1. Introduction
A major goal in biomedical research is to relate diseases or phenotypes to genotypes influenced by environmental factors. Linking genetic variants to phenotypes, or vice versa, phenotypes to genetic variants, is the first step in generating hypotheses to determine the molecular drivers underlying diseases and expressed traits. Genome-wide association studies (GWAS) and phenome-wide association studies (PheWAS) have been instrumental in generating such hypotheses in an unbiased manner for identifying the genetic basis of a disease or a trait. GWAS begins by comparing a large number of individuals with clinical manifestations or phenotypes to healthy individuals or individuals without the phenotype. Whole genome sequencing of these individuals allows for the identification of genetic variants, usually single-nucleotide variants (SNVs). Association is then determined by the increased frequency of an SNV or a genetic variant found in patients compared to healthy individuals. GWAS is a “forward genetics” approach. By contrast, PheWAS is a “reverse genetics” approach. PheWAS starts with a genetic variant and surveys a large number of different phenotypes to identify one or multiple phenotypes that statistically associate with this given genetic variant or SNV. Since the association is not causation, the holy grail in this field is to find causally or mechanistically linked genotype-phenotype relationships. Further experimental research with model organisms is required to further validate these mechanistic discoveries.
The recent emergence of medically relevant PheWAS was made possible by associating large human genetic information with electronic health records [
1,
2,
3,
4]. This required dense phenotyping of patients and collecting patient samples into biobank repositories to enable researchers to perform “omics” approaches, such as genomics, proteomics, and metabolomics [
4]. The discoveries of novel mechanisms are facilitated by integrating open-source data across species. For instance, the Monarch Initiative is a large collection of over two million genotypic-phenotypic associations that integrates data across hundreds of species from more than 30 different databases [
5].
Here, we mine the data from previous PheWAS-GWAS studies and other databases to establish the phenotypic landscape of extracellular matrix (ECM) gene variants. The ECM has emerged as a novel target for healthy aging [
6,
7], and perturbations within ECM integrity can be major drivers for either health or disease [
8,
9]. With the aim to attenuate diseases, 27 clinical trials are underway on eleven different molecular targets for modulating ECM stiffness as a primary outcome measure [
10]. Given the clinical importance of the ECM, we hypothesize that variants in ECM genes are associated with disease phenotypes and other physiological traits. To address this, we use three key approaches.
First, we surveyed published genotype-phenotype interactions for variants in ECM genes associated with the disease. All possible gene products that either form extracellular matrices or are associated with or remodel ECMs are collectively called matrisome [
11]. The human matrisome consists of 1027 proteins [
12]. The core matrisome that forms the structural components of the ECM consists of collagen, proteoglycan, and glycoproteins. The associated matrisome is mainly responsible for the maintenance, upkeep, and remodeling of the ECM and consists of ECM-related proteins, secreted factors, crosslinking proteins, and proteinases [
12].
Second, we analyzed the variants in such matrisome genes using the clinical variance diseasome list to identify unique diseases with multiple SNVs in matrisome genes. Using the integrative disease data from the Monarch Initiative, we then linked diseases with specific genes in the matrisome. Using the PheWAS approach, we then analyzed the genes implicated in phenotypic associations of disease states.
Third, we took this list of genes and diseases to create gene-disease associations based on disease categorization, from typical diseases to unexpected and age-related diseases. We identified a list of specific collagen genes that displayed the highest frequency of associations with multiple disease states per core matrisome gene. Using these associations, we then analyzed the specific predominance of mutations in said collagen genes. By cross-referencing the GWAS-PheWAS database and DrugBank repositories, we identified numerous drugs which may affect matrisome genes. Conclusively, we analyzed the rare disease databases to view any associations with genes involving the matrisome.
Thus, we use the matrisome list as a seed for screening. Taken together, our computational efforts identify targets that can be validated by experimental approaches to gain novel mechanistic insights in matrix biology with translational value.
2. Materials and Methods
Data source is referenced in the result section. Data cleaning, analysis, and visualization were performed using the statistical programming language R utilizing the key packages dplyr, purrr, and ggplot2 as well as in Python utilizing the pandas, numbat, scipy, and dask packages. All processed and output data are provided in Supplementary Tables 1–11.
To establish a robust disease-gene association, publicly-available datasets were obtained using standard web download and combined as described in [
13] (Table Updated Jan 2020). The associations include clinical variance data human diseases [
14], and the causal human disease genes from the Monarch initiative (
https://monarchinitiative.org/, diseases tab, human, causal genes).
The diseases were subsequently categorized into three groups: ECM-associated (A), other common diseases (B), and age-related illnesses (C). The grouping was performed based on substring matching using the terms listed in Supplementary Table 9.
The human matrisome gene list was obtained from Naba et al., 2016 [
12] and was utilized to subgroup the ECM-associated diseases by their matrisome division and category. The PheWAS of GWAS Catalog of SNVs was obtained from
https://phewascatalog.org/ (dataset “PheWAS of GWAS Catalog of SNPs”). Several of the graph-network visualizations were developed in Python utilizing the cytoscape and forceatlas packages.
