castlemart (both WT and defenseless-1, def-1) and Moneymaker, and bean plants (Phaseolus vulgaris L. cv Speedy) were germinated and grown (soil – Siro interior, Siro, Mira, Portugal) in a climatic chamber (photoperiod of 16:8h, 25:18°C, day:night, 50-60% relative humidity (RH)) for 28 and 10 days, respectively. On def-1 tomato plants, JA defences cannot be induced (Bergey et al., 1996; Howe et al., 1996; Li et al., 2002), making them a great tool to assess the effect of JA defences on spider mites. Experiments were carried out in a climatic chamber (photoperiod of 16:8h, 23.5±2°C, 70% RH), to which WT and def-1 tomato plants (cv Castlemart) were transferred one day before the beginning of the experiment.
2.2. Spider mites
T. urticae was sampled in the field at three locations, from here on referred to as three mite populations (ALP, DEF and MON). The populations were collected in tomato organic farms in Portugal in 2017 and were subject to a heat shock treatment, for 42 days, to remove endosymbionts such as
Wolbachia (Godinho
et al. 2020,
Table 1). These symbiont-free populations were then used to create an outbred laboratory population (Outbred), by performing inter-population, two-way (i.e., female 1 x male 2 and female 2 x male 1), controlled crosses, to normalize over-representation of genotypes. The outbred population started with more than 100 individuals, (>50 males and >50 females), of each cross combination (Godinho
et al. 2020). All field and outbred populations were reared on detached tomato leaves (cv Moneymaker) with the petioles submerged in water.
All populations were maintained in a climatized room (photoperiod of 16:8h, 23.5±2°C), inside 4L boxes with a mesh lid to allow airflow.
Benchmark populations and biomarker
T. urticae Santpoort-2 (“KMB” in Kant et al. 2008, "Santpoort-2" in Alba et al. 2015) and T. evansi Baker & Pritchard Viçosa-1 (Alba et al. 2015) were used as defence-inducer and defence-suppressor benchmarks, respectively. These populations have been widely used in studies of tomato-mite interactions (e.g., Sarmento et al. 2011a, b; Villarroel et al. 2016; Schimmel et al. 2017; Liu et al. 2020a, b) and are well-characterized inducer and suppressor populations, respectively (Alba et al. 2015). The defence-inducer benchmark was reared on detached bean plants (cv. Speedy) and the defence-suppressor benchmark was reared on detached tomato leaves (cv Castlemart). Plants infested with these benchmark populations and uninfested plants were used as controls across all assays.
T. urticae Santpoort-2 can also be used as a biomarker of JA-induced defences since this population is susceptible to JA defences (Kant et al. 2008). When on a plant pre-infested with an inducer mite, the fecundity of this biomarker is lower than on an uninfested plant or than on a plant pre-infested by a suppressor population (Kant et al. 2008, Sarmento et al. 2011a, Alba et al. 2015). As such, this is a perfect biomarker to disentangle between resistant (i.e., inducer/resistant) and suppressor strains.
2.3. Fecundity of T. urticae from field-collected populations on WT and def-1
We assessed the fecundity of each of the T. urticae populations when JA defences are induced (using WT) and in the absence of these defences (using def-1). To this end, 30 mated females (15±1 day old) from each T. evansi and T. urticae population were placed on a non-terminal leaflet, of a fully expanded leaf, of WT or def-1 tomato plants (day 0). Mite dispersal was prevented by isolating the adaxial surface of this leaflet with a 1:1 mix of entomological glue (Tanglefoot, The Scotts Company LLC, OH, USA), and lanolin (Sigma-Aldrich, St Louis, MO, USA), which was distributed around the adaxial edge of the leaflet. For each population and replicate, individual plants were used. Following four days of infestation (day 4), the number of surviving females on the leaflet and the number of eggs laid were counted. With these two measures, we calculated fecundity per female assuming linear mortality (Li & Zhang, 2022) by using: [total eggs] / [(alive females + total females)/2] and using these numbers as the average per female. There were ten to 11 and eight to nine replicates for each population, on WT and def-1, respectively. Assays on WT and def-1 plants were not performed simultaneously due to logistical constraints.
2.4. Fecundity of the biomarker strain on leaflets that had been pre-infested by mites from field-collected populations
Similar to what was described above, we placed biomarker mites on WT plants that had previously been infested, for four days, with our field and outbred populations. We first infested tomato leaflets with mites from a field-collected population for four days and then removed all mites and eggs to clean the leaflet. Cleaned leaflets were then re-infested with three biomarker individuals (15±1 days old). Biomarker survival and the number of eggs were then recorded after 48h (i.e. 6 days post the primary infestation). There were five to 11 replicates for each population.
2.5. Induction of tomato defence genes by the field-collected spider mites and expression levels of their effector 84
We analysed transcript accumulation of genes implicated in JA defences (Proteinase Inhibitor IIc, WIPI-IIc and Proteinase Inhibitor IIf, WIPI-IIf; Alba et al. 2015) and SA defences (Pathogenesis-related protein 1a, PR-1a; Alba et al. 2015) in WT plants infested with mites from the field-collected and outbred populations. We also analysed transcript accumulation of effector 84 from these same mites by using the same RNA that had been collected for assessing the expression of the plant defence genes. Ribosomal protein 49 (RP49) and Actin were used as housekeeping genes for spider mites and tomato plants, respectively (see Table S1 for primer sequences).
Material for collecting mite and plant RNA was prepared as follows: from infested leaflets, we cut out the part of the infested lamina within the glue and lanolin barrier and this was flash-frozen in liquid nitrogen and stored at -80°C. For each treatment, we obtained four to 11 replicates.
Total RNA was isolated from leaves using a protocol adapted from Verwoerd et al. (1989). Our protocol differs from it in that: (i) we used phenol at room temperature (RT) instead of hot phenol (heated to 80°C) and (ii) completed the 5-minute sample incubation step at RT, instead of 80°C. Next, 2µg of RNA were DNAse-treated with Ambion Turbo DNA-free kit (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and cDNA was synthesised with RevertAid H Minus Reverse Transcriptase (Thermo Fisher Scientific, Waltham, Massachusetts, USA). 1µL of 10-times-diluted cDNA was used as the template for a 20µL quantitative reverse-transcriptase polymerase chain reaction (RT-qPCR) using the CFX96 Real-Time system (Bio-Rad, Hercules, California, USA) with SsoFast™ EvaGreen®Supermix (Bio-Rad, Hercules, California, USA). Transcript accumulation was normalized using the ΔCt method (Alba et al. 2015). To produce a graphical representation that is easier to read in a data-neutral manner, all values were divided by the lowest average in the graph.
2.6. Statistical Analysis
All statistical analyses were performed with the software R (version 4.2.2, R Development Core Team 2022, Chichester, UK).
Generalized mixed linear mixed models (GLMM) with a normal error structure (lmer, lme4 package, Bates et al. 2015) were performed to investigate whether fecundity (at day four) varied among populations and/or host plant (WT or def-1). Each model included population (i.e., T. urticae Outbred, ALP, DEF, MON), host plant (WT or def-1) and their interaction as fixed explanatory variables, and block as a random variable. This was repeated including the benchmark controls.
To evaluate if the fecundity of the biomarker, six days post primarily infestation, was affected by pre-infestation with the T. urticae populations, we performed a GLMM model with normal error structure (lmer, lme4 package) including population as a fixed explanatory variable and block as a random variable.
The normalized transcript accumulation of effector 84, four days after infestation, was analysed by performing a GLMM assuming a Gamma distribution and a log-link function (glmmTMB, glmmTMB package, Brooks et al. 2017). The model included population as a fixed explanatory variable and block as a random factor.
To assess the impact of mite feeding on the expression of plant marker genes, independent GLMMs assuming a Gamma distribution and a log-link function (glmmTMB, glmmTMB package) were performed for the genes WIPI-IIc, WIPI-IIf and PR-1a. All models included population, time point (i.e. four days post infestation – 4dpi - or after the second infestation with the biomarker, i.e. six days post infestation – 6 dpi) and their interaction as fixed explanatory variables and block as a random variable. The models included the benchmark controls.
When significant differences were found, multiple comparisons were performed using estimated marginal means (emmeans, emmeans package) (Lenth et al. 2019) and the p-values corrected using the false discovery rate (FDR) method (α= 0.05) (Benjamini & Hochberg, 1995).
