Introduction:
Epidemiologists and population health researchers often analyze the relationship between the health and wellness of people and their surrounding environments using place-based information, often called geomarkers [
1]. Examples of geomarkers include proximity to major roadways as an estimate of traffic-related air pollution, census tract-level median household income as an estimate of community material deprivation, and the degree of imperviousness and greenness of nearby land as an estimate of nearby greenspace. In order to assign geomarkers to study participants, geocoding must be used to convert mailing addresses into geographical coordinates [
2]. There are a handful of commercial geocoding services that use varying approaches and have different costs. Some operate over the internet (Google Geocoding API [
3]), but are not designed for batch operations, and others can be operated on a local computer (e.g. ArcGIS, SAS [
3]), but require expensive software subscriptions and technical expertise. The resulting geocodes from these services still must be processed further to attach geomarker data. Our approach combines geocoding and geomarker assessment into the same software package.
The Health Information Portability and Accountability Act (HIPAA) privacy rule, the Health Information Technology for Economic and Clinical Health Act of 2009, and the Federal Policy for the Protection of Human Subjects have all contributed to the important safeguarding of personal protected health information (PHI) [
4,
5,
6]. While beneficial in terms of personal privacy, these regulations introduce obstacles when using address data. During geocoding and geomarker assessment, researchers should not transmit PHI (including mailing addresses and equivalent geographic coordinates) over the internet to a third-party service without appropriate permissions and agreements from the third-party, research subjects, the institutional review board, and the institution itself. Multi-site studies magnify this obstacle because each institution will likely have a different degree of ability to share PHI for a centralized analysis.
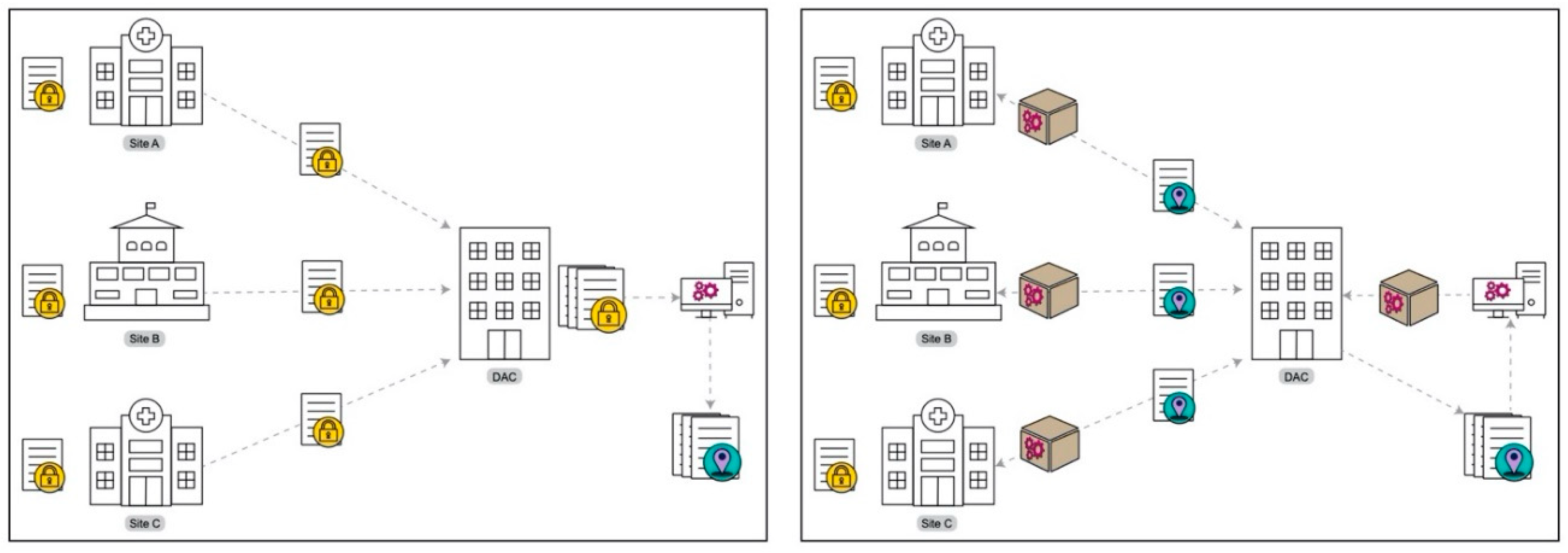
Thus, despite the wide availability of geomarker data, linking it to study participants presents significant challenges related to reproducibility and privacy that translate to problems with interoperability and accessibility. DeGAUSS (Decentralized Geomarker Assessment for Multi-Site Studies) is software that aims to provide secure, cost-effective, and easy-to-use geocoding and geomarker assessment at scale. Within a multi-site study, DeGAUSS does not require the sharing of PHI and instead can create shareable geomarker datasets that do not contain PHI (
Figure 1).