3. Results
3.1. Clinical Implications of the Human Matrisome
Previously, more than 1000 mutations in collagens were implicated in about 20 common diseases [
15,
16] (
Figure 1). To expand this, we first took advantage of comparing the 1027 human matrisome genes [
12] with genome-wide association studies (GWAS) cou-pled to phenome-wide association studies (PheWas) of curated medical health records (Resource: PheWas catalog [
17]). Out of the 3’144 single nucleotide polymorphisms (SNVs) associated with clinical phenotypes [
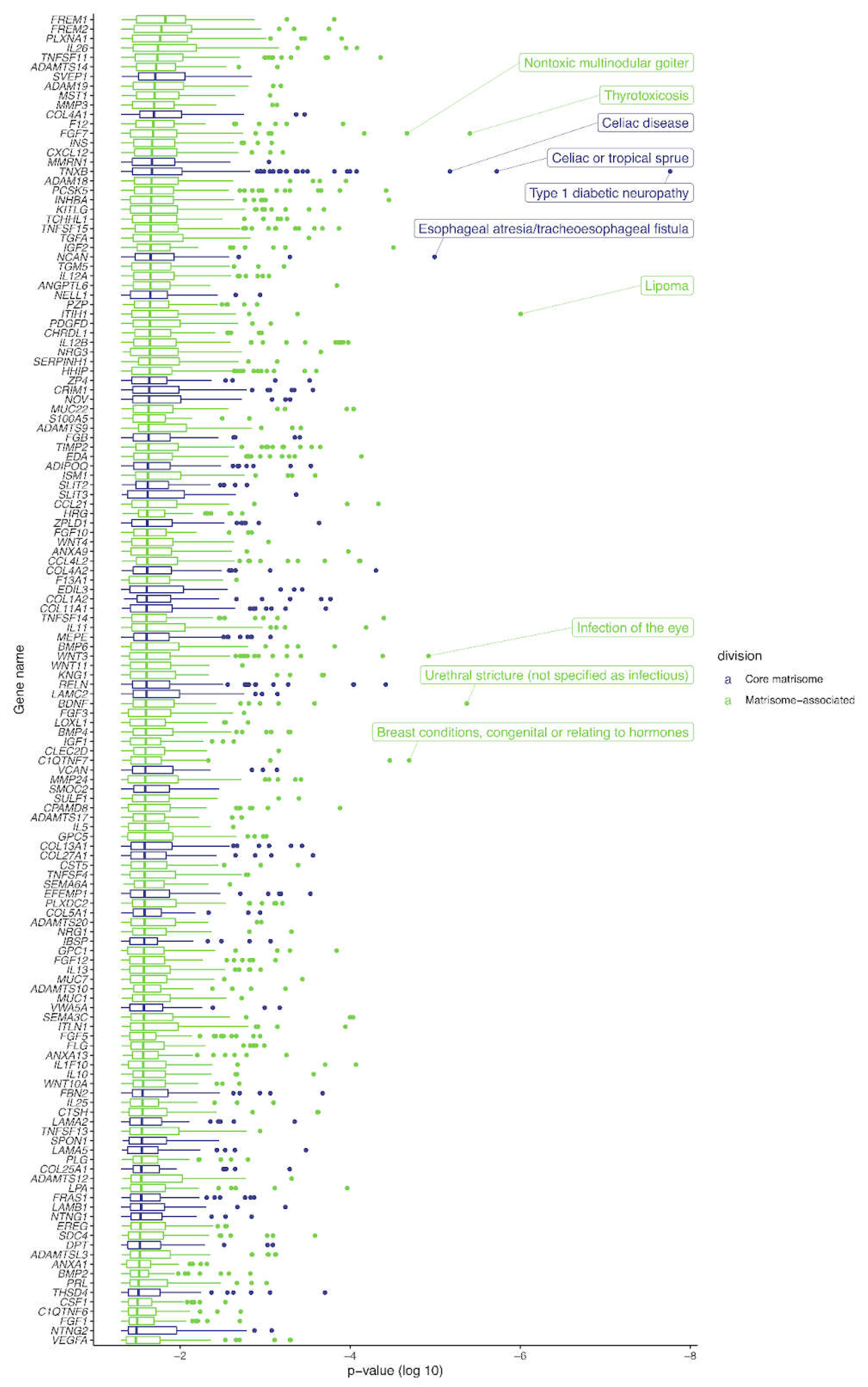
17], we uncovered 140 SNVs located within the core matrisome and matrisome-associated genes. Most notable were the striking asso-ciations of the TNXB gene, responsible for the glycoprotein tenascin XB, with Celiac or tropical sprue, Celiac’s disease, and particularly with diabetic type I Neuropathy (P value < 5x10-8). (
Figure 2, Supplementary Table 1). These matrisome proteins, containing disease-associated SNVs, function in large interactive complex ECM networks and may exhibit some interrelated pathology through signaling-related characteristics. Extracellular ma-trix proteins form protein-protein interaction complexes. Mutations in either one of these ECM proteins in a complex should manifest in a similar phenotype. Previously, the net-work of human proteins of genetic disorders has been defined, i.e., the human phenome-interactome [
18]. Out of these 506 protein-protein interaction human disease complexes [
18], 18 matrisome and adhesosome proteins are predicted to be associated with these dis-ease complexes (Supplementary Table 2). Interestingly, in addition to matrisome gene products, there were also two collagen-binding integrins (ITGB7 and ITGA2) found that might link the ECM to cellular signaling (Supplementary Table 2). To expand on this idea of using ECM complexes, we used the matrisome to screen previously predicted protein-protein interactions of the human disease network [
14]. We found 305 unique matrisome genes involved in 542 matrisome-matrisome protein interaction pairs with implications for human diseases (Supplementary Table 3). Out of the 3824 human disease network genes, we identified 161 matrisome genes associated with 270 human diseases (Supplementary Table 4), suggesting a significant contribution to diseases.
3.2. Genetic Variants in Matrisome Genes Associated with Diseases
To gain a more global view of the human disease landscape associated with SNVs in matrisome genes, we took three approaches. First, we used the clinical variance diseasome list (
www.ncbi.nlm.nih. gov/clinvar/; [
13]). In this dataset, there are 1840 unique genes associated with 9281 polymorphisms, from which we found 181 unique matrisome genes associated with 1216 polymorphisms. Within this list of polymorphisms, we found that Alport syndrome displayed the largest occurrence with 423 SNVs within the matrisome genes, followed by Marfan’s syndrome with 149 SNVs, and osteogenesis imperfecta as the third most referenced disease with 132 SNVs (Supplementary Table 5).