2.7. Cytochrome oxidase 1 (COI) and effector 84 sequencing and assembly
The genomic DNA (gDNA) of single adult females of the field-grown and laboratory populations (
Table 1) was extracted by first crushing the specimens, previously frozen at -80°C, with a plastic pestle and adding 100μL of 5% Chelex solution (Chelex 100 sodium form, Sigma-Aldrich, S
T. Louis, USA) and 5μL proteinase-K (20 mg/mL). The samples were incubated at 56°C for 60 min, followed by denaturation at 95°C for eight minutes (Walsh
et al. 1991). Amplification of
COI (seven to 14 separate individuals per population) and
effector 84 (eight to 13 separate individuals per population; see primers in Table S1) was performed using endpoint PCR and the presence of PCR product was confirmed on ethidium bromide-stained agarose gels. The remaining PCR products (1x diluted) were sent to Eurofins Genomics for Sanger sequencing.
The sequences were assembled and edited (i.e., sequence peaks were confirmed and corrected when needed) in SeqMan Pro 14 (version 14.1.0 (118), DNASTAR Lasergene 14, Madison, Wisconsin, United States) and aligned, using MUSCLE (Edgar, 2004), in MEGA (version 11, Kumar, Stecher, & Tamura, 2021). The COI alignment consisted of 115 sequences (385 bp), from which 35 were from GenBank (34 of T. urticae and one of T. evansi as an outgroup). From these, haplotypes were obtained using the DNAcollapser tool of Fabox (Villesen et al. 2007).
The reference effector 84 gDNA sequences can be found in OrcAE (Sterck et al. 2012) as tetur01g01000 (T. urticae London and T. urticae Montpellier genome). The allele and protein variants obtained for effector 84 can be found in Table S2. The ORF in the effector 84 gDNA spans two exons: the first of 42 nucleotides and the second of 697 nucleotides separated by an intron 85 nucleotides long. We amplified 635 nucleotides from the second exon (see Figure S1). The effector 84 alignment consisted of 103 sequences (635 bp, which, based on the gene model of Villaroel et al. 2016, corresponds to nucleotides 55 to 690), from which three were from GenBank (two of T. urticae and one of T. evansi as an outgroup - the latter with 637 bp). Note that since female mites are diploid, we sometimes obtained two haplotypes per individual. To resolve this, we implemented a Bayesian statistical method for phasing both haplotypes using PHASE v2.1 (Stephens et al. 2001) in DNAsp v5 (Librado & Rozas 2009).
2.8. Sequence analysis
For effector 84, DNA polymorphisms were analysed using DNASP 4.0 (Rozas et al. 2003). As a measure of DNA polymorphism within populations, five parameters were estimated: the number of polymorphic/segregating sites (S), the total number of mutations (Eta), the number of haplotypes (h), the haplotype diversity (Hd, a measure of the frequencies and number of haplotypes among individuals, varying between 0-1, Nei 1987), and the nucleotide diversity (π, average weighted sequence divergence between haplotypes, varying between 0 for no divergence to 10% for very deep divergences, Tajima 1983). Note that this analysis was performed for the complete sequence data set, but also for individual populations (i.e., ALP, DEF, MON, outbred, London_NL).
2.9. Phylogenetic analysis
We used PartitionFinder2 (Lanfear et al. 2017) and ModelFinder (Kalyaanamoorthy et al. 2017) to select the best partition arrangement and the most suitable evolutionary substitution model (COI: HKY+F+I for subset1 = 1-385\3 3-385\3, and HKY+F for subset2 = 2-385\3; Tu84: GTR+F+I+G4 for subset1 = 1-638\3 2-638\3 3-638\3). Bayesian Inference phylogenies were
Tu84 sequences were translated following the gene model of Villaroel et al. (2016). For the protein analysis, the best phylogenetic model was estimated in MEGA 11. Then, a maximum likelihood phylogenetic analysis with 1000 bootstraps, using the LG+G model with gamma shape= 0,67, was performed, and a consensus tree with bootstraps higher than 50% was obtained using MEGA. The phylogenetic trees were edited in iTOL version 6.5.8 (Letunic and Bork 2021).
2.10. Accession numbers
The sequence data from this article are available at NCBI website (
http:www.ncbi.nlm.nih.gov) and can be found under the following accession numbers:
Tu84, OQ472023 - OQ472057;
COI, OQ510005 - OQ510008.
3. Results
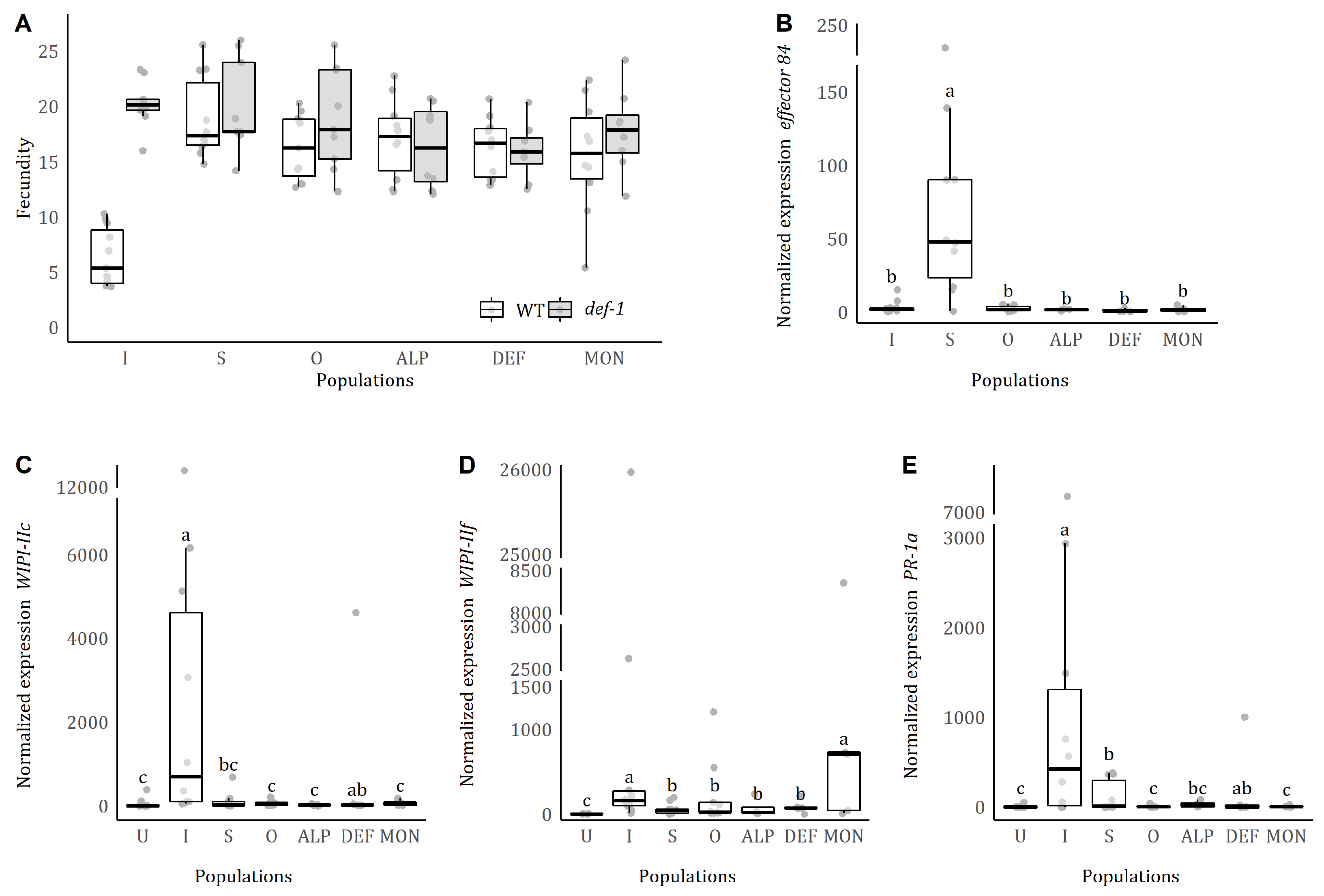
3.1. Fecundity of T. urticae from field-collected populations on WT and def-1
The field and outbred populations of
T. urticae were not significantly affected by the presence of JA-defences (
population*host plant:
= 3.971, p = 0.265;
host plant:
= 0.058, p = 0.809;
population: = 1.928, p = 0.588,
Figure 1A), with all having a similar fecundity on WT and
def-1 plants. Our benchmark controls followed a typical pattern of inducer and suppressor strains, respectively, with Santpoort-2 having a higher fecundity on
def-1 than on WT (p < 0.001) and Viçosa-1 having a similar fecundity on both WT and
def-1 (p = 0.859).
