1. Introduction
Kinetic modeling is an essential technique in systems biology, as it allows for the quantitative analysis and prediction of complex biochemical systems such as metabolic pathways and gene regulatory networks [
1]. Kinetic models are differential equation models that describe the dynamic behavior of biochemical systems based on the interactions between their molecular components. Kinetic models are stored and distributed as SBML (Systems Biology Markup Language) files [
2,
3]. In addition, there are several software tools, such as Tellurium [
4,
5], COPASI [
6,
7,
8], and CADLIVE [
9,
10,
11], that support kinetic modeling. Still, the development of kinetic models is time-consuming and expert work. First, modelers survey many research articles for known enzyme reactions, signal transduction pathways, and gene regulations contained in the system of interest and build the chemical reaction equations with kinetic parameters. Next, modelers translate them into kinetic rate equations. A few studies have been devoted to developing natural language processing-based methods that generate kinetic models from literature texts [
12,
13]. However, these studies focused on signal transduction, a specific type of biochemical systems. Moreover, their complex implementation makes it challenging for non-experts to modify or extend.
In recent years, there has been significant progress in artificial intelligence (AI) technologies, particularly in the field of generative models. Among these models, GPT (Generative Pre-trained Transformer) natural language models stand out as the most advanced. In fact, ChatGPT [
14], one of the GPT models, has gained public attention due to the ability to engage in natural conversations. It has been shown that GPT models have the capability to pass graduate-level exams [
15,
16] and write program codes [
17]. GPT models may allow researchers to automatically build kinetic models directly from research articles. To the best of our knowledge, the capabilities of GPT models for automatic kinetic model construction have not been investigated.
In this article, we aim to answer the following question: Can a GPT model generate an SBML kinetic model from a natural language description? While GPT-3 [
18] alone cannot create valid SBML models, we propose a novel approach called KinModGPT, which combines GPT-3 as a natural language interpreter and Tellurium as an SBML generator. Moreover, we demonstrate that KinModGPT can generate valid SBML models from natural language descriptions of biochemical reactions.
2. Results
2.1. GPT-3 alone cannot create valid SBML models
GPT-3 is a large language model trained on massive amounts of text data [
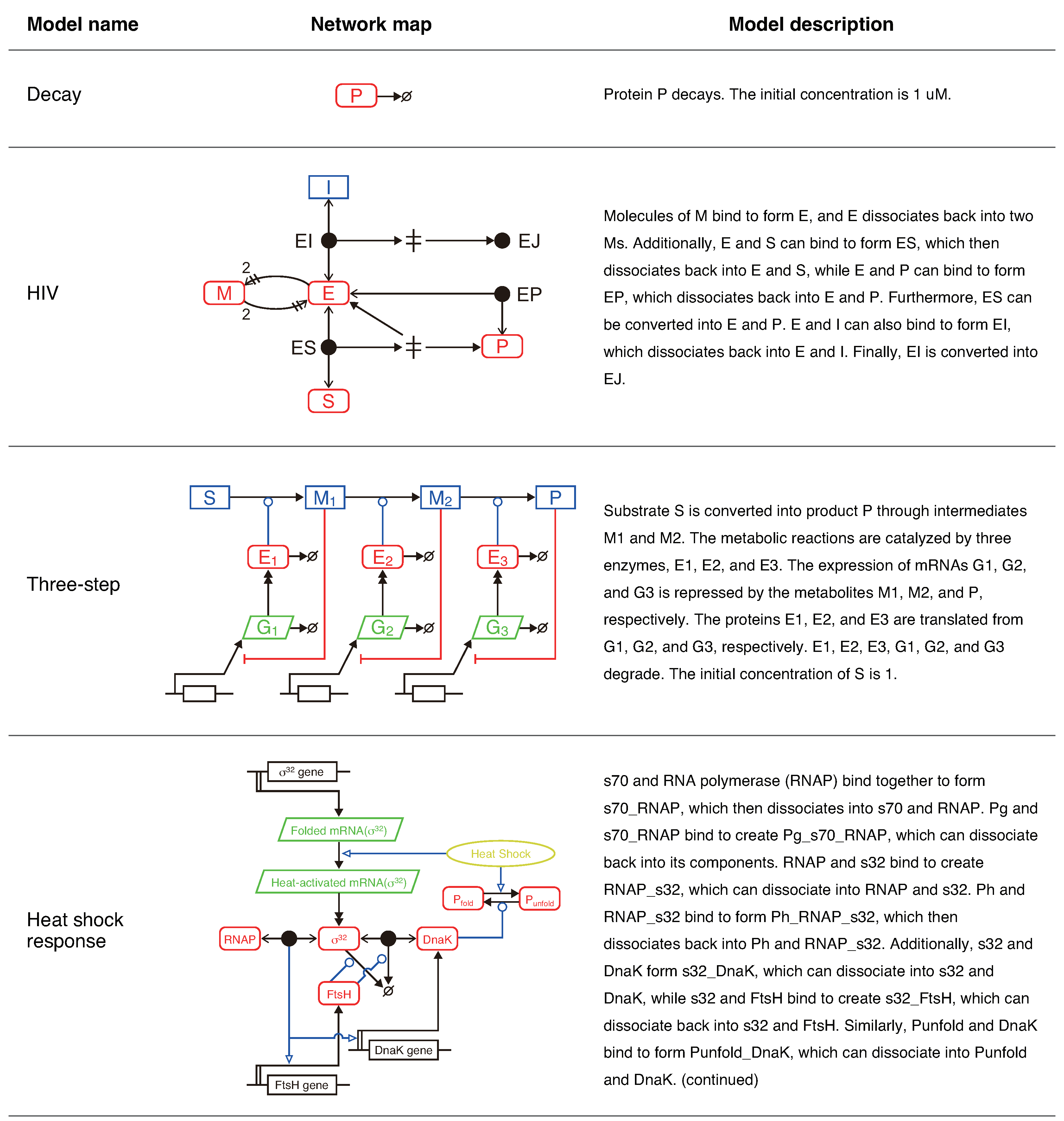
18]. When prompted with a message to start the conversation, GPT-3 generates appropriate responses by repeatedly predicting the following word in the sequence. First, we tested whether GPT-3 can directly generate an SBML model from natural language text. We employed four test problems with biochemical systems with different complexities (
Figure 1). Next, we asked GPT-3 to convert each model description to an SBML model. The instruction message for GPT-3 is shown in
Scheme A1. The results are summarized in
Table 1. For all the models tested, GPT-3 generated SBML-like models. However, upon inspecting these models using the Online SBML validator [
19], we discovered that all the generated SBML models were invalid, containing some errors, such as missing required attributes. The invalid SBML files cannot be imported by widely-used modeling tools.