Second, we used the integrative disease data from the Monarch Initiative, which accesses over 30 externally curated databases (
https://monarchinitiative.org/disease [
5]). We found 285 matrisome genes associated with 553 human diseases (Supplementary Table 6). We identified a disease association in 30 out of the total 44 collagens genes in humans resulting in 106 different pathologies (Supplementary Table 6). The collagens with the strongest known disease association are COL2A1 and COL7A1, with 19 and 13 implicated collagenopathies, respectively. The majority of disease associations comprising 19.9% of all collagen gene-to-disease links were identified in collagen COL2A1 (Supplementary Table 6). COL2A1 is predominantly linked to different forms of walking and growth disorders resulting, among others, in short stature: Spondyloepimetaphyseal Dysplasia (Strudwick Type) resulting in short stature and skeletal abnormalities, Metaphyseal Chondrodysplasia (Schmid Type) resulting in abnormally short limbs and short stature, Spondyloepimetaphyseal Dysplasia Congenita resulting in short stature and skeletal abnormalities, Kniest Dysplasia resulting in skeletal, visual, and auditory abnormalities and short stature, Hypochondrogenesis (Achondrogenesis Type II) resulting in short limbs and altered bone growth, Platyspondylic Dysplasia (Torrance Type) resulting in shortened limb formation, Spondyloperipheral Dysplasia (Short Ulna Syndrome), Dysspondyloenchondromatosis, Spondyloepiphyseal Dysplasia (Stanescu Type), Familial Avascular Necrosis of Femoral Head, and Avascular Necrosis of Femoral Head, Primary, 1 [
19].
Overlapping with the involvement in multiple skeletal phenotypes, this collagen is also involved in visual perception leading to retinal thinning (multiple epiphyseal dysplasia, Beighton type), retinal tears (autosomal dominant rhegmatogenous retinal detachment) and retinal detachment (Stickler syndrome type 1; Supplementary Table 6). By contrast, the Stickler syndrome has the most collagens linked to this syndrome (6 collagens associated: COL2A1, COL9A1, COL9A2, COL9A3, COL11A1, COL11A2; Supplementary Table 6). Followed by Ullrich congenital muscular dystrophy with four collagens associated (COL6A1, COL6A2, COL6A3, COL12A1; Supplementary Table 6).
Third, we took a PheWAS approach and searched the 1000 top hits with
P-value <10
-6 from the UK Biobank consisting of more than 2000 phenotypes associated with 3’144 GWAS from more than 500 thousand individuals [
http://big.stats.ox.ac.uk/about; [
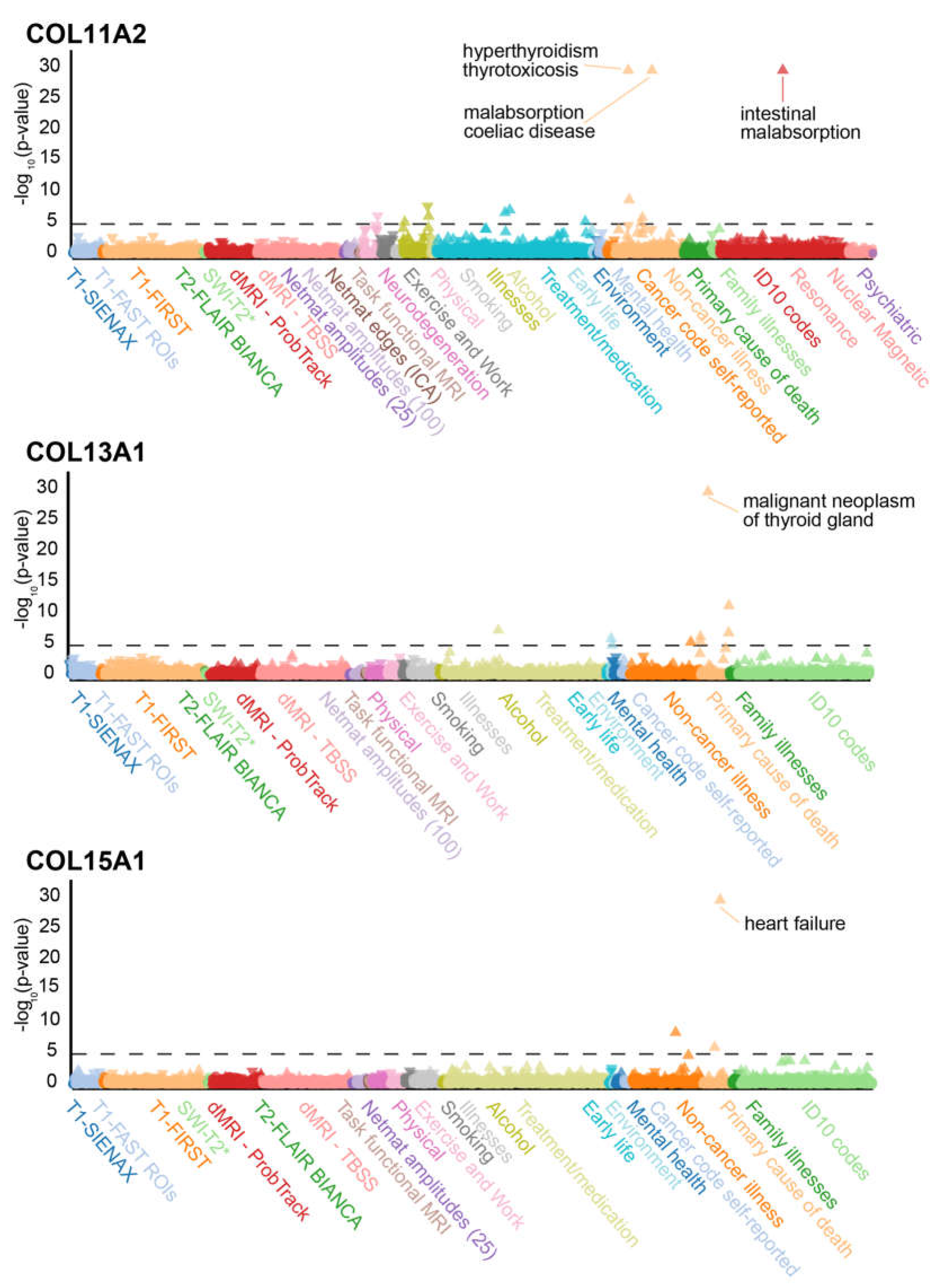
20]. We identified 61 matrisome genes in these 1000 top hits (Supplementary Table 7), including three collagens (
Figure 3). Interestingly, EYS (eyes shut homolog) displayed marked correla-tion with primary cause of death from peripheral vascular disease. As previously seen in a similar linkage with the GWAS analysis of type I diabetic neuropathy, use of insulin 1 year after diagnosis or use of an insulin product is strongly linked to TNFXB variants. The strongly linked disorders with the three collagens COL11A2, COL13A1, and COL15A1 also exhibit strong correlation with other genes: hyperthyroidism thyrotoxicosis with MUC22, malignant neoplasm of thyroid gland with SLIT2, malabsorption and coeliac dis-ease with SFTA2, and heart failure with GPC6 (Supplementary Table 7).