3.2. Induction of tomato defence genes by the field-collected spider mites and expression levels of their effector 84
The normalized transcript levels of
effector 84 were different across populations (
population: = 85.729, p<0.001,
Figure 1B). Post-hoc comparisons revealed that these differences are due to high levels of expression of this gene in
T. evansi Viçosa-1, which are 50 to 150 times higher than for
T. urticae. All
T. urticae populations, including the defence-inducer benchmark, had similar transcript levels of
effector 84.
Four days post infestation (4 dpi), the normalized transcript levels of JA and SA marker genes were different among populations (
population:
WIPI-IIc: = 49.805, p < 0.001;
WIPI-IIf: = 52.733, p < 0.001;
PR-1a: = 51.279, p < 0.001;
Figure 1C–F). Transcript levels in leaflets infested with the defence-suppressor benchmark were lower than the ones infested with the defence-inducer benchmark. Transcript accumulation levels for
WIPI-IIc and
PR-1a in plants infested with the field-collected or outbred populations were similar to the defence-suppressor benchmark and some were even similar to uninfested plants (Outbred, ALP and MON for
WIPI-IIc and
PR-1a). The transcript levels of
WIPI-IIf in leaflets infested with MON were as high as for leaflets infested with the defence-inducer benchmark.
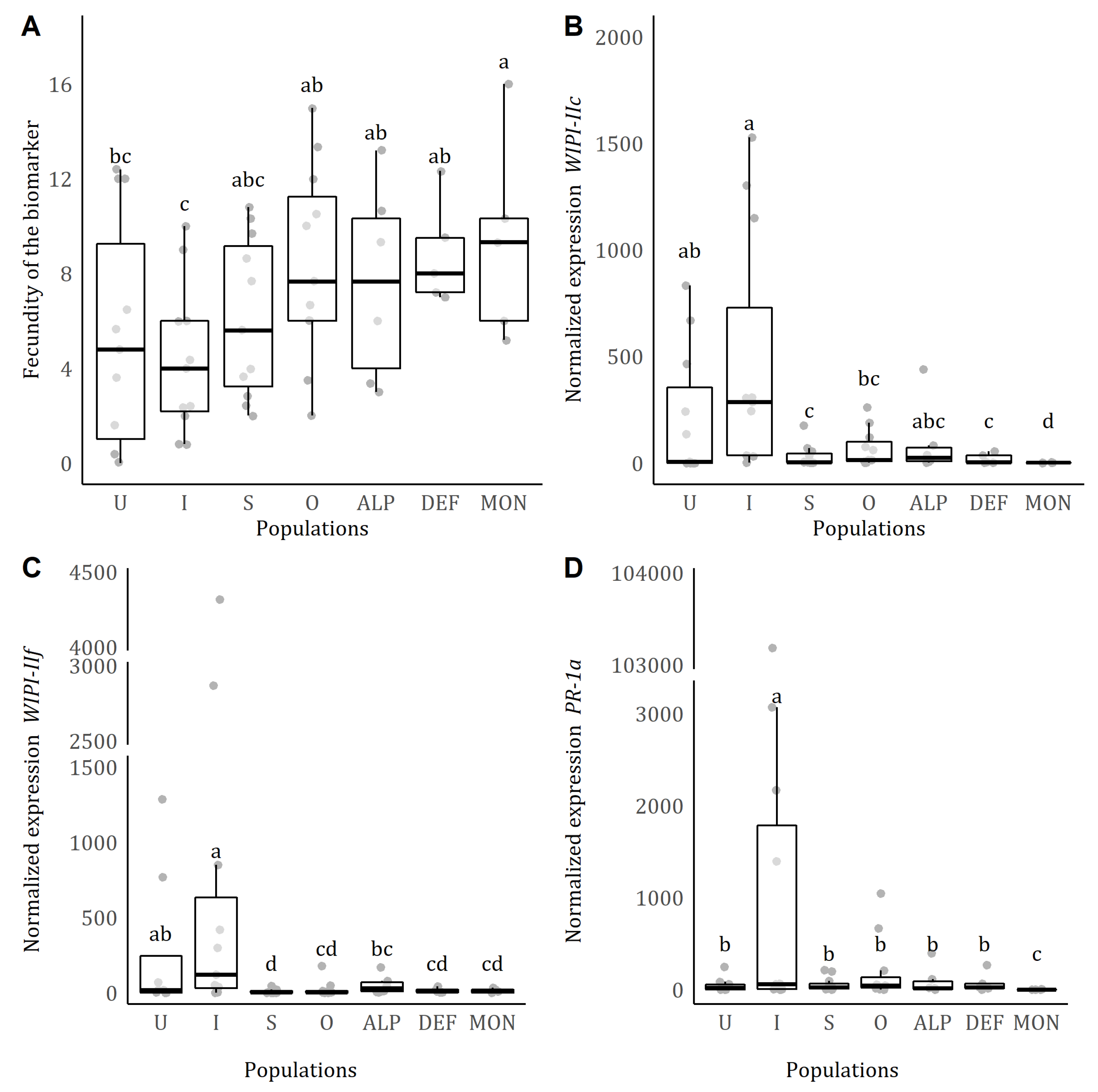
3.3. Fecundity of the biomarker strain on leaflets that had been pre-infested by mites from field-collected populations
The population pre-infesting the plant had a significant effect on the fecundity of the biomarker (
population: = 23.843, p < 0.001;
Figure 1B). Post-hoc comparisons revealed that the biomarker had higher fecundity on plants pre-infested with each of the four
T. urticae populations tested (Outbred, ALP, DEF and MON;
Figure 1B). However, there was no difference in fecundity on uninfested plants, or plants previously infested with the suppressor or the inducer benchmark.
After introducing three adult females of the biomarker strain, the transcript levels of all defence-related genes changed (
population*time point: WIPI-IIc: = 35.944, p < 0.001;
WIPI-IIf: = 49.365, p < 0.001;
PR-1a: = 13.929, p = 0.030;
Figure 2B–E). Post-hoc comparisons revealed that for all genes, except for
WIPI-IIc, transcript levels of uninfested plants were higher at 6 dpi compared to 4 dpi. However, for plants pre-infested with the field-collected, outbred and benchmark populations, transcript levels mostly remained similar to, or were lower than, 4 dpi (lower at 6 dpi for (i) defence-inducer benchmark, DEF and MON for
WIPI-IIc; (ii) defence-suppressor, Outbred and MON for
WIPI-IIf; (iii) Outbred for
PR-1a).
