Submitted:
23 February 2023
Posted:
08 March 2023
You are already at the latest version
Abstract
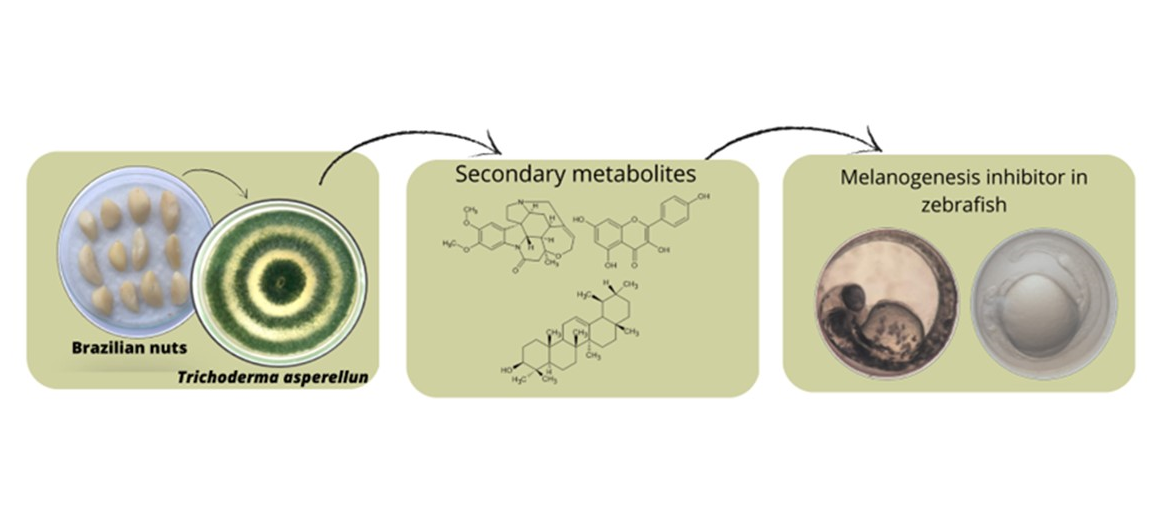
Keywords:
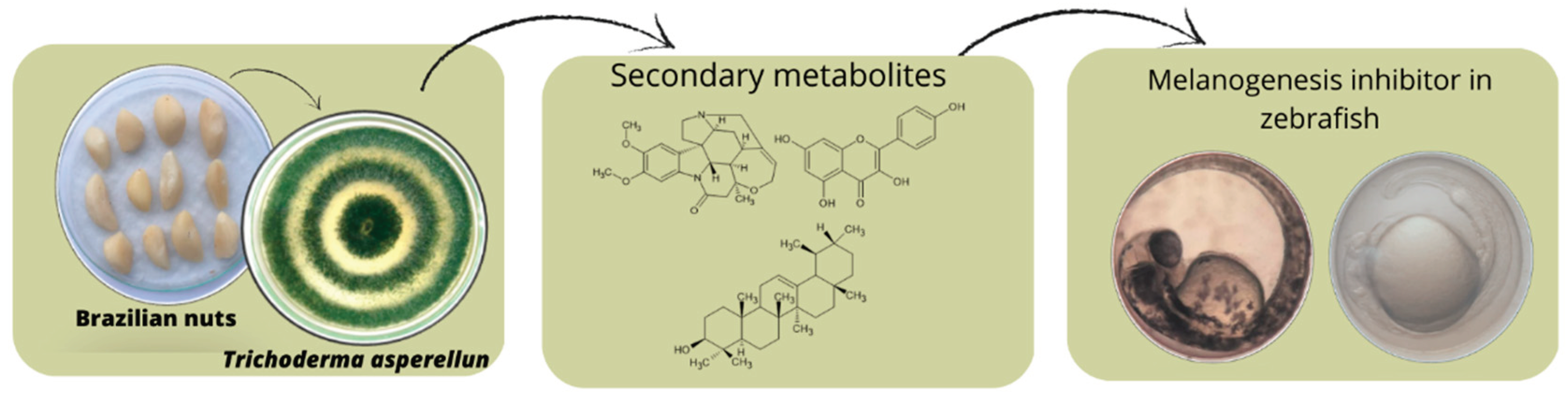
1. Introduction
2. Materials and Methods
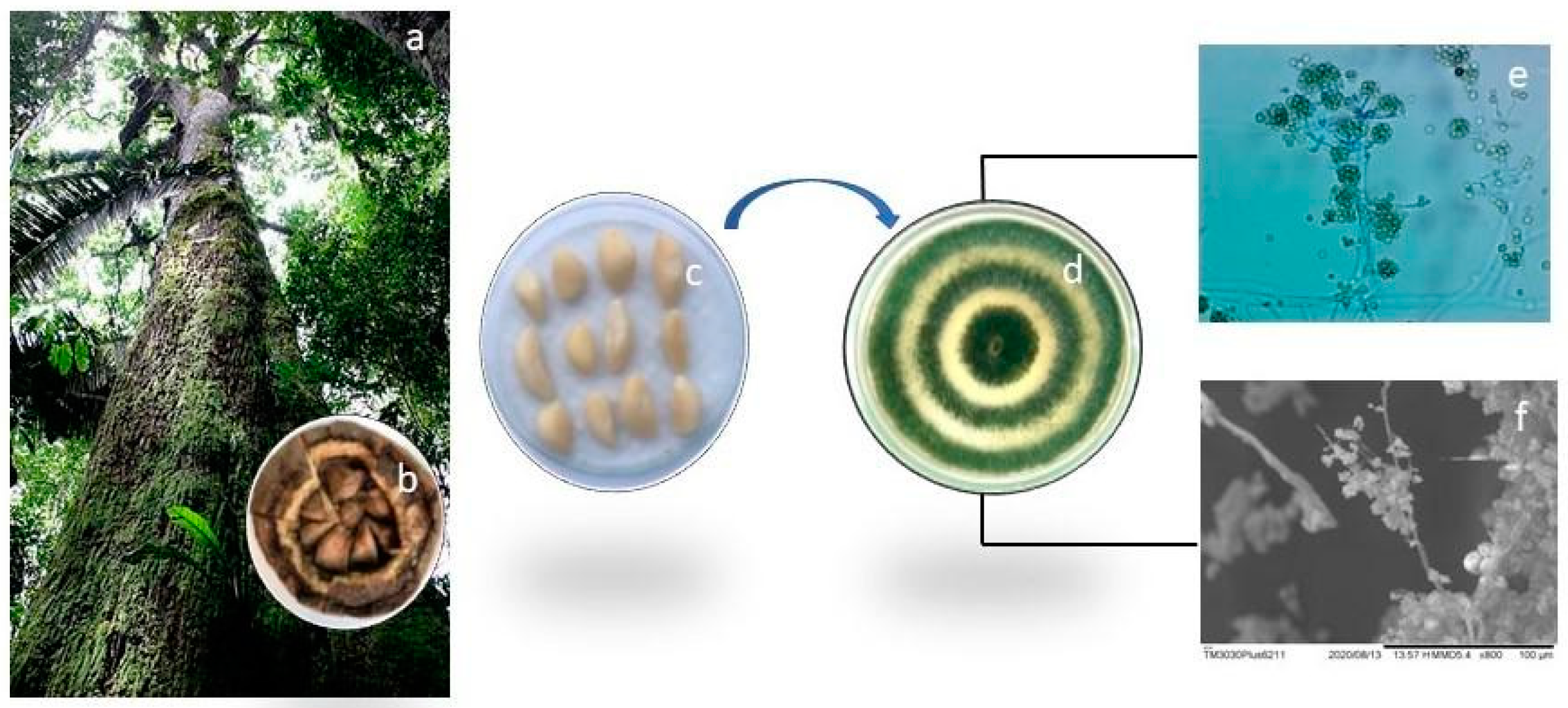
2.1. Collection of Plant Material and Isolation of Trichoderma asperellum Fungi
2.2. Fungal Extract Preparation (AM07Ac)
2.3. Fungal Species Identification
2.3.1. Fungal Genomic DNA Extraction
2.3.2. Genetic Sequencing
2.4. Chemical Profile of T. asperellum Fungal Extract (AM07Ac)
2.4.1. Characterization of Extracts (AM07Ac) by HPTLC and 1H NMR