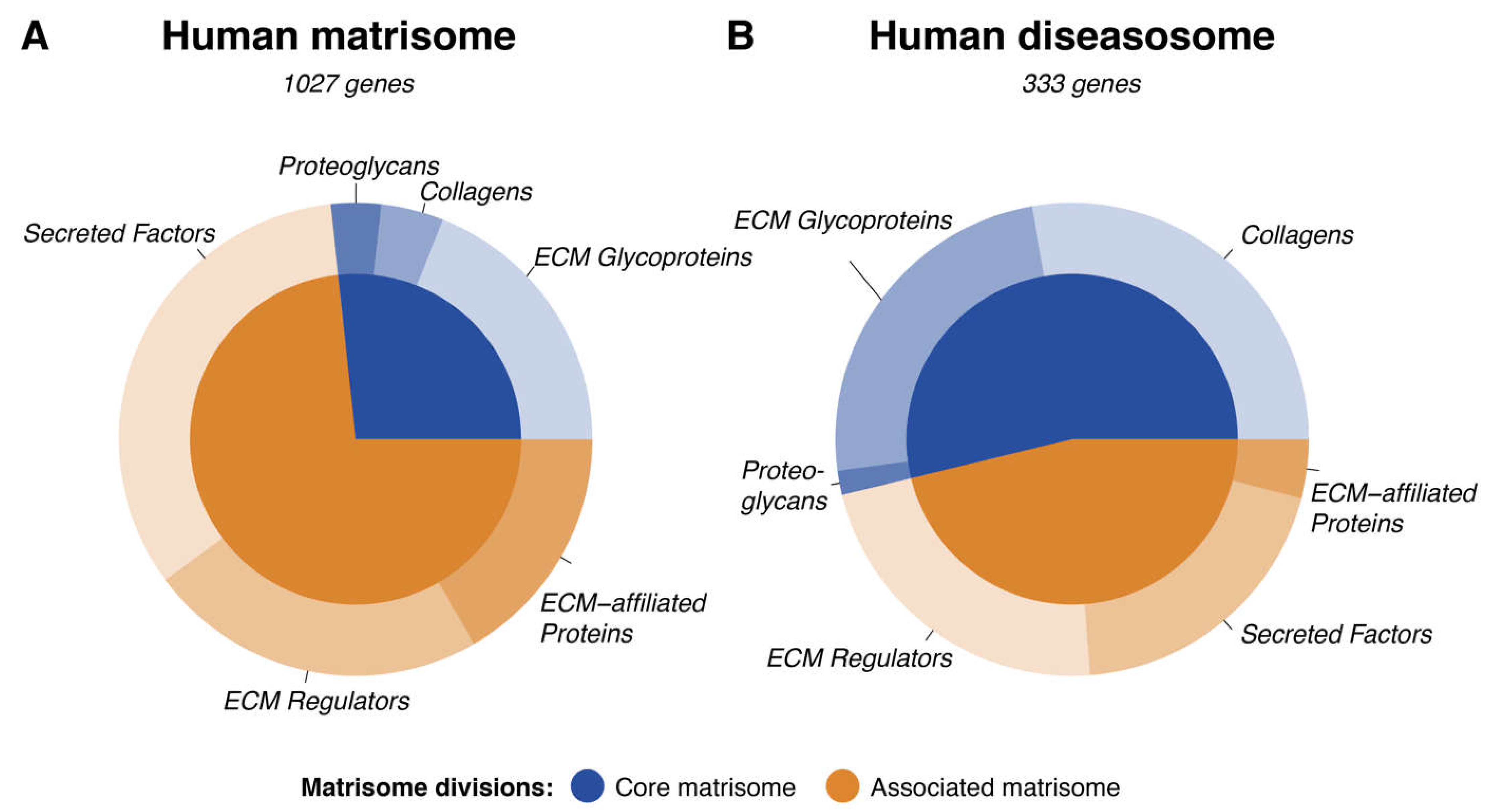
Taken together, we surveyed multiple databases and public resources (clinical variance, human diseases, and Monarch diseases) for the implication of matrisome genes with human diseases. Overall, we found 1142 human diseases associated with the matrisome (Supplementary Table 8). The human disease-matrisome consists of 333 out of the total 1027 matrisome genes (32.4%) (
Figure 4), implicating the importance of matrisome in human pathologies. The components of the core-matrisome (collagens, glycoproteins, and proteoglycans) take precedence when linked to the disease specifically, implicating the role of structural proteins in the human diseasome.
3.3. Matrisome in Age-Related Diseases
Having established the contribution of variants in matrisome genes to human diseases, we next wondered which kind of diseases are mostly affected by the ECM. To do this, we manually curated Disease-Gene-Associations from multiple data sources (see M&M; Supplementary Table 9). We curated 28920 Disease-Gene-Associations of a total of 10570 genes (Supplementary Table 9). Out of the 1027 human matrisome genes, 656 matrisome genes corresponded to 2825 matrisome gene-disease implications, which is about 10% of all our curated Disease-Gene-Associations (Supplementary Table 9). Reading through these 2825 matrisome Disease-Gene-Associations, we noticed three main disease categories.