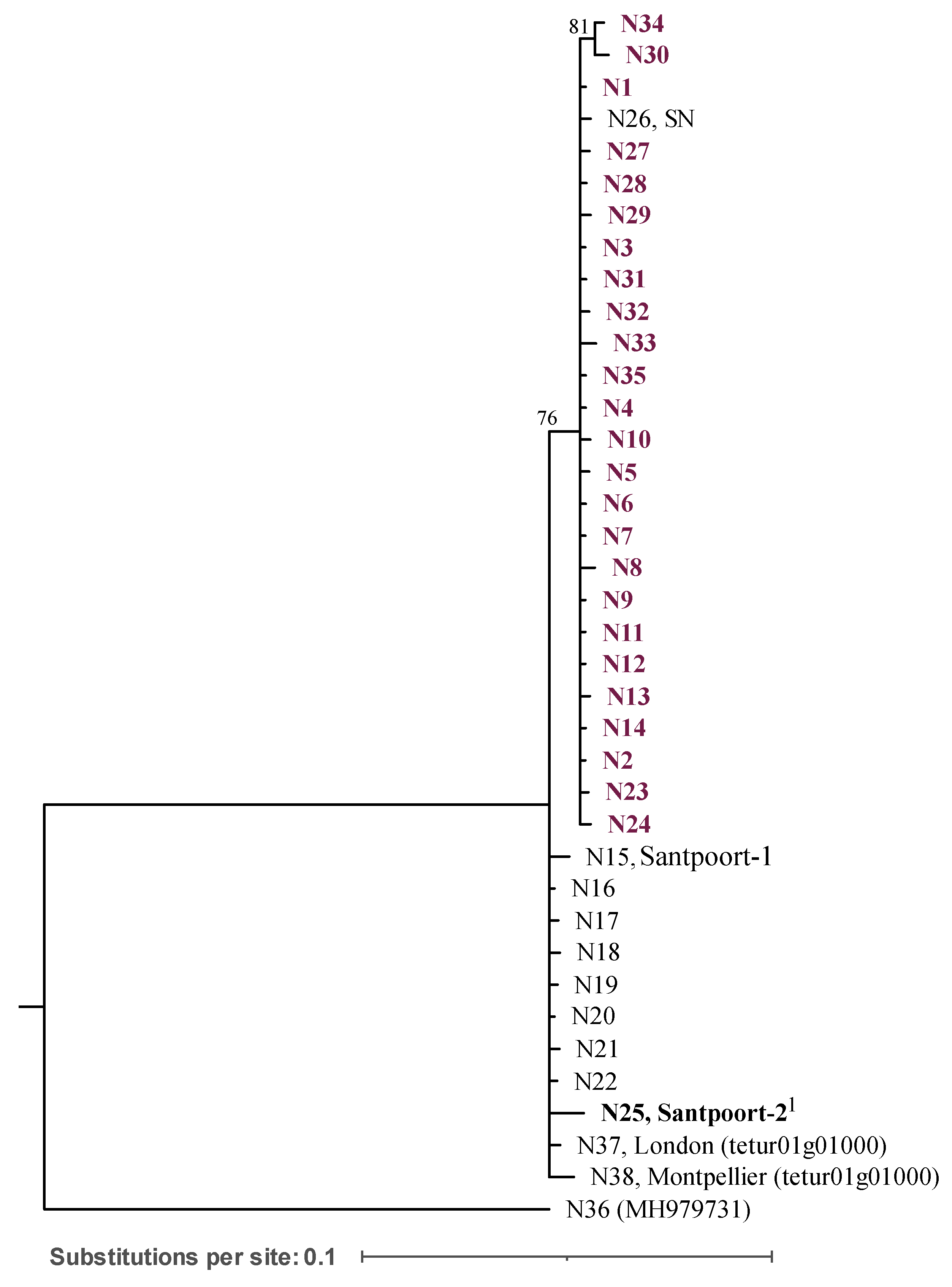
3.4. Phylogenetic analysis of COI and effector 84
Twenty mitochondrial haplotypes (M1 - M20) were found based on the COI sequences. All the populations sequenced, except Santpoort-1, belong to the same lineage of T. urticae (Figure S2).
For the
effector 84 (
Figure 3), 38 alleles (N1 - N38) were identified. The haplotype diversity (
Hd) and nucleotide diversity (
π) of all sequences were 0.910 and 0.017, respectively (
Table 2). From the populations harbouring different alleles, DEF and the outbred populations were the ones with higher
Hd and
π (
Table 2).
All the individuals sequenced from the defence-inducer population (Santpoort-2) had one allele (N25), sharing the same phylogenetic clade with Santpoort-1 (also with one allele, N15) and London_NL (with seven alleles). These populations shared the same clade with the reference alleles (N37 and N38). The population collected in Spain, SN (allele N26), and the Portuguese field and outbred populations were in a separate clade, supported by a bootstrap of 76%. The allele most shared between individuals in the field and, consequently in the outbred population, was N2 (represents 53% for ALP, 33% for DEF, 78% for MON and 37% for Outbred of all allelic variations). We found multiple alleles within ALP, DEF and MON (5, 8 and 3 alleles, respectively) with some being singletons (i.e., alleles represented by a single sequence). Twelve alleles were found in the outbred population, of which 3 were also present in the field-collected populations. The 38 alleles were found to encode 26 different proteins (P1 – P26). The same arrangement of the phylogenetic tree for the nucleotide clades was observed for the protein sequences (Figure S2).
4. Discussion
In this study, we showed that spider mites collected from populations naturally occurring on tomato plants in the field can be classified as suppressors of tomato defences. The field-collected and outbred populations had similar fecundity on WT and def-1, and they suppressed the induction of tomato defence genes by the defence-inducer benchmark strain. Also, the field-collected mites and the outbred population promoted the fecundity of the biomarker strain. Expression levels of effector 84, associated with defence suppression by mites, were similar across T. urticae lines that induce or suppress tomato defences. However, we also observed distinct genetic sequences between the effector 84 alleles of inducers and suppressors. We speculate that such allelic diversity may be subject to natural selection when these mites colonize a novel host plant and promote traits that counter plant resistance, such as defence-suppression, thereby facilitating adaptation.
Our observation aligns well with predictions that follow previous research. Previous research showed that spider mites collected in the field from non-Solanaceous hosts, notably spindle tree and deadnettle, predominantly consist of individuals that are maladapted to tomato (as a novel host) (Kant et al. 2008, Alba et al. 2015). Most of these mites were found to strongly induce JA defences in tomato plants and to be susceptible to these defences (Kant et al. 2008, Alba et al. 2015). However, in these populations individuals that can suppress or resist tomato defences were occasionally found, albeit at low frequencies (Kant et al. 2008; Villacis-Perez et al. 2022). This goes in line with what was previously shown (e.g., Magalhães et al. 2007), as it suggests that such natural spider mite populations may contain sufficient standing variation to adapt to a novel host like tomato. This was confirmed by Wybouw et al. (2015) where it was shown that spider mites from bean could adapt to tomato and that this coincided with the emergence of the ability to suppress tomato defences. Together these observations suggest that spider mites living on a non-tomato host will have a lower fitness once on tomato, but will adapt to it when natural selection favours the low-frequency traits that allow these mites to resist or suppress tomato defences.
4.1. Defence suppression of tomato defences is common in T. urticae collected from field-grown tomatoes
The defence-inducer strain was the only one in which the oviposition on
def-1 was higher than on WT (
Figure 1A). For the remaining populations, oviposition on both hosts was similar, suggesting that they can cope with induced JA defences. As observed previously, defence inducer mites have enhanced oviposition on
def-1 compared with WT (Kant
et al. 2008, Alba
et al. 2015), while for defence suppressors the oviposition is equivalent on both tomato plants (Li
et al. 2002, Kant
et al. 2008, Alba
et al. 2015). However, similar oviposition on both WT and
def-1 can be a sign of either metabolic resistance against plant defences induced by the spider mites or defence suppression (Kant
et al. 2008).
Indeed, we observed that all mites collected from populations of field-grown tomato plants tested could suppress tomato defences to varying levels and that this trait was maintained in an outbred population. For all tomato defence genes measured, normalized transcript accumulation was higher in plants infested by the defence-inducer benchmark and lower for the remaining populations (
Figure 1C–E), sometimes to levels of uninfested plants (for
WIPI-IIc and
PR-1a). However, some variation in levels of defences was observed, with some populations not differing from the defence-inducer benchmark for some genes (i.e., DEF for
WIPI-IIc and
PR-1a, and MON for
WIPI-IIf). Despite this variation, the overall results indicated that the field populations upregulate defences to much lower levels than the defence-inducer benchmark whereas, in turn, the inducer benchmark did not upregulate defences anymore in plants pre-infested with a field strain (
Figure 2B,D). These data indicate that field-collected mites can suppress inducible plant defences.
4.2. Spider mites collected from field-grown tomatoes promote fecundity and suppress defence genes induced by the biomarker strain
Previous results on the effect of suppression on the fecundity of a biomarker strain suggested that defence suppression has a persistent effect after the removal of the inducer/suppressor mites (Kant et al. 2008; Alba et al. 2015; Godinho et al. 2016; de Oliveira et al. 2016; Schimmel et al. 2017, 2018). However, if suppression of gene expression is maintained was not previously assessed (but see Teodoro-Paulo et al. 2022, Chapter 2). The results here suggest this to be the case. Similarly, it had been found previously that suppression of defence genes by the T. urticae sister species T. evansi, is retained for at least four days (de Oliveira et al. 2019). It is possible that, because mite feeding and oviposition occur in the same location, a lasting effect of suppression by the adults provides their offspring with better food (Bruessow et al. 2010). As such a longer-lasting effect of defence suppression, especially for SA marker genes, could provide a beneficial environment for offspring development. Investigating this further will give insights into the mechanism (why it is lasting), function and ecological costs (who benefits from it – offspring and/or competitors). This may help to formulate testable hypotheses regarding mite life history, community composition and adaptation.