2.2. KinModGPT can create valid SBML models
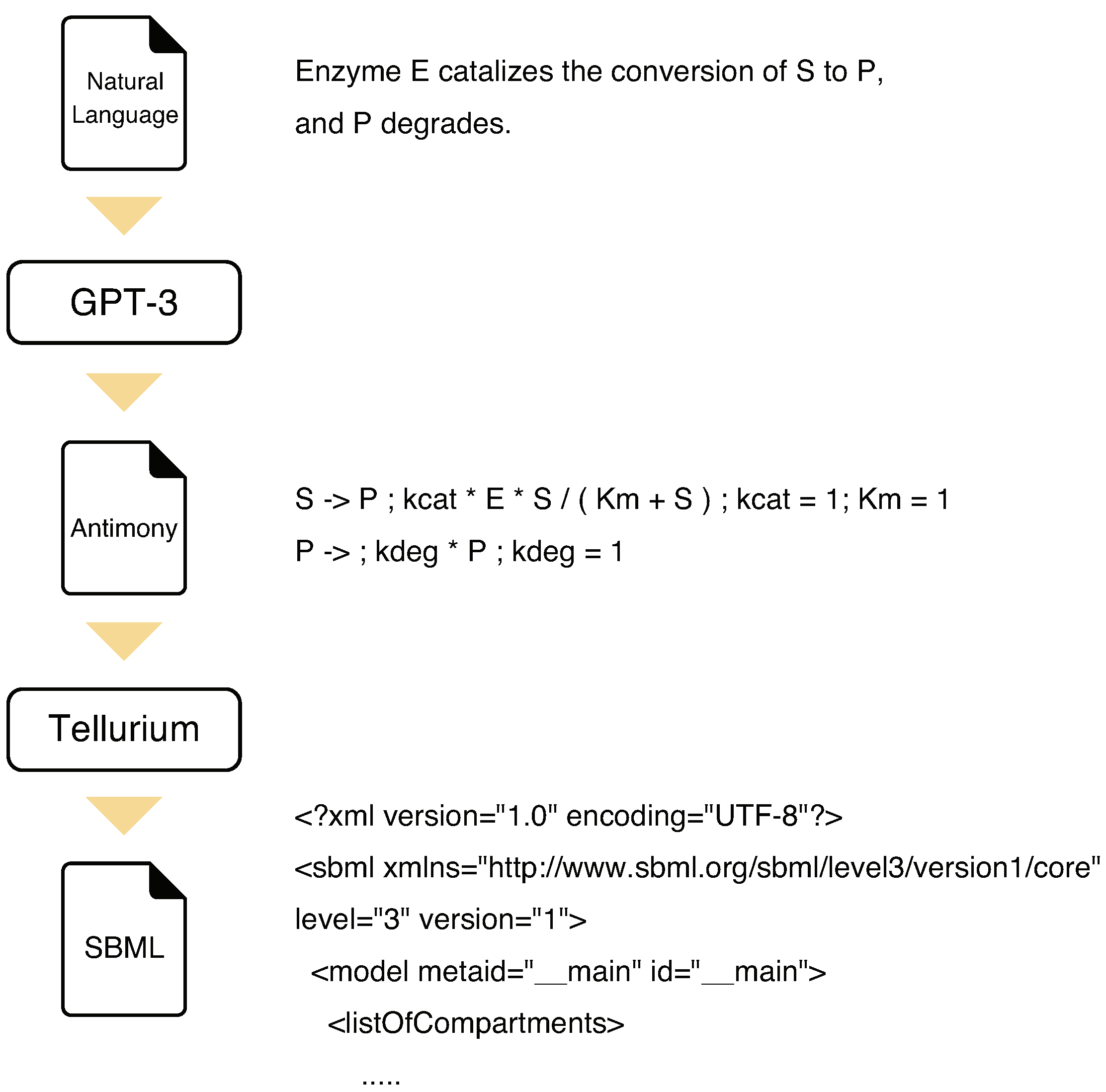
To generate an SBML model from natural language text, we introduce the strategy named KinModGPT, as outlined in
Figure 2. First, KinModGPT translates the natural language descriptions of biochemical reactions into Antimony language [
20], a human-readable model definition language. Next, KinModGPT converts the resulting Antimony model into the SBML model using Tellurium [
4,
5]. For KinModGPT, we provided the following prompt to GPT-3. First, we told GPT-3 that the task was to translate the descriptions of biochemical reactions written in natural language into Antimony. Second, we provided a conversion rule table that showed how each chemical reaction is represented in natural language and Antimony (
Scheme A2). Then, we input the natural language model descriptions. The Python code of KinModGPT and the created Antimony and SBML models are provided as Supplementary Materials.