2.4.1.2. Chemical Profiling by HPTLC
2.4.1.3. Sample Preparation and Application
2.4.1.4. Chromatographic Procedures
2.4.1.5. 1H NMR Spectrum
2.5. Melanogenesis Studies in Zebrafish Embryos
2.5.1. Experimental Animals
2.5.2. Protocol to Determine the Effect of Fungal Extract (AM07Ac) on Melanin Synthesis in Zebrafish
2.5.3. Toxicity of Melanogenic Inhibitors
2.5.4. Analysis of Pigmentation in Zebrafish
2.6. Optimization and Molecular Docking to Identify Secondary Metabolites
2.7. Statistical Analysis
3. Results
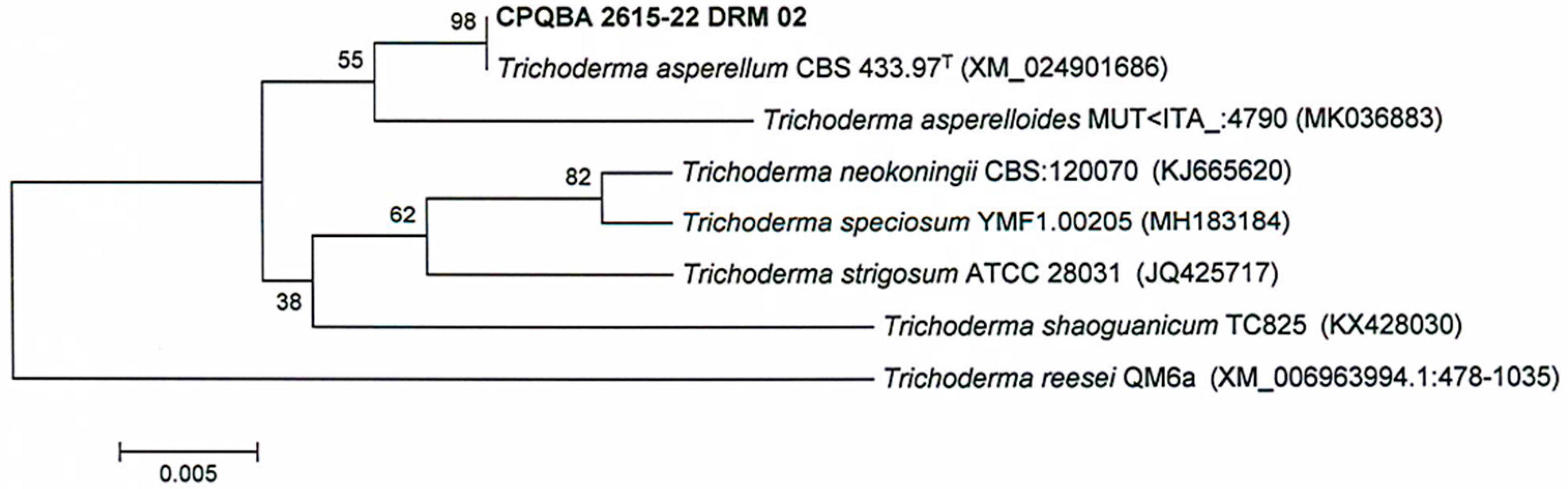
3.1. Identification and Phylogeny