In line with the defence gene induction/suppression pattern, we found that pre-infestation with mites collected from field-grown tomatoes and the outbred population increased the fecundity of the biomarker strain (
Figure 2A). Our results indicate that, although the magnitude of suppression was not uniform across the mite lines we tested, the effect on the fecundity of the biomarker was similar. In this experiment, the effect of the defence-inducer and defence-suppressor benchmarks on the biomarker strain was not significant, probably due to a large amount of variation in the uninfested control (
Figure 2A). Previous studies observed clearer effects of the induction and suppression benchmarks on the biomarker strain (Kant
et al. 2008; Alba
et al. 2015; Godinho
et al. 2016). However, other studies also failed to observe significant effects using this assay despite seeing the expected trends and this may be due to power (sample size) and timing (de Oliveira
et al. 2016; Schimmel
et al. 2017, 2018; Blaazer
et al. 2018).
4.3. There is significant genetic variation in effector 84 across T. urticae populations
Despite the variation in tomato defence suppression among the field-collected spider mites and outbred population, the expression levels of the salivary
effector 84 were similar to those of the defence-inducer benchmark (
Figure 1B).
effector 84 from both
T. urticae and
T. evansi downregulate defence gene expression in
N. benthamiana upon transient overexpression (Villarroel
et al. 2016) and it can suppress elicitor-induced cell death in both
N. benthamiana and tomato (Cui
et al. 2022). Moreover,
effector 84 expression in
T. urticae correlates negatively with the magnitude of induced defences in tomato (Schimmel
et al. 2017, b; Liu
et al. 2020a, b). However, given our
effector 84 expression data, it is certain that this effector is not the only determinant of defence suppression by
T. urticae. Probably, induction/suppression results from a complex interplay between mite effectors and elicitors (Cui
et al. 2022; Villarroel
et al. 2016; Iida
et al. 2019; Jonckheere
et al. 2016; 2018), and sequence and expression variation therein. To this end, we explored effector sequence variation at the nucleotide and amino acid level and performed a phylogenetic analysis to search for relationships between sequence variation and phenotype.
Unlike Ínak et al. 2019, which found two mitochondrial lineages of T. urticae, our analysis revealed three mitochondrial lineages (Figure S2). This might be explained by the inclusion of several COI-haplotypes not considered in the previous phylogeny, such as the Portuguese and Dutch populations, and the use of different phylogenetic methodologies. We found that the three field-collected populations, and hence the outbred population, are of the same COI-haplotype as London_NL (haplotype M6, lineage I, Figure S2). These populations shared the same lineage (lineage I) as the Spanish population (SN, haplotype M7) and the defence-inducer benchmark (Santpoort-2, haplotype M13), although these two were more closely related. Santpoort-1 belongs to a more distinct lineage (lineage II, haplotype M11, Figure S2). Although the field-collected populations shared the same COI-haplotype (at least in the fragment sequenced), they are from different origins and have different nuclear alleles (effector 84, discussed below), suggesting that these populations are genetically different.
Across all the mite lines we tested, we found 38 different alleles for
effector 84, and these translated into 26 different proteins. We do not know if some of the amino acid variations we observed affect
effector 84 functionality or structure. The alpha-fold structure prediction of
effector 84 is weak (see Uniprot (UniProt Consortium 2019): T1JPW1) and cannot be used to estimate the effect of single amino acid changes on structure and function and thus functional studies as in Villarroel et al (2016) or Cui et al (2022) will be necessary to explore this further. Across the
T. urticae lines and populations we sequenced,
effector 84 has a high haplotype diversity (0.910,
Table 2) but a very low nucleotide diversity (0.017%,
Table 2). Patterns similar to this global one were observed in several of the local populations (ALP, DEF, London). These patterns may be the result of the action of purifying selection and/or rapid population rebound following a drastic past demographic bottleneck (Grant & Bowen 1998). We identified 38 alleles across all the populations sequenced, with the field populations harbouring 15 of these, and the outbred population 12 (of which, three were shared with the field populations). This suggests high genetic differentiation between the founders of the outbred population and the individuals analysed here representing each of the three field populations. This high genetic differentiation by chance underscores the huge haplotype diversity in each of the three source populations. Nevertheless, the SNPs present in the most common allele (Figure S5), which is shared between field-collected and outbred populations, N2 (which codes for protein P1), could explain defence suppression in these populations.
4.4. Inducer and suppressor mites predominantly cluster in different effector 84 clades
We observed that inducer and suppressor populations do have different alleles and proteins. However, these differences do not segregate completely into inducer and suppressor mites (
Figure 3). Instead, the Spanish population, SN (allele N26, found in an alleged inducer population, Sato
et al. 2014) clustered with the field and outbred populations collected in Portugal (alleles N1-14, N23-24, and N27-35). Also,
T. urticae Santpoort-1 (allele N15, characterized as a suppressor population in Kant
et al. 2008) clustered with the defence-inducer benchmark, Santpoort-2, and with
T. urticae London (allele from reference genome: N25 and alleles from London_NL: N16-22). This can be explained by various factors. First, the two clusters observed in the
effector 84 gene tree may represent different geographical origins of the populations, as those collected in Portugal (ALP, DEF, MON) and Spain (SN) clustered together. Second, this clustering could be related to host adaptation. The populations collected in Portugal and Spain were collected on
Solanum hosts, while the others were collected from more distal hosts. It was previously observed that populations of
T. urticae from bean can adapt via natural selection to tomato plants, increasing their metabolite detoxification potential and their ability to reduce the production of plant defence compounds (Wybouw
et al. 2015). This suggests that defence suppression of tomato defences may rapidly emerge as a result of the selection on tomato (Blaazer
et al. 2018). Indeed, the field-collected
T. urticae populations all had been obtained from tomato plants (in the same geographic area) and were maintained in the laboratory on tomato ever since (Godinho
et al. 2020). Also, the creation of the outbred population was performed entirely on tomato plants. Hence adaptation to tomato may have acted on (low frequency) traits that, coincidentally or not, allow for suppressing tomato defences. While selection for direct resistance to tomato defences may be characterized by rapid selection for detoxification genes (Dermauw
et al. 2013), selection for suppression may depend on selection for potent effector genes. This could have implications for agricultural systems (especially in monoculture scenarios) where the ecological costs of suppression may be relatively low due to a lack of community diversity. Although R-gene resistance breeding in plants (Keith & Mitchell-Olds, 2013) may provide a temporary solution, the mere existence of multiple effector alleles in all the populations and strains, suggests that such plant resistance can be broken easily (Pilet-Nayel., 2017). We advocate that searching for effector targets to design breeding schemes to remove them or to make them invulnerable to effector manipulation (Zaidi
et al. 2018) may be a more feasible approach than sustainable crop protection.
4.5. To resist or to suppress
Our results do raise important questions regarding the outcome of adaptive processes of herbivores that have to cope with the novel defences of novel hosts such as mites on tomato.
First, the question of what determines whether suppression or resistance is favoured by natural selection. This will probably strongly depend on the standing variation in the founder population, which, in turn, is determined by selection (time), drift and gene flow on the previous host. Subsequently, it will also be determined by the physiological and ecological costs of the novel host. Physiological because the ease by which mites can degrade, modify, secrete or sequester defensive metabolites will be different across different plant hosts (Yang et al. 2001). Also, the efficiency of their effectors on the novel host, determined by the absence/presence of suitable targets as well as of R-genes, and the effector variants present in the population will determine the success of suppression as an adaptive strategy (Blaazer et al. 2018). Furthermore, also the presence of competitors will influence whether resistance may be favoured over suppression because the latter is a common good (Kant et al. 2008, Sarmento et al. 2011a, Alba et al. 2015) whereas the first is not. Finally, it will be determined by the ability of the local herbivore population to mitigate the ecological costs, as the sister species T. evansi actively excludes its competitor T. urticae from suppressed plant material via thick webbing production (Sarmento et al. 2011b), suppression localization (Schimmel et al. 2017), and reproductive interference (Sato et al. 2014).
Second, there is the question of whether resistance and suppression are complementary traits that can coexist in populations, or whether one will exclude the other or whether one will follow the other. Also, this will probably be determined by the physiology and secondary chemistry of the novel host as well as gene flow between mite populations in the vicinity of different hosts. It was suggested that suppression of defences via effectors may promote fitness less than direct resistance (i.e. may be less effective in counteracting plant defences in absolute terms, yet may operate on a wider host range, Kant et al. 2014). In that view, suppression may be favoured by selection first (Wybouw et al. 2015) and then later be replaced by resistance to mitigate the ecological costs of suppression (Blaazer et al 2018). However, this is not in line with the observation that populations from all over the world of the tomato specialist T. evansi cope with tomato, and other hosts (Paulo et al. 2018), via suppression (Knegt et al. 2020). Maybe this species lacks the necessary building blocks for evolving resistance or its active exclusion of competitors from its feeding site (Sarmento et al. 2011b; Schimmel et al. 2017; Sato et al. 2014) may buffer selection against suppression.