We tested whether KinModGPT can create valid SBML models from text model descriptions. KinModGPT successfully generated an SBML model for each test problem. We confirmed their validity using the Online SBML validator (
Table 1). Indeed, these SBML models could be imported by a widely-used modeling tool, COPASI [
6,
7,
8], demonstrating that KinModGPT can create ready-to-simulate SBML models from natural language text.
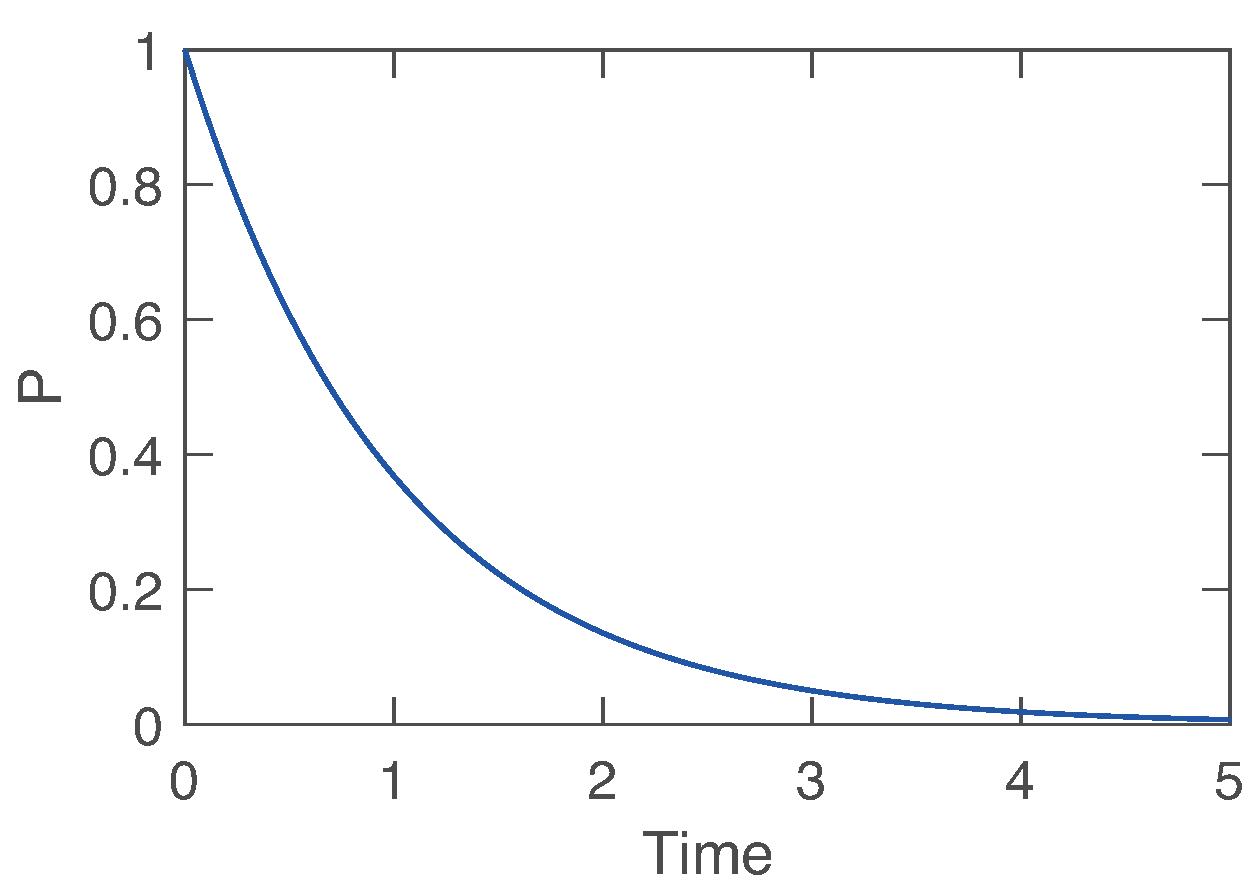
To further examine the models, we intensively analyzed each model. The Antimony model for decay is shown in
Scheme 1. It is worth noting that KinModGPT does not require an exact match between the conversion rules and model descriptions. Indeed, KinModGPT successfully interpreted “Protein P decays. The initial concentration is 1 uM.” as the combination of the two expressions: “X degrades (or decays)” and “X (concentration) is Y M (or mM or uM or nM or pM).” The generated SBML model could be simulated as it was (
Figure 3).