3.2. Chemical Profile of the Fungal Extract of Trichoderma asperellum (AM07Ac) by HPTLC and 1H NMR
3.2.1. Characterization by HPTLC
3.2.2. Characterization by 1H NMR
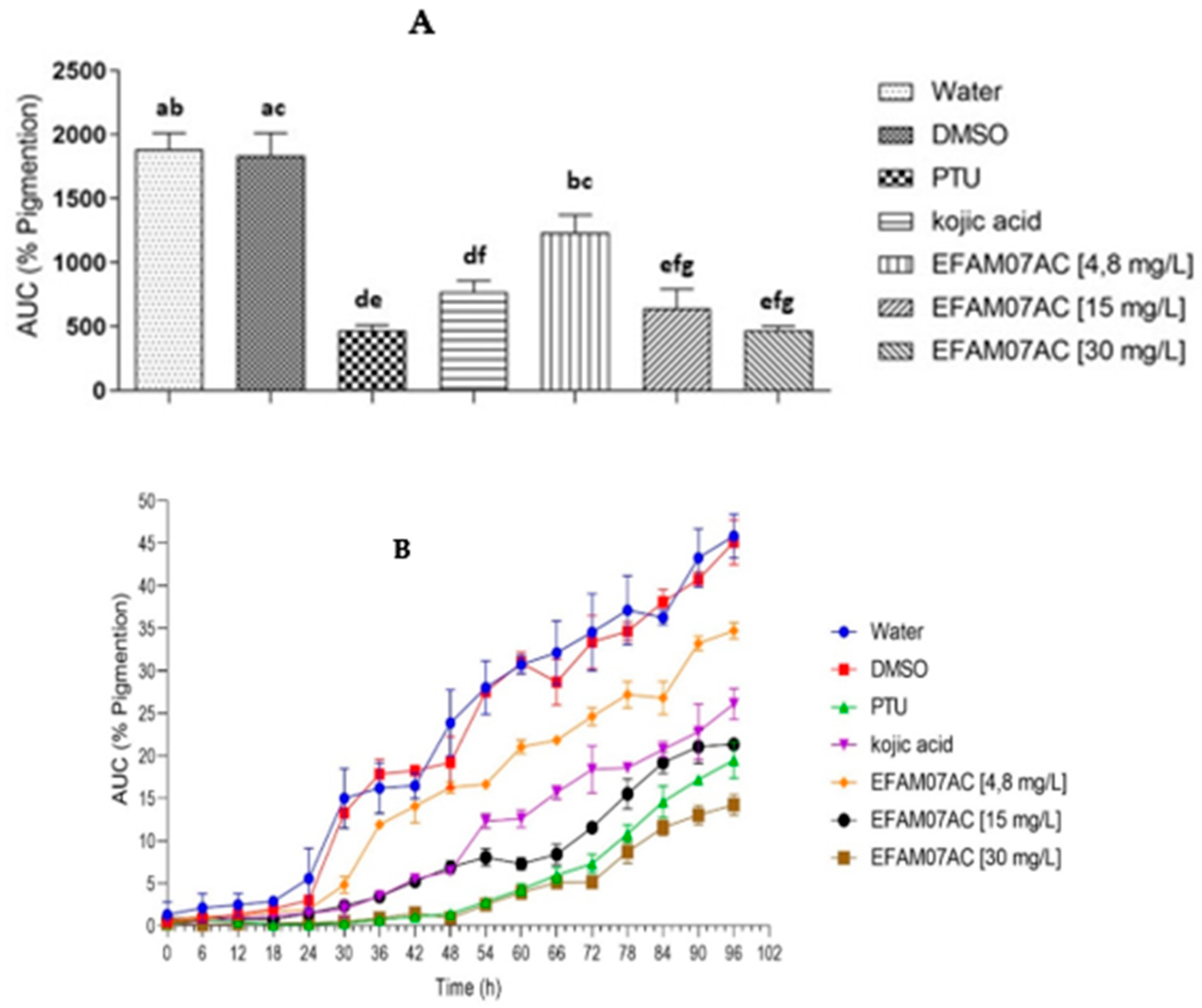
3.3. Melanogenesis Studies in Zebrafish Embryos
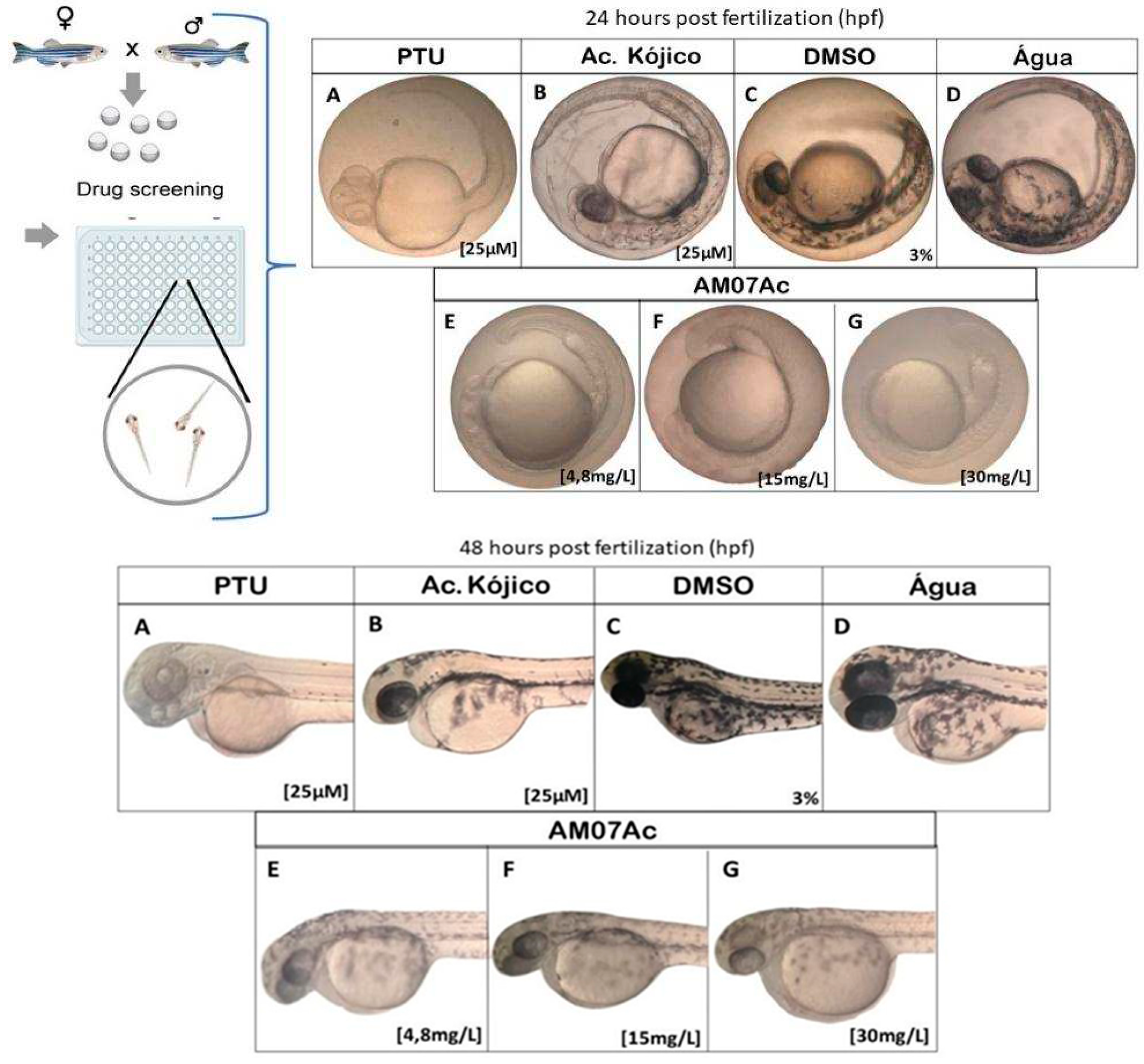
3.3.1. Effect of Treatment with Fungal Extract (AM07Ac) on Melanin Synthesis in Zebrafish
3.3.2. Toxicity of Melanogenic Inhibitors
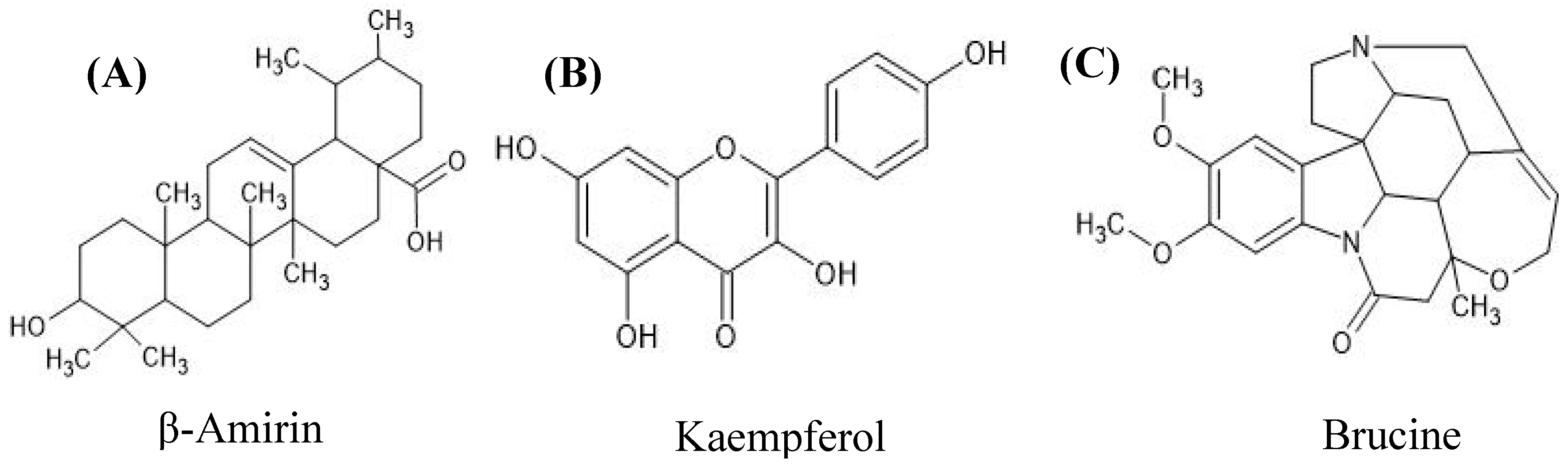
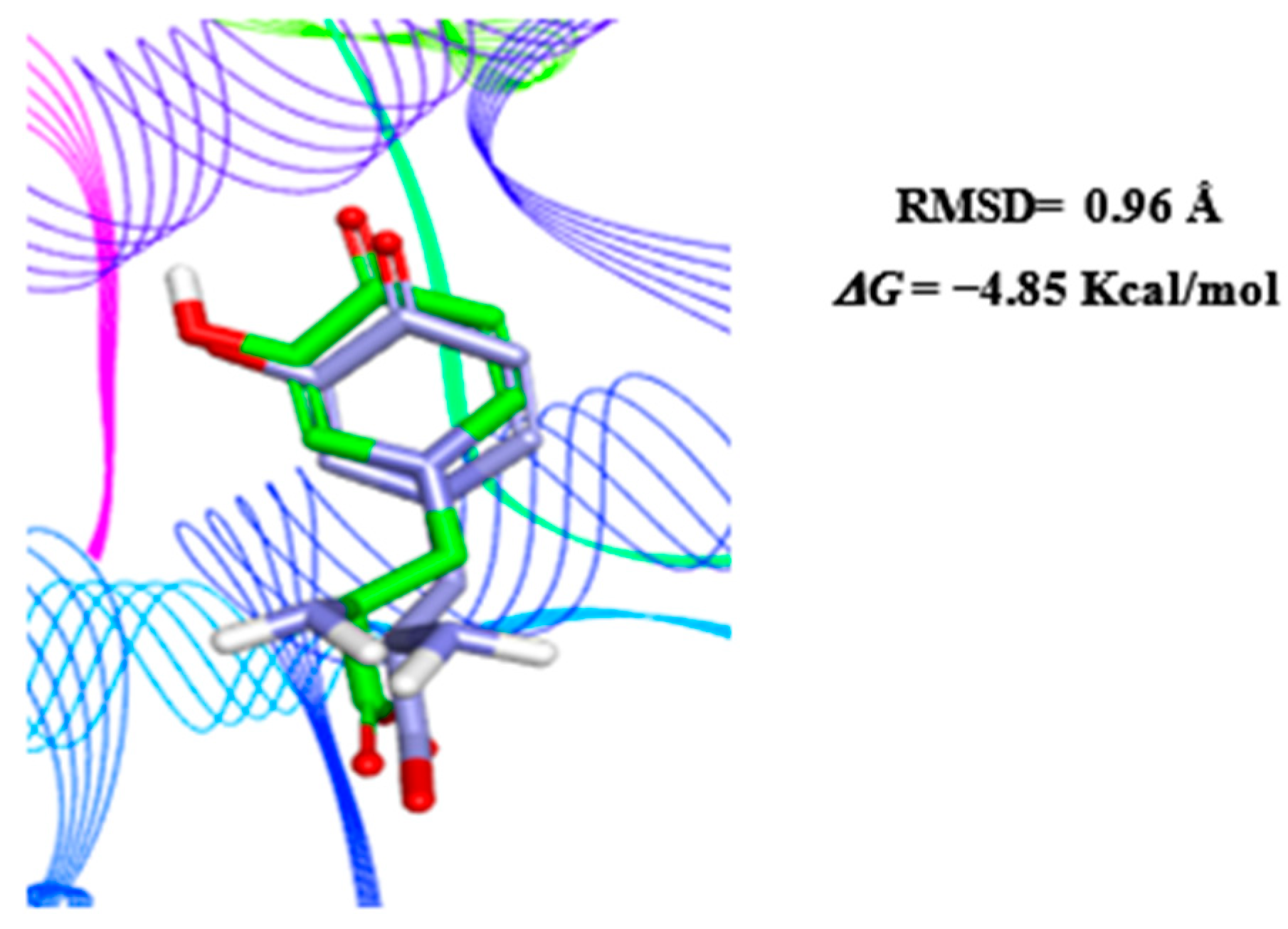
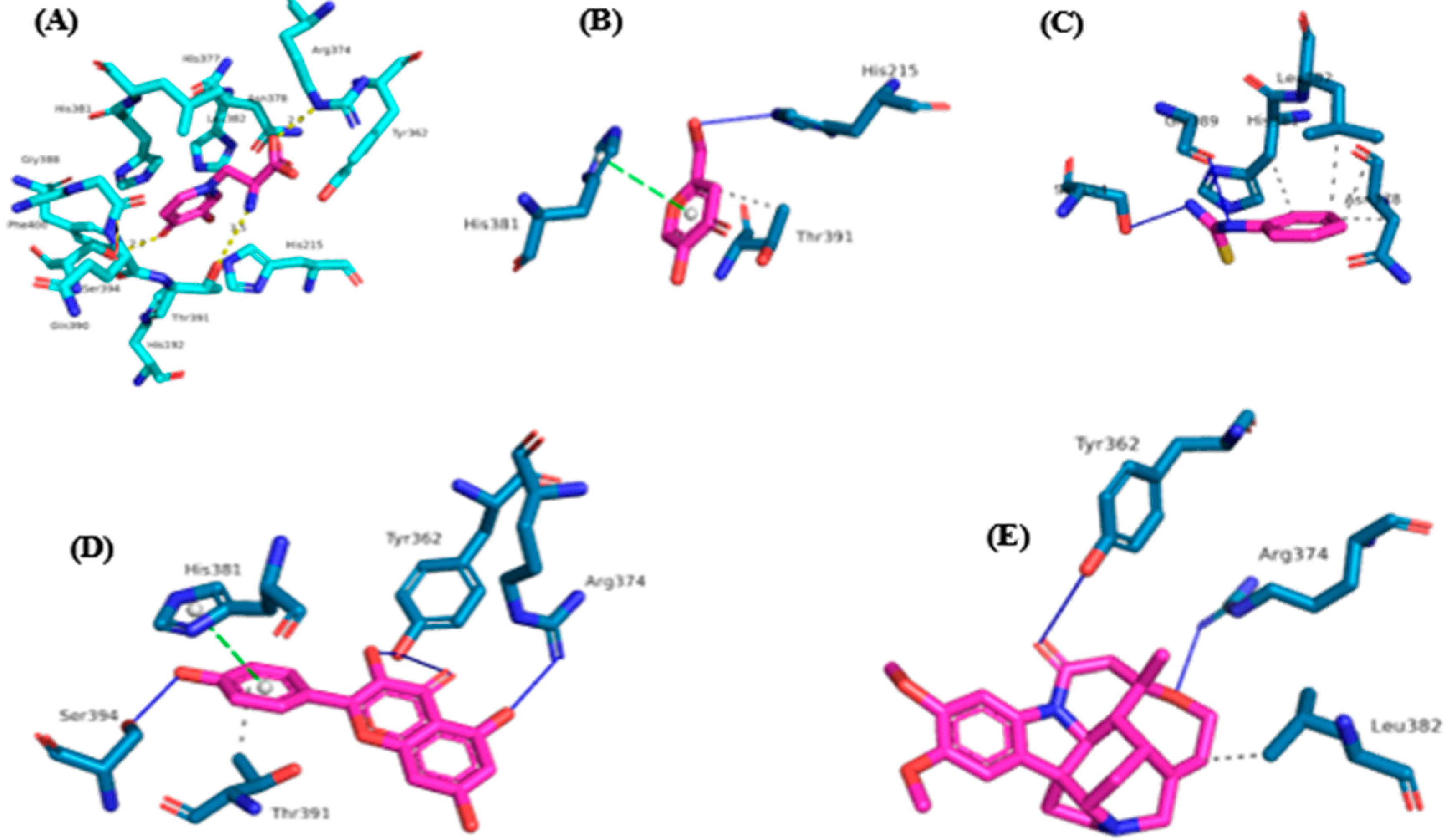
3.4. Inhibition Potential of Secondary Metabolites of AM07Ac Extract on the Enzyme Tyrosinase
5. Discussion
6. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Aly, A.; Debbab, A.; Kjer, J.; Proksch, P. Fungal endophytes from higher plants: a prolific source of phytochemicals and other bioactive natural products. Fungal Divers 2020, 41, 1-16. [CrossRef]
- Souza, J.J.D.; Vieira, I.J.; Rodrigues-Filho, E.; Braz-Filho, R. Terpenoids from Endophytic Fungi. Molecules 2011, 16, 10604-10618. [CrossRef]
- Strobel, G.; Daisy, B.; Castilho, U.; Harper, J. Natural products from endophytic microorganisms. J Nat Prod 2004, 67, 257-268.