All these hypotheses need to be addressed in future research to gain a better understanding of how herbivore intraspecific variation, gene flow, host plant identity, and community composition determine the outcome of adaptive processes of herbivores on novel hosts, and the evolution of host races, for specialists and also for generalists living on a mosaic of plant environments.
5. Conclusions & Perspectives
Our results show that the ability to suppress tomato defences in T. urticae is common among mites living on tomato in the field. We argue that this trait may emerge in populations that have dispersed from an unrelated host, given that the traits needed for suppressing tomato defences are included in its standing variation as had been shown previously (Kant et al. 2008, Alba et al. 2015, Wybouw et al. 2015). The question remains: what determines direct resistance to tomato defences to emerge and how this relates to suppression? We also argue that standing genetic variation in effectors may be a co-determinant of such adaptive processes as well as host identity. Rapid adaptation of mites to plant resistance can be of great concern and especially impact monocultures in agriculture. Since traits related to induction/suppression and resistance/susceptibility to a novel host may be retained (at low frequencies) in natural populations (Kant et al. 2008; Villacis-Perez et al. 2022), it would now be informative to transfer our tomato lines to unrelated hosts to see if a similar adaptive response can be observed under these new conditions. We feel that studies regarding effector genetic and functional variation may reveal some of the mechanisms that drive plant-herbivore co-evolution.
Author Contributions
JTP, JMA and SM conceptualized the experimental design. JTP conducted the experiments, data analyses and designed the figures. JTP and CF conducted the phylogenetic analysis. LD sequenced the samples. JTP wrote the original manuscript. ABD, JMA, CF, MRK critically revised the manuscript and contributed to its final version.
Funding
Funds were provided by an ERC consolidator grant (COMPCON, GA 725419) to SM; the Dutch Research Council (ALWOP.283; NWO-VICI 19391; U Horizon 2020 research and innovation programme (Grant 773902 — SuperPests)) to MK and FCT Doctoral grant (SFRH/BD/136416/2018) to JTP.
Ethics Statements
This article does not present research with ethical considerations.
Conflicts of Interest
We declare we have no competing interests.
References
- Agrawal, A.A., Karban, R., 1999. Why induced defenses may be favored over constitutive strategies in plants. The ecology and evolution of inducible defenses 10, 000331745. [CrossRef]
- Alba, J.M., Schimmel, B.C.J., Glas, J.J., Ataíde, L.M.S., Pappas, M.L., Villarroel, C.A., Schuurink, R.C., Sabelis, M.W., Kant, M.R., 2015. Spider mites suppress tomato defenses downstream of jasmonate and salicylate independently of hormonal crosstalk. New Phytologist 205, 828–840. [CrossRef]
- Ali, J.G., Agrawal, A.A., 2012. Specialist versus generalist insect herbivores and plant defense. Trends in plant science 17, 293–302. [CrossRef]
- Ament, K., Kant, M.R., Sabelis, M.W., Haring, M.A., Schuurink, R.C., 2004. Jasmonic Acid Is a Key Regulator of Spider Mite-Induced Volatile Terpenoid and Methyl Salicylate Emission in Tomato. Plant Physiology 135, 2025–2037. [CrossRef]
- Azandeme-Hounmalon, G.Y., Fellous, S., Kreiter, S., Fiaboe, K.K., Subramanian, S., Kungu, M., Martin, T., 2014. Dispersal behavior of Tetranychus evansi and T. urticae on tomato at several spatial scales and densities: implications for integrated pest management. PloS one 9, e95071. [CrossRef]
- Bates, D., Kliegl, R., Vasishth, S., Baayen, H., 2015. Parsimonious mixed models. arXiv preprint. arXiv:1506.04967.
- Benjamini, Y., Hochberg, Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society series b-methodological 57, 289–300. [CrossRef]
- Berenbaum, M.R., 1986. Target Site Insensitivity in Insect-Plant Interactions 257–272. [CrossRef]
- Bergey, D.R., Howe, G.A., Ryan, C.A., 1996. Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proceedings of the National Academy of Sciences of the United States of America 93, 12053–12058. [CrossRef]
- Blaazer, C.J.H., Villacis-Perez, E., Chafi, R., Van Leeuwen, T., Kant, M.R., Schimmel, B.C.J., 2018. Why Do Herbivorous Mites Suppress Plant Defenses. Frontiers in Plant Science 9, 1057–1057. [CrossRef]
- Bowers, M.D., 2022. Sequestered Caterpillar Chemical Defenses: From “Disgusting Morsels” to Model Systems, in: Marquis, R.J., Koptur, S. (Eds.), Caterpillars in the Middle: Tritrophic Interactions in a Changing World. Springer International Publishing, Cham, pp. 165–192. [CrossRef]
- Brooks, M.E., Kristensen, K., van Benthem, K.J., Magnusson, A., Berg, C.W., Nielsen, A.W., Nielsen, A., Skaug, H.J., Mächler, M., Bolker, B.M., 2017. glmmTMB Balances Speed and Flexibility Among Packages for Zero-inflated Generalized Linear Mixed Modeling. R Journal 9, 378–400. [CrossRef]
- Bruessow, F., Gouhier-Darimont, C., Buchala, A., Metraux, J.-P., Reymond, P., 2010. Insect eggs suppress plant defence against chewing herbivores. The Plant Journal 62, 876–885. [CrossRef]
- Chen, D.-S., Jin, P.-Y., Zhang, K.-J., Ding, X.-L., Yang, S.-X., Ju, J.-F., Zhao, J.-Y., Hong, X.-Y., 2014. The complete mitochondrial genomes of six species of Tetranychus provide insights into the phylogeny and evolution of spider mites. PloS one 9, e110625. [CrossRef]
- Cianfrogna, J., Zangerl, A., Berenbaum, M., 2002. Dietary and developmental influences on induced detoxification in an oligophage. Journal of chemical ecology 28, 1349–1364. [CrossRef]
- Cui, J.-R., Bing, X.-L., Tang, Y.-J., Liu, F., Ren, L., Zhou, J.-Y., Liu, H.-H., Wang, M.-K., Hoffmann, A.A., Hong, X.-Y., 2022. A conserved protein disulfide isomerase enhances plant resistance against herbivores. Plant Physiol kiac489. [CrossRef]
- de Mendonça, R.S., Navia, D., Diniz, I.R., Auger, P., Navajas, M., 2011. A critical review on some closely related species of Tetranychus sensu stricto (Acari: Tetranychidae) in the public DNA sequences databases. Experimental and Applied Acarology 55, 1–23. [CrossRef]
- de Oliveira, E.F., Pallini, A., Janssen, A., 2019. Herbivore performance and plant defense after sequential attacks by inducing and suppressing herbivores. Insect Science 26, 108–118. [CrossRef]
- de Oliveira, E.F., Pallini, A., Janssen, A., 2016. Herbivores with similar feeding modes interact through the induction of different plant responses. Oecologia 180, 1–10. [CrossRef]
- Dermauw, W., Wybouw, N., Rombauts, S., Menten, B., Vontas, J., Grbić, M., Clark, R.M., Feyereisen, R., Van Leeuwen, T., 2013. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proceedings of the National Academy of Sciences 110, E113–E122.
- Després, L., David, J.-P., Gallet, C., 2007. The evolutionary ecology of insect resistance to plant chemicals. Trends in Ecology and Evolution 22, 298–307. [CrossRef]
- Edgar, R.C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32, 1792–1797. [CrossRef]
- Farmer, E.E., Johnson, R.R., Ryan, C.A., 1992. Regulation of Expression of Proteinase Inhibitor Genes by Methyl Jasmonate and Jasmonic Acid. Plant Physiology 98, 995–1002. [CrossRef]
- Folmer, O. et al. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. - Molecular Marine Biology and Biotechnology 3: 294–299.