In the model description of the HIV model (
Figure 1), five sentences describe ten reactions. Indeed, the second sentence contains four reactions. KinModGPT extracted the necessary information from these complex sentences and generated a valid SBML model. The Antimony model is provided in
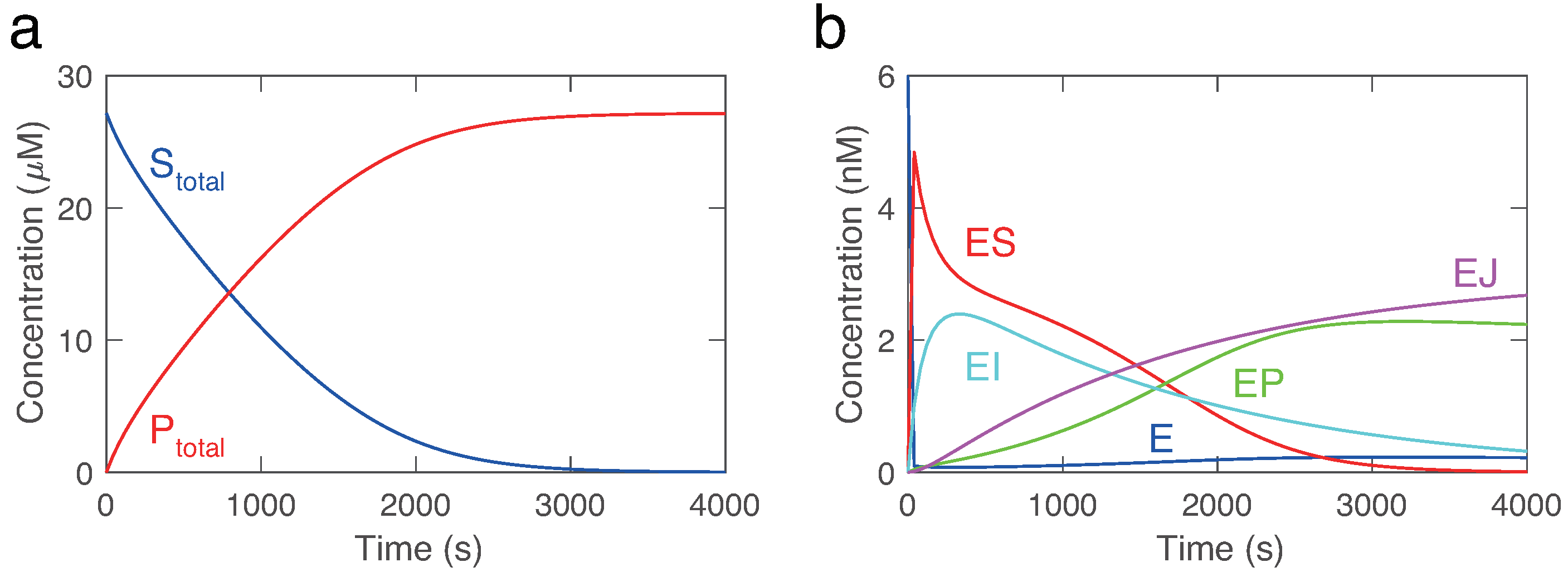
Scheme 2. The SBML model could be simulated by modeling tools without any modifications. However, to check whether the SBML model reproduces the behavior of the HIV proteinase system, we manually set known realistic values to kinetic parameters. As shown in
Figure 4, the substrate (S) is converted into the product (P) over time. Moreover, the inactive enzyme-inhibitor complex (EJ) gradually increases. This behavior is consistent with the network map (
Figure 1), model description, and literature [
21,
22,
23].
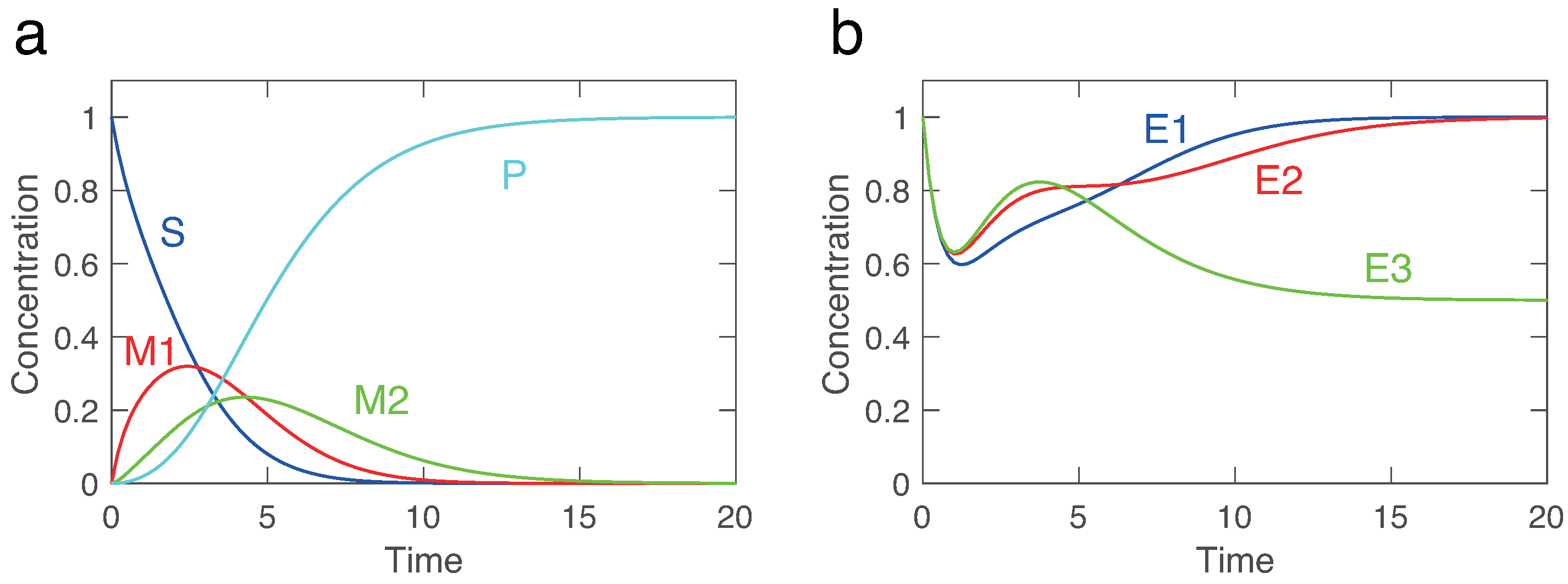
The three-step problem highlights the remarkable capability of KinModGPT. The model description’s first two complex sentences, "Substrate S is converted into product P through intermediates M1 and M2. The metabolic reactions are catalyzed by three enzymes, E1, E2, and E3. ", contain four metabolites and three enzyme reactions. These sentences may be challenging even for experienced modelers to interpret. However, KinModGPT successfully interpreted and translated the sentences into the Antimony model (
Scheme 2). Next, we tested whether the created SBML model could reproduce a reasonable behavior. As shown in