- Jay, J.M. Microbiologia de alimentos, 6rd ed.; Artmed: Porto Alegre, Brasil; 2005.
- Keller, N.P.; Turner, G.; Bennett, J.W.MFungal secondary metabolism frombiochemistry to genomics. Nat Rev Microbiol 2005, 937-947.
- Martín, J.F; Casqueiro, J.; Liras, P. Secretion systems for secondary metabolites: how producer cells send out messages of intercellular communication. COMICR 2005, 8, 3282-293. [CrossRef]
- Silva, A.; Silveira, B.K.S.; Freitas, B.V.M.; Hermsdorff, H.H.M.; Bressan, J. Foods 2022, 11, 2925.
- Martins, M.; Pacheco, A.M.; Lucas, A.C.S.; Andrello, A.C.; Appoloni, C.R.; Xavier, J.J.M. Brazil Nuts: Determination of Natural Elements and Aflatoxin. Acta Amaz 2012, 42, 157–164. [CrossRef]
- Carvalho, A. L. S.; Martelli, M.C.; Nascimento, S.C.C. Brasil, D.S.B. Óleo de Castanha-do-Brasil: métodos de extração e aplicações na indústria Res Soc Dev 2022, 11, e29511427256.
- Taniwaki, M.H.; Pitt, J.i.; Iamanaka, b.t.; Sartori, d.; Copetti, M.V.; Balajee, A.; Fungaro, M.H.P.; Frisvad. J.C. Aspergillus bertholletius sp. nov. from Brazil Nuts. Plos One 2012, 7, 42480. [CrossRef]
- Ioca, L.P.; Allard, P.; Berlinck, R.G.S. Thinking big about small beings – the (yet) underdeveloped microbial natural products chemistry in Brazil. Nat Prod Rep 2014, 31, 646–675. [CrossRef]
- Chowdhary, B.; Mahajan, V.K.; Mehta, K.S.; Chauhan, P.S.; Sharma, V.; Sharma, A.; Vashist, S.; Kumar, P. Therapeutic efficacy and safety of oral tranexamic acid 250 mg once a day versus 500 mg twice a day: a comparative study. Arch Dermatol Res 2020, 313, 109-117. [CrossRef]
- Imada, C.; Sugimoto, Y.; Makimura, T.; Kobayashi, T.; Hamada, N.; Watanabe, E. Isolation and characterization of tyrosinase inhibitor producing microorganisms from marine environment. Fish Sci 2001, 67, 1151–1156. [CrossRef]
- Matos, M.J.; Santana, L.; Uriarte, E.; Delogu, G.; Corda, M.; Fadda, M. B.; Era, B.; Fais, A. New halogenated phenylcoumarins as tyrosinase inhibitors. Bioorg Med Chem Lett 2011, 21, 3342–3345. [CrossRef]
- Mackintosh, J.A. The antimicrobial properties of melanocytes, melanosomes and melanin and the evolution of black skin. J Theor Biol 2001, 211, 101-113. [CrossRef]
- Hwang, K.-S.; Yang, J.Y.; Lee, J.; Lee, Y.-R.; Kim, S.S.; Kim, G.R.; Chae, J.S.; Ahn, J.H.; Shin, D.-S.; Choi, T.-Y.; et al. A novel anti-melanogenic agent, KDZ-001, inhibits tyrosinase enzymatic activity. J Dermatol Sci 2018, 89, 165–171. [CrossRef]
- Orlow, S.J. Melanosomes are specialized members of the lysosomal lineage of organelles. J Invest Dermatol 1995, 105, 3–7. [CrossRef]
- Raposo, G.; Marks, M. S. Melanosomes - Dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol 2007, 8, 786–797. [CrossRef]
- Wasmeier, C.; Hume, A.N.; Bolasco, G.; Seabra, M.C. Melanosomes at a glance. Journal of Cell Science 2008,121, 3995–3999.
- Fernandes, M.S.; Kerkar, S. Microorganisms as a source of tyrosinase inhibitors: A review. Ann. Microbiol 2017, 67, 343– 358. [CrossRef]
- Hosoi, J.; Abe, E.; Suda, T., Kuroki, T. Regulation of melanin synthesis of B16 mouse melanoma cells by 1 alpha, 25-dihydroxyvitamin D3 and retinoic acid. Cancer Res 1985, 45, 1474–1478.
- Yang, Z.; Zhang, Y.; Sun, L.; Wang, Y.; Gao, X.; Cheng, Y. An ultrafiltration highperformance liquid chromatography coupled with diode array detector and mass spectrometry approach for screening and characterising tyrosinase inhibitors from mulberry leaves. Anal Chim Acta 2012, 719, 87–95. [CrossRef]
- Harman, G.E.; Howell, C.R.; Viterbo, A.; Chet, I.; Lorito, M. Trichoderma species opportunistic, avirulent plant symbionts. Nat Rev Microbiol 2004, 2, 43–56.
- Yedidia, I.; Shoresh, M.; Kerem, Z.; Benhamou, N.; Kapulnik, Y.; Chet, I. Concomitant induction of systemic resistance to Pseudomonas syringae pv. lachrymans in cucumber by Trichoderma asperellum (T-203) and accumulation of phytoalexins. Appl Environ Microbiol 2003,69, 7343–7353.
- Gasowska, B.; Kafarski, P.; Wojtasek, H. Interaction of mushrCoom tyrosinase with aromatic amines, o-diamines and oaminophenols. Biochim Biophys Acta 2004, 3,170–177.