- Godinho, D.P., Cruz, M.A., De La Masseliere, M.C., de la Masselière, M.C., Teodoro-Paulo, J., Eira, C., Fragata, I., Rodrigues, L.R., Zélé, F., Magalhães, S., 2020. Creating outbred and inbred populations in haplodiploids to measure adaptive responses in the laboratory. Ecology and Evolution 10, 7291–7305. [CrossRef]
- Godinho, D.P., Janssen, A., Dias, T., Cruz, C., Magalhães, S., 2016. Down-regulation of plant defence in a resident spider mite species and its effect upon con- and heterospecifics. Oecologia 180, 161–167. [CrossRef]
- Gotoh, T., Araki, R., Angham Boubou, Angham Boubou, Boubou, A., Migeon, A., Ferragut, F., Ferragut, F., Navajas, M., 2009. Evidence of co-specificity between Tetranychus evansi and Tetranychus takafujii (Acari: Prostigmata, Tetranychidae): comments on taxonomic and agricultural aspects. International Journal of Acarology 35, 485–501. [CrossRef]
- Grant, W., Bowen, B.W., 1998. Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of heredity 89, 415–426. [CrossRef]
- Grbić, M., Van Leeuwen, T., Clark, R.M., Rombauts, S., Rouzé, P., Grbić, V., Osborne, E.J., Dermauw, W., Thi Ngoc, P.C., Ortego, F., others, 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492. [CrossRef]
- Groen, S.C., Whiteman, N.K., 2022. Ecology and Evolution of Secondary Compound Detoxification Systems in Caterpillars, in: Marquis, R.J., Koptur, S. (Eds.), Caterpillars in the Middle: Tritrophic Interactions in a Changing World. Springer International Publishing, Cham, pp. 115–163. [CrossRef]
- Hai-Jian Huang, Huang, H.-J., Cui, J.-R., Chen, L., Zhu, Y.-X., Hong, X.-Y., 2019. Identification of Saliva Proteins of the Spider Mite Tetranychus evansi by Transcriptome and LC-MS/MS Analyses. Proteomics 19, 1800302. [CrossRef]
- Heckel, D.G., 2014. Insect detoxification and sequestration strategies 77–114. [CrossRef]
- Helle, W., Sabelis, M.W., 1985. Spider mites: their biology, natural enemies and control. Elsevier Amsterdam.
- Howe, G.A., Lightner, J., Browse, J., Ryan, C.A., 1996. An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. The Plant Cell 8, 2067–2077. [CrossRef]
- Iida, J., Desaki, Y., Hata, K., Uemura, T., Yasuno, A., Islam, M., Maffei, M.E., Ozawa, R., Nakajima, T., Galis, I., others, 2019. Tetranins: new putative spider mite elicitors of host plant defense. New Phytologist 224, 875–885. [CrossRef]
- İnak, E., Alpkent, Y.N., Sultan Çobanoğlu, Dermauw, W., Van Leeuwen, T., Van Leeuwen, T., 2019. Resistance incidence and presence of resistance mutations in populations of Tetranychus urticae from vegetable crops in Turkey. Experimental and Applied Acarology 78, 343–360. [CrossRef]
- Isman, M.B., Duffey, S.S., 1982. Toxicity of tomato phenolic compounds to the fruitworm, Heliothis zea. Entomologia experimentalis et applicata 31, 370–376. [CrossRef]
- Jonckheere, W., Dermauw, W., Khalighi, M., Pavlidi, N., Reubens, W., Baggerman, G., Tirry, L., Menschaert, G., Kant, M.R., Vanholme, B., Van Leeuwen, T., Van Leeuwen, T., 2018. A gene family coding for salivary proteins shot of the polyphagous spider mite tetranychus urticae exhibits fast host dependent transcriptional plasticity. Molecular Plant-microbe Interactions 31, 112–124. [CrossRef]
- Jones, A.C., Felton, G.W., Tumlinson, J.H., 2022. The dual function of elicitors and effectors from insects: reviewing the “arms race” against plant defenses. Plant Mol Biol 109, 427–445. [CrossRef]
- Kallure, G.S., Kumari, A., Shinde, B.A., Giri, A.P., 2022. Characterized constituents of insect herbivore oral secretions and their influence on the regulation of plant defenses. Phytochemistry 193, 113008. [CrossRef]
- Kalyaanamoorthy, S., Minh, B.Q., Wong, T.K.F., von Haeseler, A., Jermiin, L.S., 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14, 587–589. [CrossRef]
- Kant, M., Jonckheere, W., Knegt, B., Lemos, F., Liu, J., Schimmel, B., Villarroel, C., Ataide, L., Dermauw, W., Glas, J., others, 2015. Mechanisms and ecological consequences of plant defence induction and suppression in herbivore communities. Annals of botany 115, 1015–1051. [CrossRef]
- Kant, M.R., Ament, K., Sabelis, M.W., Haring, M.A., Schuurink, R.C., 2004. Differential Timing of Spider Mite-Induced Direct and Indirect Defenses in Tomato Plants. Plant Physiology 135, 483–495. [CrossRef]
- Kant, M.R., Sabelis, M.W., Haring, M.A., Schuurink, R.C., 2008. Intraspecific variation in a generalist herbivore accounts for differential induction and impact of host plant defences. Proceedings of The Royal Society B: Biological Sciences 275, 443–452. [CrossRef]
- Karban, R., Myers, J.H., 1989. Induced Plant Responses to Herbivory. Annual Review of Ecology, Evolution, and Systematics 20, 331–348. [CrossRef]
- Keith, R., Mitchell-Olds, T., 2013. Genetic variation for resistance to herbivores and plant pathogens: hypotheses, mechanisms and evolutionary implications. Plant Pathology 62, 122–132. [CrossRef]
- Knegt, B., Meijer, T.T., Kant, M.R., Kiers, E.T., Egas, M., 2020. Tetranychus evansi spider mite populations suppress tomato defenses to varying degrees. Ecology and Evolution 10, 4375–4390. [CrossRef]
- Lanfear, R., Frandsen, P.B., Wright, A.M., Senfeld, T., Calcott, B., 2016. PartitionFinder 2: New Methods for Selecting Partitioned Models of Evolution for Molecular and Morphological Phylogenetic Analyses. Molecular Biology and Evolution 34, 772–773. [CrossRef]
- Lenth, R.V., 2022. emmeans: Estimated Marginal Means, aka Least-Squares Means R package version 1.7.3.
- Letunic, I., Bork, P., 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic acids research 49, W293–W296. [CrossRef]
- Li, C., Williams, M.M., Loh, Y.-T., Lee, G.I., Howe, G.A., 2002. Resistance of Cultivated Tomato to Cell Content-Feeding Herbivores Is Regulated by the Octadecanoid-Signaling Pathway. Plant Physiology 130, 494–503. [CrossRef]
- Li, G.-Y., Zhang, Z.-Q., 2022. Age-specific mortality and fecundity of a spider mite under diet restriction and delayed mating. Insect Science 29, 889–899. [CrossRef]
- Librado, P., Rozas, J., 2009. DnaSP v5. Bioinformatics 25, 1451–1452. [CrossRef]
- Liu, J., Chafi, R., Legarrea, S., Alba, J.M., Meijer, T.T., Menken, S.B.J., Kant, M.R., 2020a. Spider Mites Cause More Damage to Tomato in the Dark When Induced Defenses Are Lower. Journal of Chemical Ecology 46, 631–641. [CrossRef]
- Liu, J., Legarrea, S., Alba, J.M., Dong, L., Chafi, R., Menken, S.B.J., Kant, M.R., 2020b. Juvenile Spider Mites Induce Salicylate Defenses, but Not Jasmonate Defenses, Unlike Adults. Frontiers in Plant Science 11, 980. [CrossRef]
- Musser, R.O., Hum-Musser, S.M., Herb Eichenseer, Eichenseer, H., Peiffer, M., Gary Ervin, Ervin, G.N., Murphy, J.B., Felton, G.W., 2002. Herbivory: caterpillar saliva beats plant defences. Nature 416, 599–600. [CrossRef]
- Navajas, M., Boursot, P., 2003. Nuclear ribosomal DNA monophyly versus mitochondrial DNA polyphyly in two closely related mite species: the influence of life history and molecular drive. Proceedings of The Royal Society B: Biological Sciences 270, 124–127. [CrossRef]
- Nei, M., 1987. Molecular evolutionary genetics. Columbia university press.
- Paulo, J.T., Godinho, D.P., da Silva, A.B., Branquinho, C., Magalhães, S., 2018. Suppression of Plant Defenses by Herbivorous Mites Is Not Associated with Adaptation to Host Plants. International Journal of Molecular Sciences 19, 1783. [CrossRef]
- Pilet-Nayel, M.-L., Moury, B., Caffier, V., Montarry, J., Kerlan, M.-C., Fournet, S., Durel, C.-E., Delourme, R., 2017. Quantitative resistance to plant pathogens in pyramiding strategies for durable crop protection. Frontiers in Plant Science 8, 1838. [CrossRef]
- R Core Team, 2022. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria.