Figure 5, the substrate (S) is converted into the product (P) through two intermediate metabolites (M1 and M2). The expression of the third enzyme (E3) is repressed compared to the first and second enzymes because the accumulated P represses the expression of E3. This behavior matches the network map and model description shown in
Figure 1.
Scheme 3.
The three-step model in Antimony language, created by KinModGPT.
Scheme 3.
The three-step model in Antimony language, created by KinModGPT.
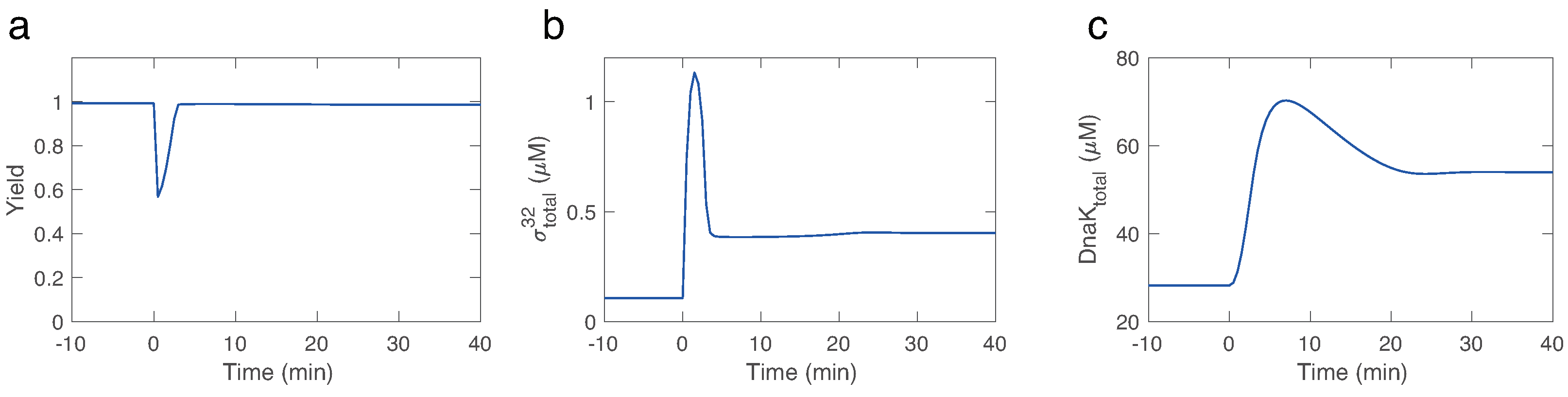
Finally, KinModGPT has successfully created an SBML model for the heat shock response model that consists of 25 variables and 50 rate equations, demonstrating its potential for developing complex, realistic kinetic models. The Antimony model is provided in
Scheme A4 and
Scheme A5. To test the created SBML model, we assigned realistic parameter values and simulated its behavior. Upon heat shock, proteins are rapidly denatured, and thus the yield (the fraction of folded proteins in the total protein pool) decreases (
Figure 6). However, the
transcription factor is produced, which then initiates the expression of the chaperone protein DnaK. Denatured proteins are then quickly refolded by DnaK, and thus the yield is recovered. This behavior is consistent with literature [
24,
25,
26].
Figure 6.
Simulation of the created SBML model for the the heat shock response model. Heat shock occurs at 0 min and is implemented through an increase in the rate constant for protein denaturing. Yield is the fraction of folded proteins in a pool of total proteins, i.e., . We tuned the kinetic parameters before simulation.
Figure 6.