This protection of PHI is part of what makes DeGAUSS so useful for clinical research. Because the container image has all the required code, geospatial data, and geospatial software libraries, PHI does not need to leave the user’s machine and is never exposed over the internet. Rather than requiring individual study sites to share address data with a central data analysis center (DAC), DeGAUSS can be run at each site, geocoding addresses in batch, appending geomarker information, and removing PHI before sharing the data with the DAC (
Figure 2).
Implementation:
DeGAUSS was created using a derivative of Geocoder::US 2.0, which uses TIGER/Line address range files [
7]. These files, provided by the US Census Bureau, are used to convert addresses into geographical coordinates. Generally, a street address is matched to a TIGER/Line street range that is used to interpolate a location. DeGAUSS has previously been shown to have a similar geocoding accuracy and match rate compared to other conventional range-based geocoders (e.g. SAS, ArcGIS) [
7]. In addition to address geocoding, DeGAUSS provides containers to assess geomarker data at the supplied locations. The DeGAUSS library currently contains 12 publicly available images and several other purpose-built, private images.
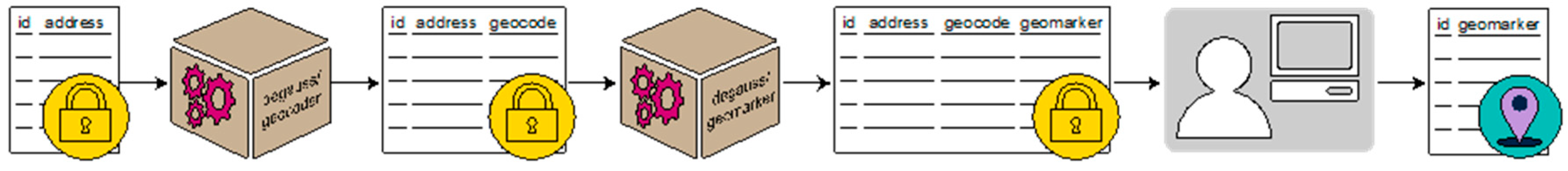
DeGAUSS was built using containerized R code that utilizes geospatial software libraries, like gdal and geos. A container is a standard unit of software that packages all code and applications within it. The containerized code also includes any necessary geographic data and is hosted and deployed using the Docker platform. Docker, briefly, delivers containers to end users via operating system-level virtual machines [
8]. The end user accesses the containers through Docker’s platform Docker Hub using command line prompts. With one command, a DeGAUSS container can be downloaded and run to append geocoding and geomarker information to an existing CSV file.
Use:
DeGAUSS is used through Docker, a free and open source piece of software used to build, share, and run software containers. After installing Docker, a DeGAUSS image can be downloaded and run as a container in one command. Docker and DeGAUSS commands are given through a command-line interface shell, such as “Terminal” on macOS or “PowerShell”, “Cmd”, or “Windows Subsystem for Linux” on Microsoft Windows. The command is made up of several components, but the user will only change the name (and version) of the DeGAUSS image to use and the file name of the input csv file in the current working directory (
Figure 3).