- Selinheimo, E.; Kruus, K.; Buchert, J.; Hopia, A.; Autio, K. Effects of laccase, xylanase and their combination on the rheological properties of wheat doughs. J Cereal Sci 2006, 43, 152–159. [CrossRef]
- Selinheimo, E.; Saloheimo, M.; Ahola, E.; Westerholm-Parvinen, A.; Kalkkinen, N.; Buchert, J.; Kruus, K. Production and characterization of a secreted, C-terminally processed tyrosinase from the filamentous fungus Trichoderma reesei. FEBS J 2006, 273, 4322–4335.
- Kang, D.W.; Kim, K.-M.; Kim, Y.-S.; Seo, Y.-J.; Song, D.-Y.; Oh, D.-Y.; Choi, S.-O.; Hwang, J.-H.; Kim, S.W.; Bang, K.H.; et al. Inhibition of Tyrosinase by Metabolites Originating from Thrichoderma atroviride. J Life Sci 2021, 31, 47–51.
- Kima, Y.-J.; Uyama, H. Tyrosinase inhibitors from natural and synthetic sources: structure, inhibition mechanism and perspective for the future. Cell Mol Life Sci 2005, 62, 1707-23. [CrossRef]
- Chang, T.S. An Updated Review of Tyrosinase Inhibitors. Int J Mol Sci 2009, 10, 2440–2475. [CrossRef]
- Postlethwait, J.; Chung, B.-C. Characterization of duplicated zebrafish cyp19. Genes J Exp Zool 2001, 290, 709-714.
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; Mclaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503.
- Silveira, T. R.; Schneider, A.C.; Hammes, T.O. Zebrafish: modelo consagrado para estudos de doenças humanas. Cienc Cult 2012, 64, 4-5. [CrossRef]
- Chen, E.; Ekker, S.C. Zebrafish as a Genomics Research Model. Curr Pharm Biotechnol 2004, 5, 409-413.
- McGrath, P.; Li, C-Q. Zebrafish: a predictive model for assessing drug-induced toxicity. Drug Discovery Today 2008, 13, 394-401. [CrossRef]
- Ferreira; A.M.; Souza,A.A.; Koga, R.C.R.; Sena, I.S.; Matos, M.J.S.M.; Tomazi, R.; Ferreira, I.M.; Carvalho, J.C.T. Anti-Melanogenic Potential of Natural and Synthetic Substances: Application in Zebrafish Model. Molecules 2023, 28, 1053. [CrossRef]
- Holanda; F.H.; Willian G. B.; Morais, E.M.; Sena, I.S.; Ferreira, A.M.; Faustino, S.M.M.M.; Solon, L.G.S.; Porto, A.L.M.; Ferreira, I.M. Study of biodegradation of chloramphenicol by endophytic fungi isolated from Bertholletia excelsa (Brazil nuts). Biocatal Agric Biotechnol 2019, 20, 101200. [CrossRef]
- Aamir, S.; Sutar S., Singh, S.K.; Baghela, A. (A rapid and efficient method of fungal genomic DNA extraction, suitable for PCR based molecular methods. Plant Pathol Quar J Fungal Biolog 2015, 5, 74-81.
- Thompson, J. D.; Gibson, T. J.; Plewniak, F.; Jeanmougin, F.; Higgins, D. G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997,24, 4876-4882.
- Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 1999, 41, 95-98.
- Tamura K, Stecher G, Peterson D, Filipski A, and Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0 Mol Biol Evol 2013, 30, 2725-2729.
- Kimura, M. A simple model for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. Journal of Molecular Evolution, 1980, 16, 111-120.
- Saitou, N.; Nei, M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987, 4, 406-425. [CrossRef]
- Pinheiro, W.B.S.; Pinheiro Neto J.R.; Botelho, A.S. ; Santos, K.I.P.; Silva, G.A.; Muribeca, A.J.B.; Pamplona, S.G.S.R.; Fonseca, S.S.S. ; Silva, M.N ; Arruda, M.S.P. The use of Bagassa guianensis aubl. Forestry waste as an alternative for obtaining bioproducts and bioactive compounds. Arab J Chem 2022, 15, 103813. [CrossRef]
- 4Feng,Y.; Qiu, W.; Li, R.; Hu,J.; Luo, S.; Zhang, T.; He,X.; Zheng, C. Genome-wide identification of the interactions between key genes and pathways provide new insights into the toxicity of bisphenol F and S during early development in zebrafish. Chemosphere 2018, 213, 559-567. [CrossRef]
- Kagotani, K.; Nakayama, H.; Zang, L.; Fujimoto, Y.; Hayashi, A.; Sono, R.; Nishimura, N.; Shimada, Y. Lecithin-Based Dermal Drug Delivery for Anti-Pigmentation Maize Ceramide. Molecules 2020, 25, 1595. [CrossRef]
- Kim, D-C.; Kim, S.; Hwang, K-S.; Kim, C-H. P-Coumaric Acid Potently Down-Regulates Zebrafish Embryo Pigmentation: Comparison ofin VivoAssay and Computational Molecular Modeling with Phenylthiourea. Biomed Sci Lett 2017. [CrossRef]
- Zhang, W.; Pei, J.; Lai, L. Computational Multitarget Drug Design. J Chem Inf Model 2017, 57, 403−412. [CrossRef]
- Brooks; B.R.; Bruccoleri, R.E.; Olafson, B.D.; States, D.J.; Swaminathan, S.; Karplus, M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983, 4, 187–217. [CrossRef]
- Laxmi, K. Chemsketch study of phenobarbital: An antiepileptic drug. Int J Comput Theor Chem 2017, 5, 25.
- Mweene, P.; Muzaza, G. Implementation of interactive learning media on chemical materials. JEV 2020, 1, 8-13. [CrossRef]
- Schrödinger, L.; Delano, W. PyMOL, 2020. Accessed on http://www.pymol.org/pymol.
- Samuels, G.J.; Dodd, S.L.; Gams, W.; Castlebury, L.A.; Petrini, O. (2002) Trichoderma species associated with the green 650 mold epidemic of commercially grown Agaricus bisporus. Mycologia 2002, 94, 146-170. [CrossRef]
- Lemes; G.F.; Ferri, P.H.; Lopes, M.N. Constituintes Químicos de Hyptidendron canum (Pohl ex Benth.) R. Harley (LAMIACEAE). Quim Nova 2011, 34, 39-42.
- Ayers; S.; Deborah L. Zink, Kenneth Mohn, Joanne S. Powell, Christine M. Brown, Terry Murphy, Robert Brand, Seef Pretorius, Dennis Stevenson, Donald Thompson, Sheo B. Singh. Flavones from Struthiola argentea with anthelmintic activity in vitro. Phytochemistry 2008, 69, 541-545. [CrossRef]
- Semenov, V.A.; Samultsev, D.O.; Krivdin, L.B. Calculation of 15N NMR Chemical Shifts in a Diversity of Nitrogen-Containing Compounds Using Composite Method Approximation at the DFT, MP2, and CCSD Levels. J Phy. Chem A 2019, 123, 8417−8426.
- Peterson, S.W.; Ito, Y.; Horn, B.W.; Goto, T. Aspergillus bombycis, a new aflatoxigenic species and genetic variation in its sibling species, A. nomius. Mycologia 2001, 93, 689-703.