Category (A) are expected and typical ECM diseases, such as Ehler-Danlos syndrome, Osteogenesis imperfecta, Marfan syndrome, Alport syndrome, Fraser syndrome, Von Willebrand disease, etc.
In category (B), we grouped common diseases that are more or less unlikely to be affected by ECM, such as diabetes type 1, asthma, autism, lissencephaly, schizophrenia, seizures, muscular dystrophy, obesity, stroke, etc.
Since we notice several chronic and age-related pathologies in the 2825 matrisome-gene-disease-associations, we grouped age-related diseases in category (C), such as arthritis, Alzheimer’s disease, cancer, diabetes type 2, chronic obstructive pulmonary disease, fibrosis, Parkinson’s disease, cirrhosis, osteoporosis, hypertension, etc.
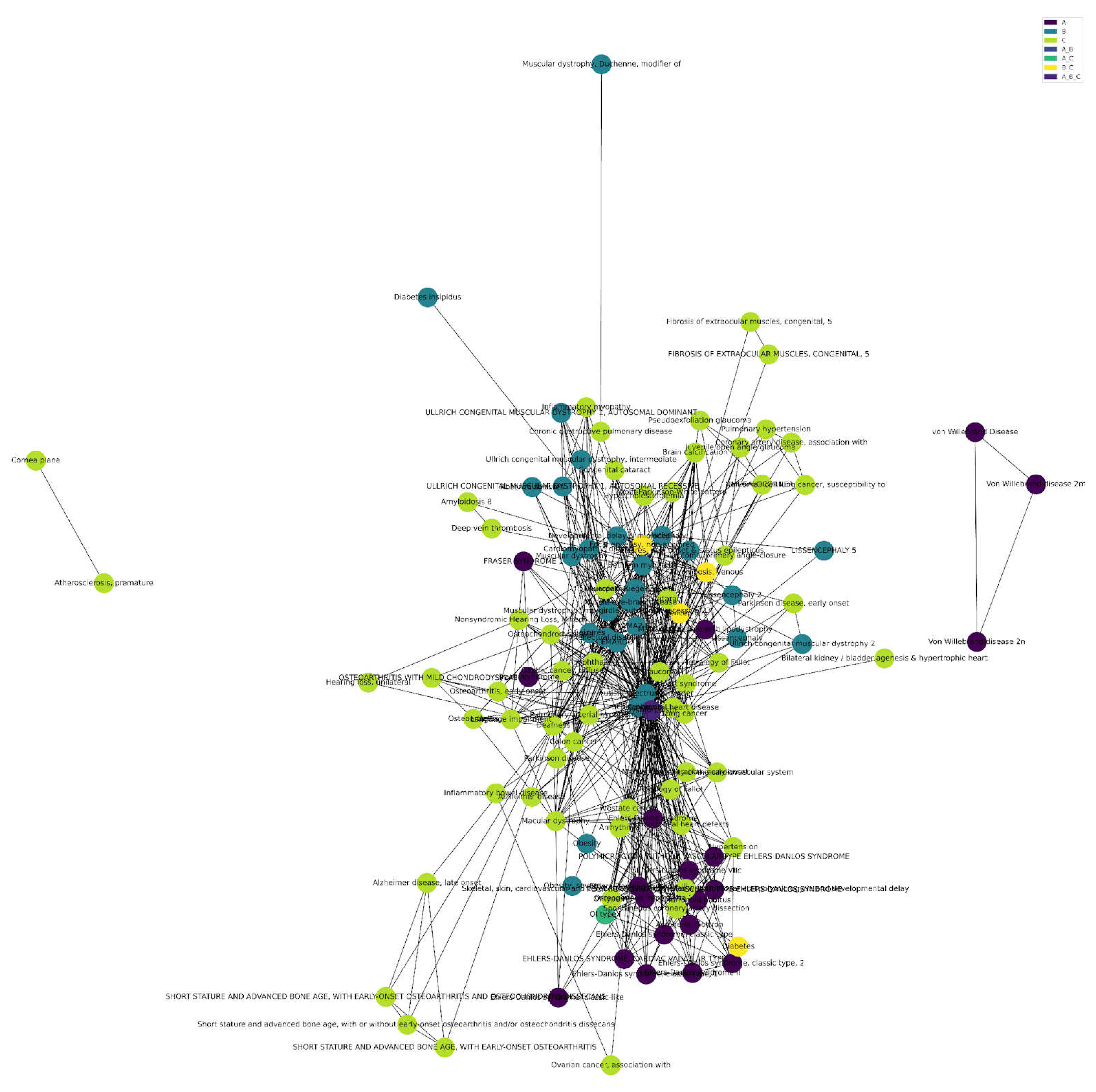
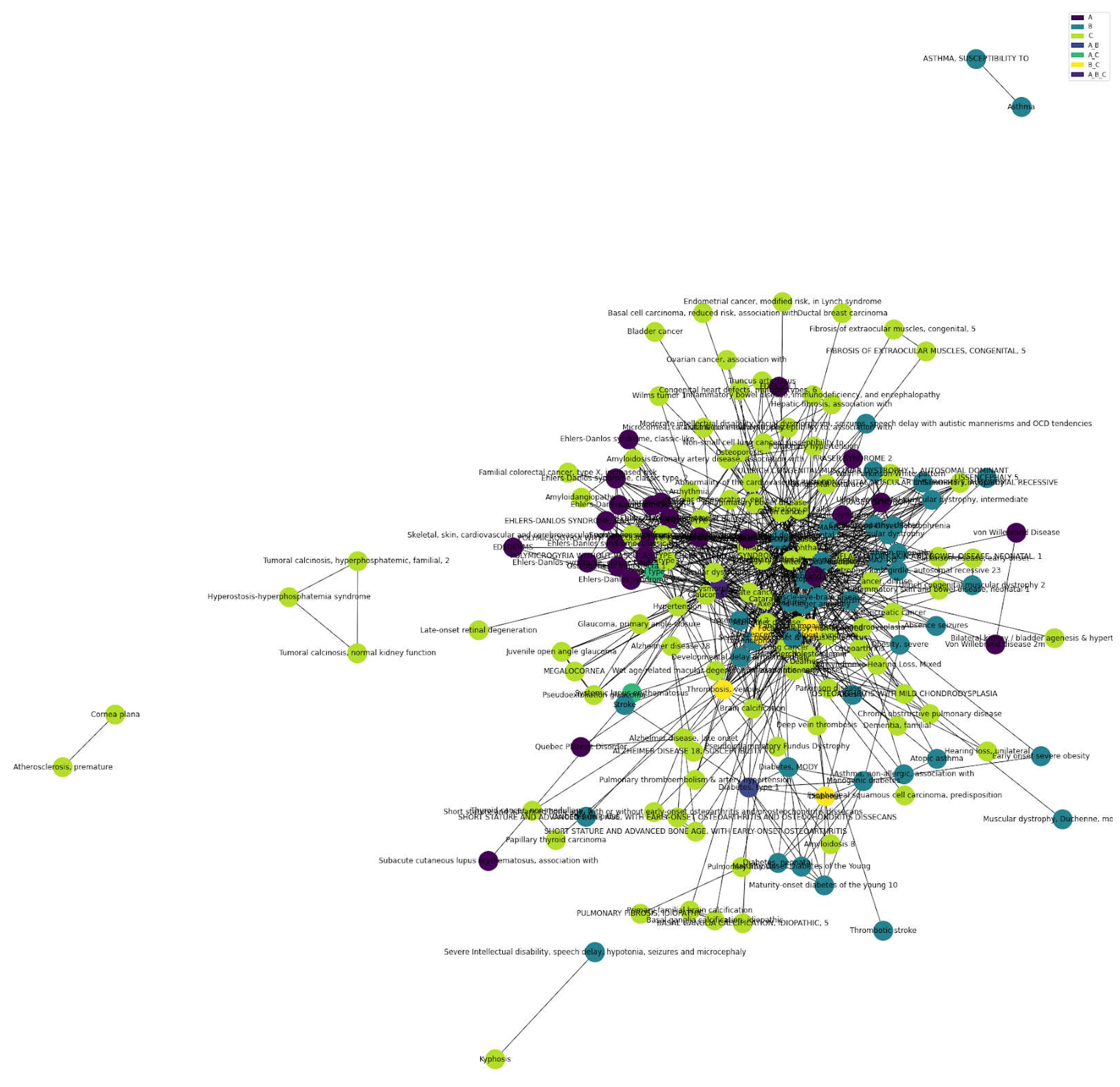
Surprisingly, age-related diseases form the predominant category for all matrisome and also just the core-matrisome (
Figure 5, Supplementary Figure 1, Supplementary Table 9). Out of the 2825 matrisome Disease-Gene-Associations, we found 1250 age-related dis-eases (category C), including overlapping disease categories (A_C, B_C, and A_B_C;