Simulation of the created SBML model for the the heat shock response model. Heat shock occurs at 0 min and is implemented through an increase in the rate constant for protein denaturing. Yield is the fraction of folded proteins in a pool of total proteins, i.e., . We tuned the kinetic parameters before simulation.
3. Discussion
In this article, we explored the possibilities of using GPT models for kinetic modeling automation. We developed KinModGPT by integrating GPT-3 [
18] and Tellurium [
4,
5]. KinModGPT successfully converted a kinetic model written in a natural language text into the SBML model. Furthermore, all the created SBML models could be imported by widely-used modeling tools. To our knowledge, this work presents the first method applying GPT models to kinetic modeling, contributing to advances in systems biology.
How did GPT-3 fail to generate valid SBML files without any help from a modeling tool? Despite the effectiveness of language models, such as GPT-3, in generating natural sentences, their output may lack the precision required to generate accurate models. In contrast, even a single missing tag in SBML can lead to errors in the model. GPT-3 could not generate any valid SBML models, not even for the simplest models with one variable and one reaction. It is challenging to fix the generated invalid SBML models, as it requires manual review of the SBML files in XML format.
Instead of creating SBML models directly from natural language texts, KinModGPT employs Antimony language as an intermediate representation. Since Antimony language is simpler than SBML, GPT-3 can translate a natural language model description into Antimony language without errors. Then, the modeling tool, Tellurium, creates an SBML model from the Antimony model. For all four test problems, there were no conversion errors. However, even if there were some errors, they could be easily corrected by using modeling tools or by directly editing the Antimony models. This is another advantage of KinModGPT over the GPT-3 only approach.
Despite its promising results, KinModGPT has some limitations. Firstly, the current version of the natural language-Antimony conversion rule table covers only a part of biochemical reactions. However, the table can easily be expanded or tailored to a specific application domain. Moreover, we may be able to eliminate the need for manual rule definition by fine-tuning or by retraining GPT-3 with a large number of “Rosetta stones,” i.e., natural language model descriptions and their Antimony counterparts. Another limitation is that the current version of KinModGPT cannot automatically set appropriate kinetic parameter values. Thus, kinetic parameters must be tuned in the downstream modeling process to create a realistic model that complies with experimental data [
27,
28,
29,
30].
Gyori et al. developed the Integrated Network and Dynamical Reasoning Assembler (INDRA), which automatically builds kinetic models from natural language texts [
12]. While INDRA focuses on modeling cell signaling pathways, its applicability to other types of biochemical systems is uncertain. In contrast, we have demonstrated that KinModGPT can effectively model various biochemical systems, including metabolic pathways, protein-protein interactions, and gene regulations. KinModGPT also offers the advantage of extensibility. Due to its simple implementation, modelers can easily customize the conversion rules for translating natural language texts to Antimony. For example, we modeled enzyme reactions using the simplest irreversible Michaelis-Menten equation in the three-step model; however, by modifying the conversion rule table (written in a simple text file), modelers can easily switch to a reversible equation. In addition, as GPT-3 is multilingual, the conversion rules can be written in languages other than English.
It should be noted that the purpose of this article is not to provide a perfect solution for kinetic modeling automation but rather to demonstrate the potential of GPT models in this field. With the continued refinement of KinModGPT, AI may be able to extract information from many relevant articles to generate a kinetic model automatically. Such a development could significantly accelerate model development and improve an understanding of complex biological systems.
4. Materials and Methods
4.1. GPT-3
We chose GPT-3 (text-davinci-003) [
18], one of OpenAI’s GPT series with an open API. Trained on massive amounts of text data, GPT-3 is a powerful tool for natural language processing tasks, including language translation, text summarization, and question answering. By predicting the next word in a sequence given preceding words, GPT-3 can generate human-like natural language text. In GPT-3, the parameter called
temperature determines the randomness of responses. In this study, we set
temperature = 0 for reproducibility.
4.2. Tellurium
Tellurium [
4,
5] is a Python library for modeling and simulating biochemical systems. For model development, Tellurium employs Antimony [
20], a human-readable, text-based language that facilitates the creation of kinetic models. Additionally, Tellurium can convert Antimony models to SBML models.
4.3. Test problems
We utilized four test problems to benchmark KinModGPT (
Figure 1). The first problem, decay, is the simplest scenario in which an SBML model is generated from just two sentences. This model has a single variable and a single rate equation. The HIV model represents the mechanism of irreversible inhibition of HIV proteinase [
21,
22,
23]. It is comprised of 9 variables and 10 rate equations, and its model description consists of 5 sentences. The three-step model describes a hypothetical metabolic pathway with 3 enzyme reactions and 3 gene regulations. This model consists of 10 variables and 15 rate equations, described in 5 sentences. Finally, the heat shock response model represents the realistic, complex regulatory mechanisms that confer robustness to heat shock in
Escherichia coli [
24,
25,
26]. This model consists of 25 variables and 50 rate equations, described in 20 sentences.
Author Contributions
Conceptualization, K.M.; software, K.M.; investigation, K.M. and H.K.; writing—original draft preparation, K.M. and H.K. All authors have read and agreed to the submitted version of the manuscript.
Funding
This work was supported by Grant-in-Aid for Scientific Research (C) (22K12247) and Grant-in-Aid for Scientific Research (B) (22H03688) from the Japan Society for the Promotion of Science. This work was further supported by JST PRESTO (JPMJPR20K8).