- Choi, T.Y.; Kim, J.H.; Ko, D.H.; Kim, C.H.; Hwang, J.S.; Ahn, S.; Kim, S.Y.; Kim, C.D.; Lee, J.H.; Yoon, T.J. Zebrafish as a new model for phenotype-based screening of melanogenic regulatory compounds. Pigment Cell Res 2007, 20; 120–127. [CrossRef]
- Ramos, R. S.; Costa, J.S.; Silva, R.C.; Costa, G.V.; Rodrigues, A.B.L.; Rabelo, E.M.; Souto, R.N.P.; Taft, C.A.; Silva, C.H.T.P.D.; Rosa, J.M.C.; Santos, C.B.R.D.; Macêdo, W.J.D.C. Identification of Potential Inhibitors from Pyriproxyfen with Insecticidal Activity by Virtual Screening. Pharmaceuticals 2019, 12, 20. [CrossRef]
- Ramos, R.S.; Macêdo, W.J.C.; Costa, J.S.; Silva, C.H.T.P.; Rosa, J.M.C.; Cruz, J.N.; Oliveira, M.S.; Aguiar Andrade, E.H.; Silva, R.B.L.; Souto, R.N.P.; Santos, C.B.R. Potential Inhibitors of the Enzyme Acetylcholinesterase and Juvenile Hormone with Insecticidal Activity: Study of the Binding Mode via Docking and Molecular Dynamics Simulations. J Biomol Struct Dyn 2020, 38, 4687–4709. [CrossRef]
- Ramos, R.S.; Borges, R.S.; Souza, J.S.N.; Araujo, I.F.; Chaves, M.H.; Santos, C.B.R. Identification of Potential Antiviral Inhibitors from Hydroxychloroquine and 1,2,4,5-Tetraoxanes Analogues and Investigation of the Mechanism of Action in SARS-CoV-2. Int J Mol Sci 2022, 23, 1781.
- Luo, Y.; Wang, J.; Li, S.; Wu,Y.; Wang, Z.; Chenm S.; Chen, H. Discovery and identification of potential anti-melanogenic active constituents of Bletilla striata by zebrafish model and molecular docking. BMC Complement Med Ther 2022, 22, 1-14. [CrossRef]
- Zeilinger, S.; Gruber, S.; Bansal, R.; Mukherjee, P. K. Secondary metabolism in Trichoderma–chemistry meets genomics. Fungal Biol Rev 2016, 30, 74-90. [CrossRef]
- Al-Brakati, A.; Albarakati, A.J.A.; Lokman, M.S.; Theyab, A.; Algahtani, M.; Menshawi, S.; AlAmri, O.D.; Al Omairi, N.E.; Essawy, E.A.; Kassab, R.B.; Abdel Moneim, A.E. Possible role of Kaempferol inreversing oxidative damage, inflammation, and apoptosis-mediated cortical injury following cadmium exposure. Neurotox Res 2021, 39, 198-209. [CrossRef]
- Tu, Y-H.; Liu, F.; Guo, D.D.; Fan, L.J.; Zhu, S-X.; Xue, Y-R.; Gao, Y.; Guo, M-L. Molecular characterization of flavanone 3-hydroxylase gene and flavonoid accumulation in two chemotyped safflower lines in response to methyl jasmonate stimulation. BMC Plant Biology 2016, 16, 132. [CrossRef]