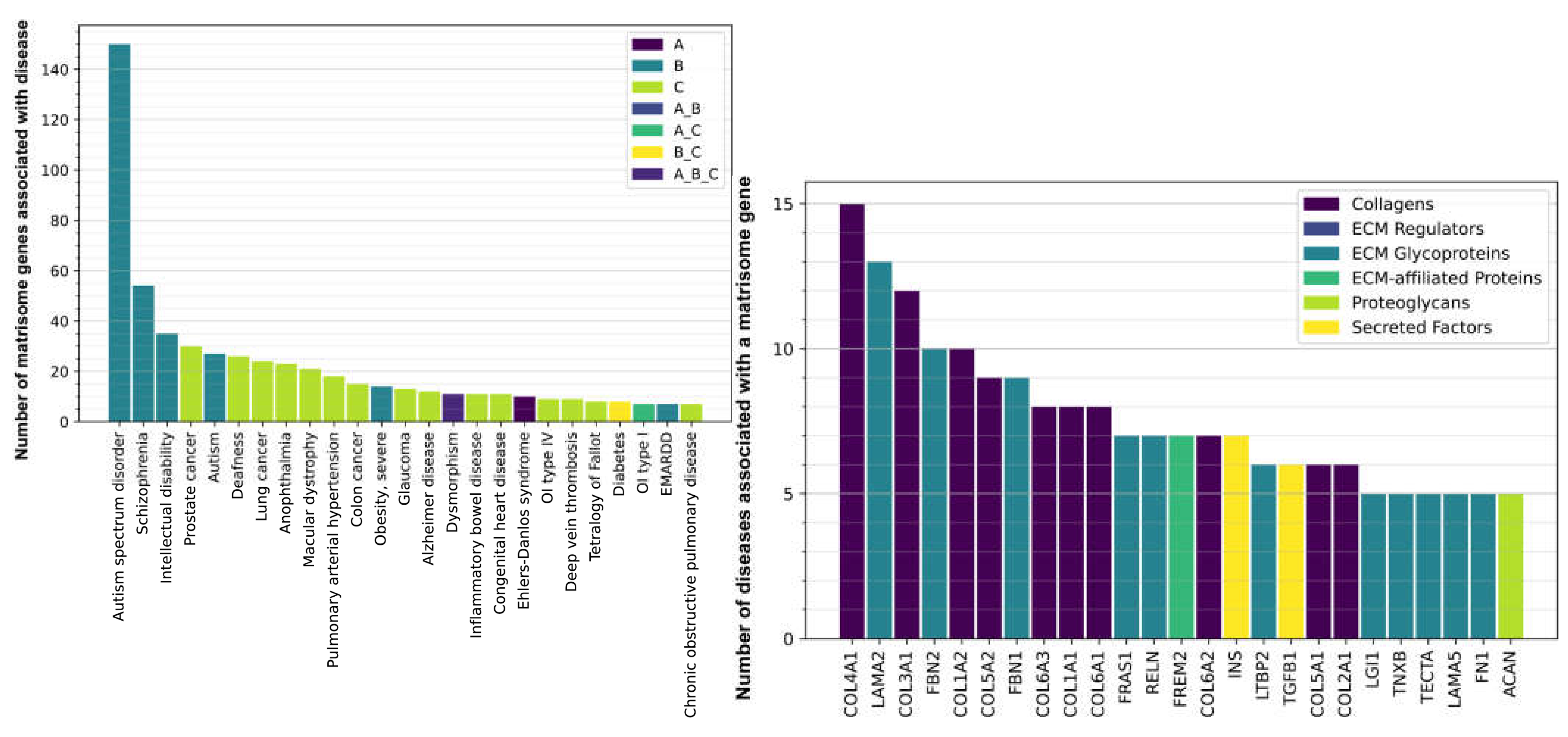
Figure 5, Supplementary Table 9). For agerelated diseases, about 15-30 matrisome genes are as-sociated each with different cancers, such as prostate cancer, lung cancer, colon cancer, and glaucoma (
Figure 6A, Supplementary Table 9). A number of matrisome genes (5-20 genes) each are associated with age-related diseases, such as macular dystrophy, cardiovascular diseases, metabolic diseases (Diabetes Type II and severe obesity), Alzheimer’s disease, and fibrotic lung diseases (
Figure 6A, Supplementary Table 9). Most surprisingly, about 150 matrisome genes are associated with autism, about 50 with schizophrenia, and 35 with intellectual disabilities (
Figure 6A, Supplementary Table 9). The sheer predominance of frequency of matrisome genes involved in type B (unexpected) and type C (age-related) diseases strongly implicates ECM proteins in disease association, suggesting that each individual gene may be associated with a plethora of different disease states.