- Rasmy, A.H., 1985. The biology of the two-spotted spider mite Tetranychus urticae as affected by resistant solanaceous plants. Agriculture, ecosystems & environment 13, 325–328. [CrossRef]
- Rozas, J., Sánchez-DelBarrio, J.C., Messeguer, X., Rozas, R., 2003. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. [CrossRef]
- Sarmento, R. de A., Lemos, F., Bleeker, P.M., Schuurink, R.C., Pallini, A., Maria Goreti de Almeida Oliveira, de Almeida Oliveira, M.G., Lima, E.R., Kant, M.R., Sabelis, M.W., Janssen, A., 2011. A herbivore that manipulates plant defence. Ecology Letters 14, 229–236. [CrossRef]
- Sarmento, R.A., Lemos, F., Dias, C.R., Kikuchi, W.T., Rodrigues, J.C., Pallini, A., Sabelis, M.W., Janssen, A., 2011. A herbivorous mite down-regulates plant defence and produces web to exclude competitors. PLoS One 6, e23757.
- Sato, Y., Alba, J.M., Sabelis, M.W., 2014. Testing for reproductive interference in the population dynamics of two congeneric species of herbivorous mites. Heredity 113, 495–502. [CrossRef]
- Schimmel, B.C.J., Alba, J.M., Wybouw, N., Glas, J.J., Meijer, T.T., Schuurink, R.C., Kant, M.R., 2018. Distinct Signatures of Host Defense Suppression by Plant-Feeding Mites. International Journal of Molecular Sciences 19, 3265. [CrossRef]
- Schimmel, B.C.J., Ataíde, L.M.S., Chafi, R., Villarroel, C.A., Alba, J.M., Schuurink, R.C., Kant, M.R., 2017. Overcompensation of herbivore reproduction through hyper-suppression of plant defenses in response to competition. New Phytologist 214, 1688–1701. [CrossRef]
- Sharma, Rakesh K., Sharma, Rakesh K., Sharma, R. K., Sharma, R., Bhullar, M.B., Bhullar, M.B., Singh, S., 2019. Mitochondria COI-Based Molecular Characterization and Genetic Analysis of the Fenazaquin Selected Resistant Strain of Two-Spotted Spider Mite, Tetranychus urticae Koch. International Journal of Current Microbiology and Applied Sciences 8, 2508–2517. [CrossRef]
- Stephens, M., Smith, N.M.J., Donnelly, P., 2001. A new statistical method for haplotype reconstruction from population data. American Journal of Human Genetics 68, 978–989. [CrossRef]
- Sukumaran, J., Holder, M.T., 2010. DendroPy: a Python library for phylogenetic computing. Bioinformatics 26, 1569–1571. [CrossRef]
- Tajima, F., Tajima, F., 1983. Evolutionary relationship of DNA sequences in finite populations. Genetics 105, 437–460. [CrossRef]
- Tamura, K., Stecher, G., Kumar, S., 2021. MEGA11: molecular evolutionary genetics analysis version 11. Molecular biology and evolution 38, 3022–3027. [CrossRef]
- Teodoro-Paulo, J., Alba, J., Charlesworth, S., Kant, M., Magalhães, S., Duncan, A., 2022. Intraspecific variation for host immune activation by an arthropod herbivore. [CrossRef]
- Tornero, P., Gadea, J., Conejero, V., Vera, P., 1997. Two PR-1 genes from tomato are differentially regulated and reveal a novel mode of expression for a pathogenesis-related gene during the hypersensitive response and development. Molecular Plant-Microbe Interactions 10, 624–634. [CrossRef]
- UniProt Consortium, 2019. UniProt: a worldwide hub of protein knowledge. Nucleic acids research 47, D506–D515. [CrossRef]
- van Kan, J.A.L., Joosten, M.H.A.J., Matthieu H. A. J. Joosten, Wagemakers, C.A.M., van den Berg-Velthuis, G.C.M., G.C.M. van den Berg-Velthuis, van den Berg-Velthuis, G., de Wit, P.J.G.M., 1992. Differential accumulation of mRNAs encoding extracellular and intracellular PR proteins in tomato induced by virulent and avirulent races of Cladosporium fulvum. Plant Molecular Biology 20, 513–527. [CrossRef]
- Van Leeuwen, T., Dermauw, W., 2016. The Molecular Evolution of Xenobiotic Metabolism and Resistance in Chelicerate Mites. Annual Review of Entomology 61, 475–498. [CrossRef]
- Van Leeuwen, T., Vanholme, B., Van Pottelberge, S., Van Nieuwenhuyse, P., Nauen, R., Tirry, L., Denholm, I., 2008. Mitochondrial heteroplasmy and the evolution of insecticide resistance: Non-Mendelian inheritance in action. Proceedings of the National Academy of Sciences of the United States of America 105, 5980–5985. [CrossRef]
- Vega-Muñoz, I., Duran-Flores, D., Fernández-Fernández, Á.D., Heyman, J., Ritter, A., Stael, S., 2020. Breaking bad news: dynamic molecular mechanisms of wound response in plants. Frontiers in Plant Science 11, 610445. [CrossRef]
- Verwoerd, T.C., Dekker, B., Hoekema, A., 1989. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic acids research 17, 2362. [CrossRef]
- Villacis-Perez, E., Alba, J.M., Cotte, J., Van Loon, Z., Breeuwer, J.A., Van Leeuwen, T., 2022. Interactions with plant defences isolate sympatric populations of an herbivorous mite. Frontiers in Ecology and Evolution 346. [CrossRef]
- Villarroel, C.A., Jonckheere, W., Alba, J.M., Glas, J.J., Dermauw, W., Haring, M.A., Van Leeuwen, T., Schuurink, R.C., Kant, M.R., 2016. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant Journal 86, 119–131. [CrossRef]
- Villesen, P., 2007. FaBox: an online toolbox for fasta sequences. Molecular Ecology Notes 7, 965–968. [CrossRef]
- Vlot, A. Corina, A. C. Vlot, Vlot, A. C., Dempsey, D.A., Klessig, D.F., 2009. Salicylic Acid, a multifaceted hormone to combat disease. Annual Review of Phytopathology 47, 177–206. [CrossRef]
- Walling, L.L., 2000. The Myriad Plant Responses to Herbivores. Journal of Plant Growth Regulation 19, 195–216. [CrossRef]
- Walsh, P.S., Metzger, D.A., Higuchi, R., 1991. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513.
- Wasternack, C., Wasternack, Claus, Hause, Bettina, Hause, B., 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [CrossRef]
- Wybouw, N., Zhurov, V.V., Zhurov, V., Martel, C., Bruinsma, K., Hendrickx, F., Grbic, V., Van Leeuwen, T., 2015. Adaptation of a polyphagous herbivore to a novel host plant extensively shapes the transcriptome of herbivore and host. Molecular Ecology 24, 4647–4663. [CrossRef]
- Yang, X., Margolies, D.C., Zhu, K.Y., Buschman, L.L., 2001. Host plant-induced changes in detoxification enzymes and susceptibility to pesticides in the twospotted spider mite (Acari: Tetranychidae). Journal of economic entomology 94, 381–387. [CrossRef]
- Zaidi, S.S.-A., Mukhtar, M.S., Mansoor, S., 2018. Genome editing: targeting susceptibility genes for plant disease resistance. Trends in Biotechnology 36, 898–906. [CrossRef]
- Zhang, D., Gao, F., Jakovlić, I., Zou, H., Zhang, J., Zhang, J., Li, W.X., Wang, G.T., 2020. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Molecular Ecology Resources 20, 348–355. [CrossRef]
- Zhurov, V.V., Zhurov, V., Navarro, M., Bruinsma, K., Arbona, V., Vicente Arbona, Santamaria, M.E., Santamaria, E.M., Cazaux, M., Wybouw, N., Osborne, E.J., Ens, C., Rioja, C., Vermeirssen, V., Rubio-Somoza, I., Krishna, P., Diaz, I., Schmid, M., Gómez-Cadenas, A., Van de Peer, Y., Grbic, M., Clark, R.M., Van Leeuwen, T., Grbic, V., 2014. Reciprocal Responses in the Interaction between Arabidopsis and the Cell-Content-Feeding Chelicerate Herbivore Spider Mite. Plant Physiology 164, 384–399. [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).