Below is a step-by-step example of DeGAUSS being used to estimate the length and proximity of major roadways as well as nearby greenness for a set of addresses.

On your machine, open a command-line shell and navigate to the folder where the address file is stored. This is often completed using the “cd” command followed by the name of the target directory. Generally, navigation-based commands are the same across different shells and operating systems, but there can be small differences. For DeGAUSS commands, specific instructions for different operating systems are provided on the DeGAUSS website.
- 2.
Use a DeGAUSS Docker command to geocode the addresses using version 3.0.2 of “degauss/geocoder”:
docker run --rm -v $PWD:/tmp degauss/geocoder:3.0.2 sample_addresses.csv
If you have not previously used this version of this image, Docker will first download it, which can take several minutes, depending on the size of the image and internet speeds. Docker will then create and run a container to geocode the addresses. DeGAUSS relies on parallel processing where possible so length of time required to geocode will vary based on the host machine’s number of cores it has made available to Docker. For example, 50,000 addresses were geocoded in about 30 minutes using Docker Desktop with 6 cores and 10 GB of RAM on a 15-inch, 2019 MacBook Pro with a 2.6 GHz Intel Core i7 processor.
- 3.
-
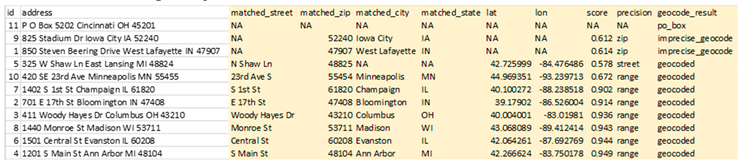
The results file, called “sample_addresses_geocoded_v3.0.2.csv”, will be written to the same folder where the input CSV file is located.
This file is the same as the input CSV file, but with appended columns for matched address components, geocoding score and precision, latitude, longitude, and a categorical geocoding result. The geocoder will recognize invalid street addresses, such as PO boxes, known foster addresses and non-address text, and alert the user in the “geocode_result” column. The “matched_” columns can be used to verify the accuracy of results, while the “score” and “precision” columns are used to determine the category of the geocode result, precise or imprecise. A geocode will be categorized as imprecise if the precision level is one of intersection, zip or city and/or the score, which is the percentage of text match between the input address and matched address, is less than 0.5. However, the user can specify a custom score threshold by including a number argument between 0 and 1 in their Docker command, placing it after the name of the address file.
- 4.
Now that we have geocoded addresses, we can use DeGAUSS to add a geomarker. In this example we will use the DeGAUSS images for the proximity to major roadways and greenspace, DeGAUSS/roads version 0.1 and DeGAUSS/greenspace version 0.2. The programs can either be run in parallel on the geocoded file or they can be run sequentially, creating one file with both geomarkers. Here, we first added the roadway geomarker and then add greenspace to that result. This is done using the following commands while in the directory of the geocoded .csv file:
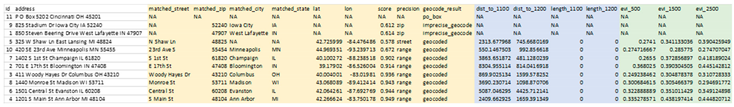
- 5.
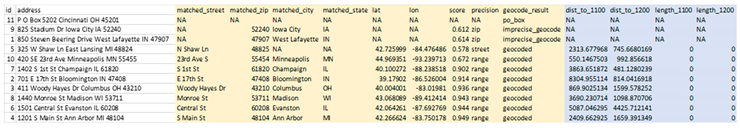
These two DeGAUSS containers append new columns to our dataset with their respective geomarkers, while keeping intact our original dataset. Now that we have added our geomarkers, we can remove the addresses to create a geomarker dataset without geographic PHI.