Data Availability Statement
The program code for the computational experiments and the created Antimony and SBML models are provided as Supplementary Materials.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| GPT |
Generative Pre-trained Transformer |
| SBML |
Systems Biology Markup Language |
| COPASI |
COmplex PAthway SImulator |
| CADLIVE |
Computer-Aided Design of LIVing systEms |
| INDRA |
Integrated Network and Dynamical Reasoning Assembler |
Appendix A
Scheme A1.
Instruction message for the GPT-3 only approach. This instruction is followed by a model description (see
Figure 1).
Scheme A1.
Instruction message for the GPT-3 only approach. This instruction is followed by a model description (see
Figure 1).
Scheme A2.
Instruction message for KinModGPT. This instruction is followed by a model description (see
Figure 1).
Scheme A2.
Instruction message for KinModGPT. This instruction is followed by a model description (see
Figure 1).
Scheme A3.
Full model description for the heat shock response model.
Scheme A3.
Full model description for the heat shock response model.
Scheme A4.
The first half of the heat shock response model in Antimony language, created by KinModGPT.
Scheme A4.
The first half of the heat shock response model in Antimony language, created by KinModGPT.
Scheme A5.
The second half of the heat shock response model in Antimony language, created by KinModGPT.
Scheme A5.
The second half of the heat shock response model in Antimony language, created by KinModGPT.
References
- Kitano, H. , Systems biology: a brief overview. Science 2002, 295, 1662–1664. [Google Scholar] [CrossRef] [PubMed]
- Hucka, M.; Finney, A.; Sauro, H. M.; Bolouri, H.; Doyle, J. C.; Kitano, H.; Arkin, A. P.; Bornstein, B. J.; Bray, D.; Cornish-Bowden, A.; Cuellar, A. A.; Dronov, S.; Gilles, E. D.; Ginkel, M.; Gor, V.; Goryanin, II; Hedley, W. J.; Hodgman, T. C.; Hofmeyr, J. H.; Hunter, P. J.; Juty, N. S.; Kasberger, J. L.; Kremling, A.; Kummer, U.; Le Novere, N.; Loew, L. M.; Lucio, D.; Mendes, P.; Minch, E.; Mjolsness, E. D.; Nakayama, Y.; Nelson, M. R.; Nielsen, P. F.; Sakurada, T.; Schaff, J. C.; Shapiro, B. E.; Shimizu, T. S.; Spence, H. D.; Stelling, J.; Takahashi, K.; Tomita, M.; Wagner, J.; Wang, J.; Forum, S. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics 2003, 19, 524–531. [Google Scholar] [CrossRef]
- Keating, S. M.; Waltemath, D.; Konig, M.; Zhang, F.; Drager, A.; Chaouiya, C.; Bergmann, F. T.; Finney, A.; Gillespie, C. S.; Helikar, T.; Hoops, S.; Malik-Sheriff, R. S.; Moodie, S. L.; Moraru, II; Myers, C. J.; Naldi, A.; Olivier, B. G.; Sahle, S.; Schaff, J. C.; Smith, L. P.; Swat, M. J.; Thieffry, D.; Watanabe, L.; Wilkinson, D. J.; Blinov, M. L.; Begley, K.; Faeder, J. R.; Gomez, H. F.; Hamm, T. M.; Inagaki, Y.; Liebermeister, W.; Lister, A. L.; Lucio, D.; Mjolsness, E.; Proctor, C. J.; Raman, K.; Rodriguez, N.; Shaffer, C. A.; Shapiro, B. E.; Stelling, J.; Swainston, N.; Tanimura, N.; Wagner, J.; Meier-Schellersheim, M.; Sauro, H. M.; Palsson, B.; Bolouri, H.; Kitano, H.; Funahashi, A.; Hermjakob, H.; Doyle, J. C.; Hucka, M.; members, S. L. C. SBML Level 3: an extensible format for the exchange and reuse of biological models. Mol Syst Biol 2020, 16, e9110. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.; Medley, J. K.; Konig, M.; Stocking, K.; Smith, L.; Gu, S.; Sauro, H. M. , Tellurium: An extensible python-based modeling environment for systems and synthetic biology. Biosystems 2018, 171, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Medley, J. K.; Choi, K.; Konig, M.; Smith, L.; Gu, S.; Hellerstein, J.; Sealfon, S. C.; Sauro, H. M. , Tellurium notebooks-An environment for reproducible dynamical modeling in systems biology. PLoS Comput Biol 2018, 14, e1006220. [Google Scholar] [CrossRef] [PubMed]
- Hoops, S.; Sahle, S.; Gauges, R.; Lee, C.; Pahle, J.; Simus, N.; Singhal, M.; Xu, L.; Mendes, P.; Kummer, U. , COPASI–a COmplex PAthway SImulator. Bioinformatics 2006, 22, 3067–3074. [Google Scholar] [CrossRef] [PubMed]
- Mendes, P.; Hoops, S.; Sahle, S.; Gauges, R.; Dada, J.; Kummer, U. , Computational modeling of biochemical networks using COPASI. Methods Mol Biol 2009, 500, 17–59. [Google Scholar] [PubMed]