Next, we asked which matrisome gene is associated with several different diseases. Basement membrane-forming collagen type IV (COL4A1) and laminin (LAMA2), which form ECM around organs [
21], are each associated with 15 and 13 different diseases, respectively (
Figure 6B, Supplementary Table 9). Next are fibril-forming collagens collagen type III (COL3A1), collagen type I (COL1A1, COL1A2), collagen type V (COL5A1, COL5A2), and collagen type II (COL2A1), which form the connective tissue to support muscles, joints, skin, and other organs [
21] (
Figure 6B, Supplementary Table 9). Fibrillin (FBN1, FBN2), which are components of elastic fibers in the cardiovascular system, are implicated in more than 8 diseases (
Figure 6B, Supplementary Table 9), but fibrillins also attach to the large latent protein complex (LTBP2) and TGF-1β (TGF1B) (
Figure 6B, Supplementary Table 9), which is important for collagen production and ECM remodeling [
22]. Thus, we find a strong association of collagen-forming ECM with several diseases.
3.4. Collagens and Diseases
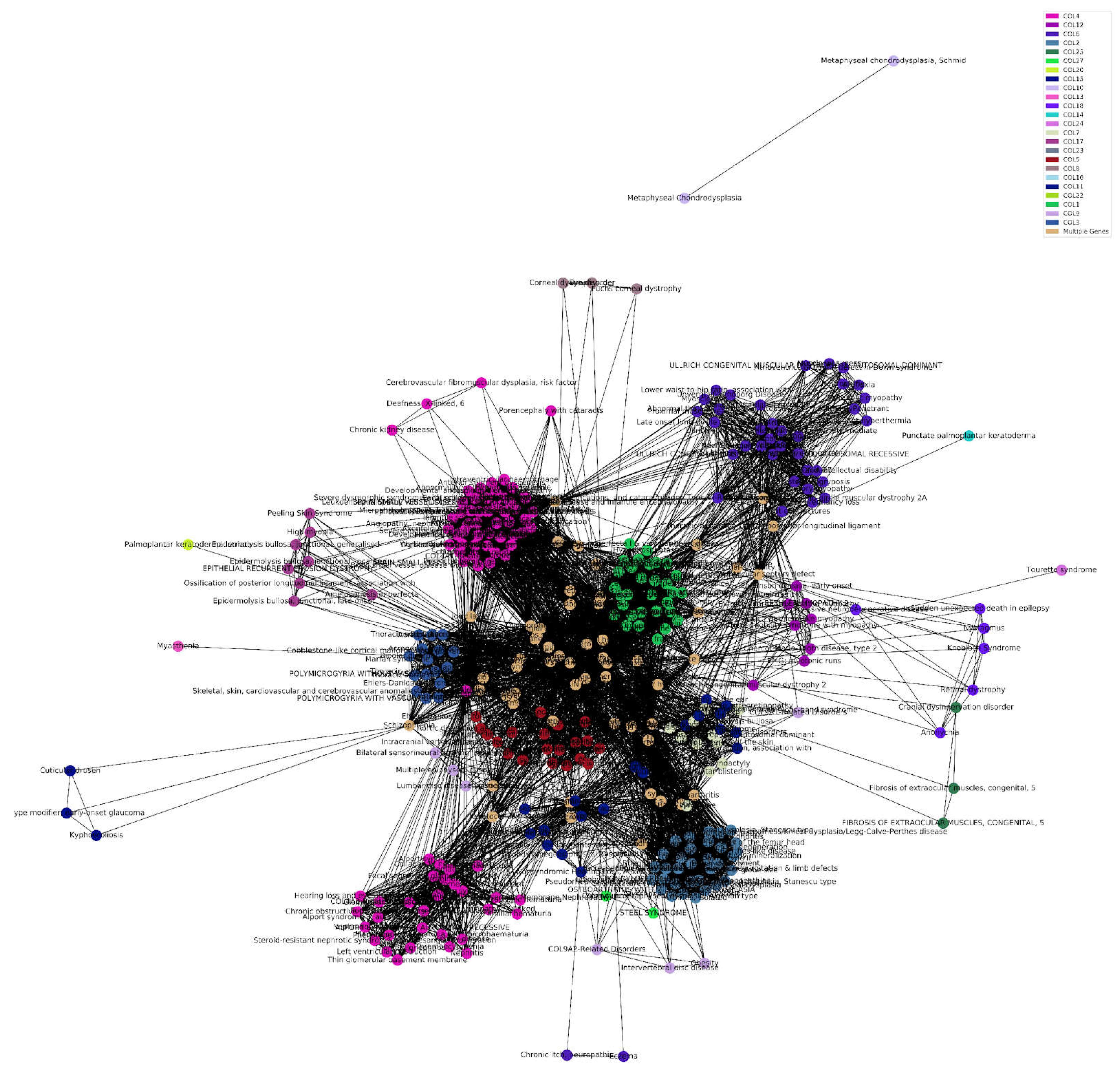
To gain a better understanding of the relationship between collagens and diseases, we performed a cluster analysis. We found an expected correlation of COL4 with Alport syndrome, and an unexpected correlation with hearing loss and chronic kidney disease. Lower waist-to-hip ratio is associated with COL6, as is expected due to the associations with adipose tissue fibrosis or metabolic dysregulation [
23], and predictor of all-cause mortality in type 2 diabetes and microalbuminuria [
24]. Surprisingly, COL12 was linked with early onset PD, and COL18 with progressive neurodegenerative disease. Osteoarthritis and Schizophrenia additionally link to variants in multiple types of collagen (
Figure 7, Supplementary Table 9).