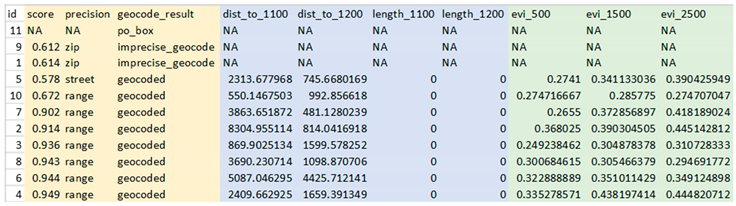
In these steps we have taken 10 street addresses and 1 PO box, geocoded the possible addresses, appended relevant geomarkers and removed PHI without exposing any PHI over the internet.
For further support, the developers host troubleshooting resources and instructional articles at the DeGAUSS website:
https://degauss.org/. In addition to those resources, the developers are reachable via email and GitHub at
https://github.com/degauss-org where users can open issues, request assistance and contribute to DeGAUSS.
Discussion
DeGAUSS is a compelling software option for geocoding protected address information and estimating geomarkers because of its low cost, ease of use, and scalability. Additionally, using a containerization framework ensures reproducibility by preventing differences in results due to operating systems, software versions, or methodology.
DeGAUSS is undergoing regular maintenance and development. The current library of DeGAUSS images is hosted at
https://degauss.org/available_images.html and includes neighborhood deprivation, distance to roadways, greenspace, drivetime, pollution models, daily weather data and land cover usage. DeGAUSS is developed publicly and is designed to be a framework for open contribution of other geomarkers by software users and developers.
While useful in many ways, there are limitations related to the usability of DeGAUSS. The installation of Docker Desktop is required to use Docker and DeGAUSS on a Windows or Mac computer and may require administrative access to install. However, once this process is initially complete, it is not required again to use any other DeGAUSS images. In preliminary survey data, 87.5% (7/8) of respondents said it is “somewhat easy”, “easy”, or “very easy” to both install and use Docker.
A second limitation is the use of the command line to execute the program, which is not something all users are familiar with. The user is required to navigate to the folder with their data and execute the DeGAUSS command all via the command line. Additionally, there are differences in the way the command line is used between Windows and Mac operating systems. However, the lack of a graphical user interface allows for truly cross-platform reproducible software without having to develop different versions for different operating systems.
Ultimately, DeGAUSS aims to be an impactful tool in any scientist’s toolkit. Indeed, DeGAUSS has been used by researchers in several multi-site studies funded by the National Institutes of Health (NIH) to overcome key privacy and scalability obstacles. By providing transparent and reproducible geocoding, DeGAUSS is a compelling alternative to conventional geocoding programs that are otherwise costly, require specialized technical expertise, and may necessitate institutional agreements to share PHI.
References
- Beck, A.F., et al., Housing code violation density associated with emergency department and hospital use by children with asthma. Health Aff (Millwood), 2014. 33(11): p. 1993-2002.
- Goldberg, D.W., J.P. Wilson, and C.A. Knoblock, From text to geographic coordinates: the current state of geocoding. URISA journal, 2007. 19(1): p. 33-46.
- Lemke, D., et al., [Who Hits the Mark? A Comparative Study of the Free Geocoding Services of Google and OpenStreetMap]. Gesundheitswesen, 2015. 77(8-9): p. e160-5.
- Act, A., Health insurance portability and accountability act of 1996. Public law, 1996. 104: p. 191.
- Redhead, C.S. The Health Information Technology for Economic and Clinical Health (HITECH) Act. 2009. Congressional Research Service, Library of Congress.
- Porter, J.P., Protecting Human Research Subjects: The Office for Protection from Research Risks. Kennedy Institute of Ethics Journal, 1992. 2(3): p. 279-282.
- Brokamp, C., et al., Decentralized and reproducible geocoding and characterization of community and environmental exposures for multisite studies. J Am Med Inform Assoc, 2018. 25(3): p. 309-314.
- Boettiger, C., An introduction to Docker for reproducible research. ACM SIGOPS Operating System Review, 2015. 49(1): p. 71-79.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).