- Calderon-Montano, J.M.; Burgos-Moron, E.; Perez-Guerrero, C.; Lopez-Lazaro, M. A review on the dietary flavonoid kaempferol. Mini Rev Med Chem 2011, 11, 298–344. [CrossRef]
- Lu, L.; Huang, R.; Ye Wu1,Y.; Jin, J-M.; Chen, H.Z.; Zhang, L-J.; Luan, X. Brucine: a review of phytochemistry, pharmacology, and toxicology. Front Pharmacol 2020, 11, 377. [CrossRef]
- Rosa, G.P.; Palmeira, A.; Resende, D.I.S.P. ; Almeida, I.F. ; Kane-Pagès, A.; Barreto, M.C. ; E. Sousa, E.; Pinto, M.M.M. Xanthones for melanogenesis inhibition: Molecular docking and QSAR studies to understand their anti-tyrosinase activity. Bioorg Med Chem 2021, 29, 115873. [CrossRef]
- Ferreira, A.M.; de Souza, A.A.; Koga, R.d.C.R.; Sena, I.d.S.; Matos, M.d.J.S.; Tomazi, R.; Ferreira, I.M.; Carvalho, J.C.T. Anti-Melanogenic Potential of Natural and Synthetic Substances: Application in Zebrafish Model. Molecules, 2023, 28, 1053, 2023.
- Rho, H.S.; Ahn, S.M.; Lee, B.C.; Kim, M.K.; Ghimeray, A.K..; Jin, C.W.; Cho, D.H. Alterações no conteúdo de flavonoides e na atividade inibitória da tirosinase no extrato de folhas de kenaf após tratamento com infravermelho distante. Bioorg Med Chem 2010, 20, 7534-7536.
- Kubo, I.; Kinst-Hori, I. Flavonols from saffron flower: tyrosinase inhibitory activity and inhibition mechanism. J Agric Food Chem 1999, 47, 4121-4125. [CrossRef]
- Kurpejović, E.; Wendisch, V.F.; Sariyar Akbulut, B. Tyrosinase-based production of L-DOPA by Corynebacterium glutamicum. Appl Microbiol Biotechnol 2021, 105, n. 24, 9103-9111. [CrossRef]
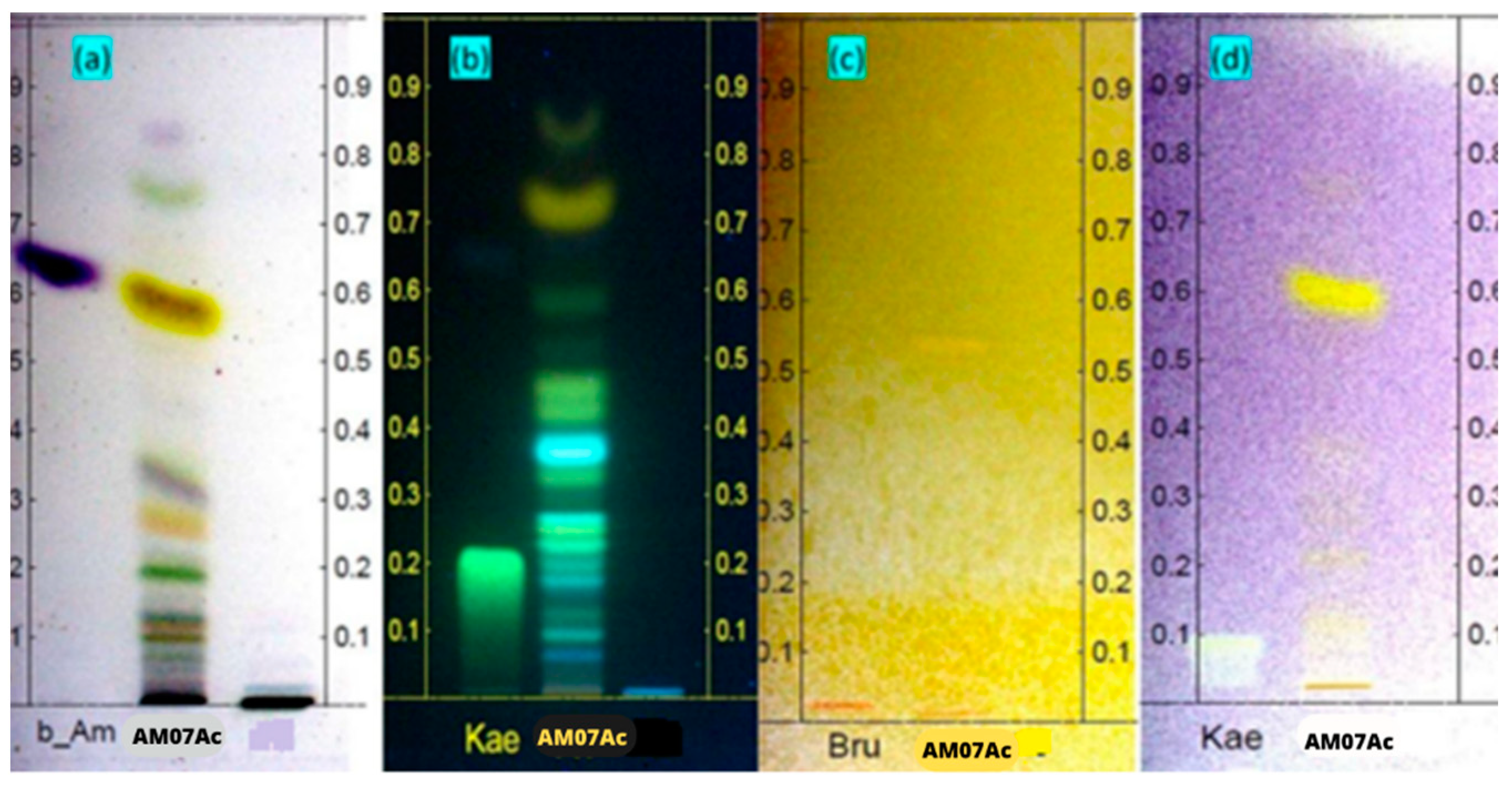
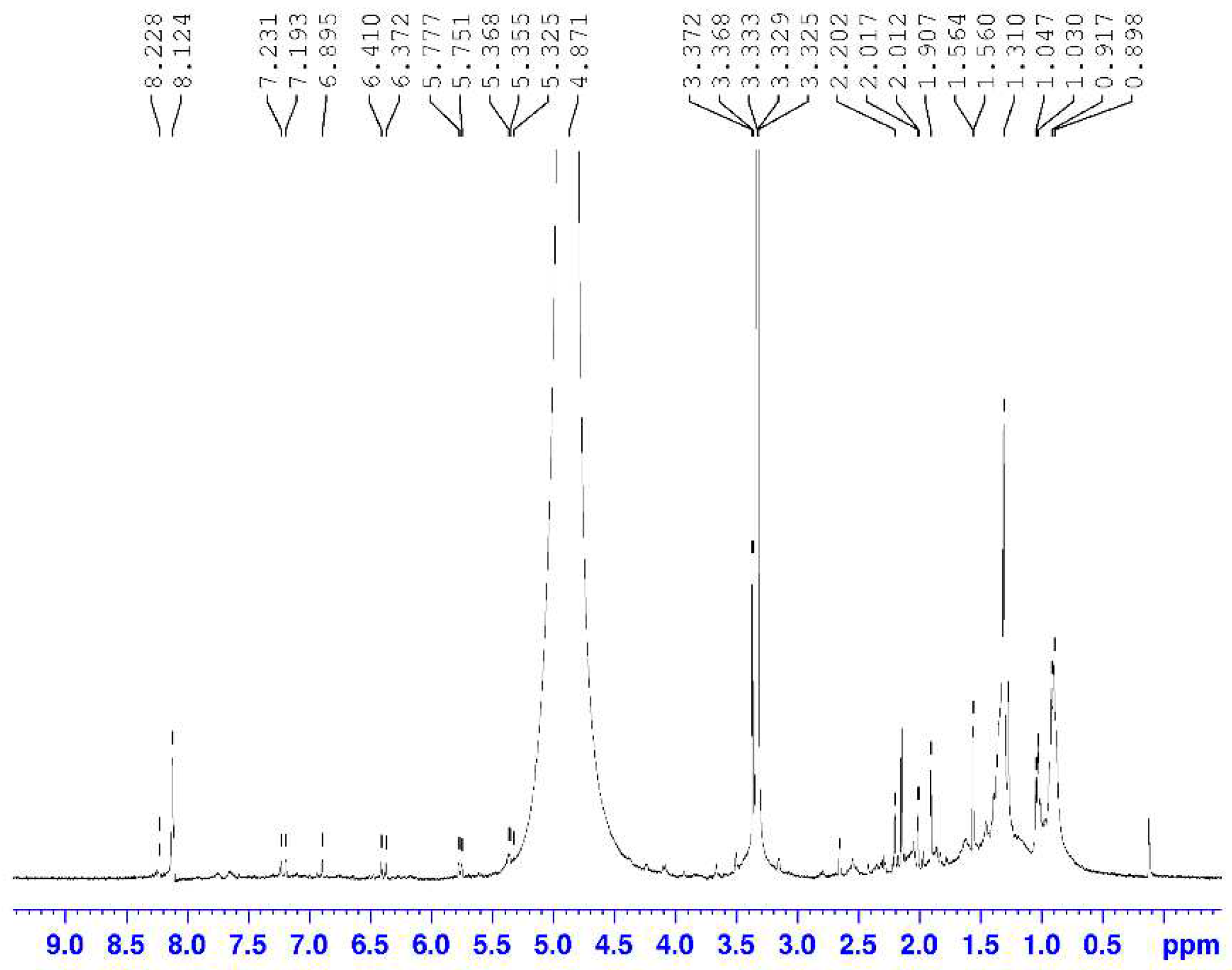
| Chemical Shift (ppm) | Assignments | Area (%) |
| 0.5 - 1.5 | -CHn; -CHn | 0.16 |
| 1.5 - 3.0 | CHn-C=O; CHn-N; Ar-CHn; Ar-CHn- | 0.35 |
| 3.0 - 4.5 | CHn-C=O; -CHn-O-; -CHn-N- | 2.06 |
| 4.5 - 6.0 | Ph-O-CHn; HC=C- (non-conjugated) | 15.00 |
| 6.0 - 9.5 | Ph-H; Ph-CH=O | 0.13 |
| Protease | Binder | Binding Affinity (Kcal/mol) | H-Bond | Lipophilic interactions |
| Tyrosinase | MMS | -5.9 | Arg374, Thr391, Ser394 | His192, His215, Leu382, His377, Tyr362, Asn378, His381, Gly388, Thr391, Asn378, Phe400, |
| PTU | -5.7 | Gly388, Gly 389, Thr391 | His381, Leu382 | |
| Kojic acid | -5.7 | His215 | His381, Thr391 | |
| Kaempferol | -6.8 | Tyr362, Arg374, Ser394 | His381 | |
| Brucine | -5.7 | Tyr362, Arg374 | Leu382 | |
| β-Amyrin | -4.5 | - | Gln390 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