- Bergmann, F. T.; Hoops, S.; Klahn, B.; Kummer, U.; Mendes, P.; Pahle, J.; Sahle, S. , COPASI and its applications in biotechnology. J Biotechnol 2017, 261, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Kurata, H.; Matoba, N.; Shimizu, N. , CADLIVE for constructing a large-scale biochemical network based on a simulation-directed notation and its application to yeast cell cycle. Nucleic Acids Res 2003, 31, 4071–4084. [Google Scholar] [CrossRef] [PubMed]
- Kurata, H.; Masaki, K.; Sumida, Y.; Iwasaki, R. , CADLIVE dynamic simulator: direct link of biochemical networks to dynamic models. Genome Res 2005, 15, 590–600. [Google Scholar] [CrossRef]
- Kurata, H.; Inoue, K.; Maeda, K.; Masaki, K.; Shimokawa, Y.; Zhao, Q. , Extended CADLIVE: a novel graphical notation for design of biochemical network maps and computational pathway analysis. Nucleic Acids Res 2007, 35, e134. [Google Scholar] [CrossRef] [PubMed]
- Gyori, B. M.; Bachman, J. A.; Subramanian, K.; Muhlich, J. L.; Galescu, L.; Sorger, P. K. , From word models to executable models of signaling networks using automated assembly. Mol Syst Biol 2017, 13, 954. [Google Scholar] [CrossRef] [PubMed]
- Todorov, P. V.; Gyori, B. M.; Bachman, J. A.; Sorger, P. K. , INDRA-IPM: interactive pathway modeling using natural language with automated assembly. Bioinformatics 2019, 35, 4501–4503. [Google Scholar] [CrossRef] [PubMed]
- Roose, K. , The Brilliance and Weirdness of ChatGPT. New York Times, Dec 26, 2022. [Google Scholar]
- Terwiesch, C. Would Chat GPT Get a Wharton MBA? A Prediction Based on Its Performance in the Operations Management Course; Mack Institute for Innovation Management at the Wharton School, University of Pennsylvania: 2023.
- Choi, J. H.; Hickman, K. E.; Monahan, A.; Schwarcz, D. , ChatGPT Goes to Law School. SSRN 2023. [Google Scholar] [CrossRef]
- Bussler, F. , Will GPT-3 Kill Coding? Jul 21, 2020. Available online: https://towardsdatascience.com/will-gpt-3-kill-coding-630e4518c04d (accessed on Mar 4, 2023).
- Brown, T.; Mann, B.; Ryder, N.; Subbiah, M.; Kaplan, J. D.; Dhariwal, P.; Neelakantan, A.; Shyam, P.; Sastry, G.; Askell, A. , Language models are few-shot learners. Advances in neural information processing systems 2020, 33, 1877–1901. [Google Scholar]
- Bergmann, F. T.; Hucka, M.; Bornstein, B. J.; Jouraku, A. Online SBML Validator. Available online: https://synonym.caltech.edu/ validator_servlet/ (accessed on Mar 4, 2023).
- Smith, L. P.; Bergmann, F. T.; Chandran, D.; Sauro, H. M. , Antimony: a modular model definition language. Bioinformatics 2009, 25, 2452–2454. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Xu, Y. , libSRES: a C library for stochastic ranking evolution strategy for parameter estimation. Bioinformatics 2006, 22, 124–126. [Google Scholar] [CrossRef] [PubMed]
- Kuzmic, P. , Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Analytical biochemistry 1996, 237, 260–273. [Google Scholar] [CrossRef]
- Mendes, P.; Kell, D. , Non-linear optimization of biochemical pathways: applications to metabolic engineering and parameter estimation. Bioinformatics 1998, 14, 869–883. [Google Scholar] [CrossRef] [PubMed]
- Kurata, H.; El-Samad, H.; Yi, T.-M.; Khammash, M.; Doyle, J. In Feedback Regulation of the Heat Shock Response in E. coli, Conference on Decision and Control, Orlando, Florida, USA, 2001; Orlando, Florida, USA, 2001; pp 837-42.
- El-Samad, H.; Kurata, H.; Doyle, J. C.; Gross, C. A.; Khammash, M. , Surviving heat shock: control strategies for robustness and performance. Proc Natl Acad Sci U S A 2005, 102, 2736–2741. [Google Scholar] [CrossRef] [PubMed]
- Kurata, H.; El-Samad, H.; Iwasaki, R.; Ohtake, H.; Doyle, J. C.; Grigorova, I.; Gross, C. A.; Khammash, M. , Module-based analysis of robustness tradeoffs in the heat shock response system. PLoS Comput Biol 2006, 2, e59. [Google Scholar] [CrossRef] [PubMed]
- Jaqaman, K.; Danuser, G. , Linking data to models: data regression. Nat Rev Mol Cell Biol 2006, 7, 813–819. [Google Scholar] [CrossRef]
- Banga, J. R. , Optimization in computational systems biology. BMC Syst Biol 2008, 2, 47. [Google Scholar] [CrossRef] [PubMed]
- Ashyraliyev, M.; Fomekong-Nanfack, Y.; Kaandorp, J. A.; Blom, J. G. , Systems biology: parameter estimation for biochemical models. FEBS J 2009, 276, 886–902. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Boogerd, F. C.; Kurata, H. , RCGAToolbox: A Real-coded Genetic Algorithm Software for Parameter Estimation of Kinetic Models. IPSJ Transactions on Bioinformatics 2021, 14, 30–35. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).