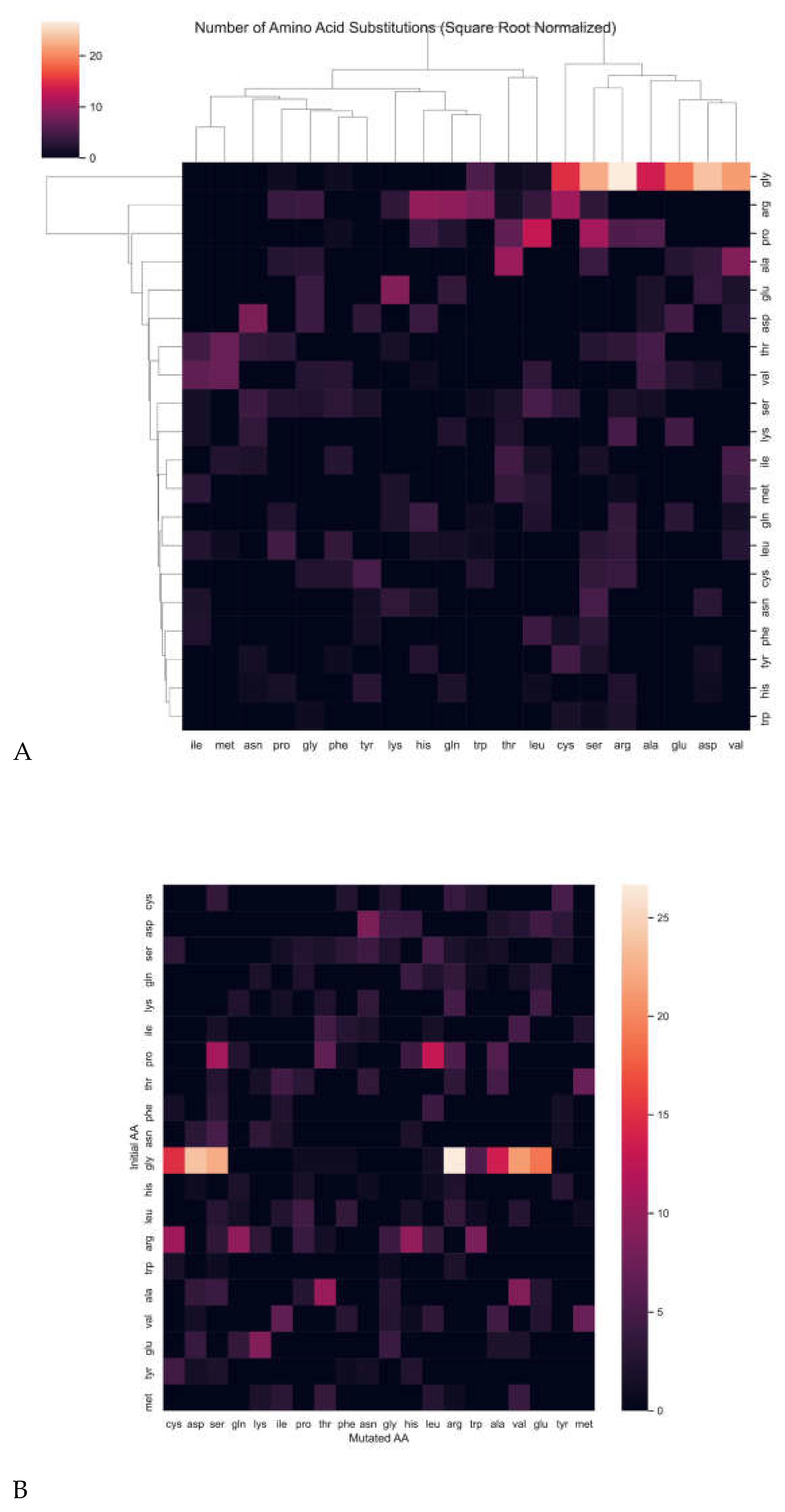
Next, we asked which amino acid in collagens is most frequently mutated and associated with diseases. Glycine was by far the most disease-associated mutation, followed by proline and arginine (
Figure 8A, Supplementary Table 9). For steric reasons, glycine is the smallest amino acid and is required to be at the third position of the (Gly-X-Y) repeats, where X is often proline and Y is often hydroxyproline [
21]. Proline in the endoplasmic reticulum is posttranslationally modified to hydroxyproline, important for stabilizing the collagen triple helix [
21]. Interestingly, the most frequent substitution of an initial glycine is by arginine, followed by aspartic acid, serine, and valine (
Figure 8B, Supplementary Table 9).
3.5. Potential Strategiues Using Matrisome for Drug Repurposing
Given the large contribution of the matrisome to diseases, we wondered whether the matrisome would provide targets for drug repurposing strategies. The key to drug repositioning is to cross-reference GWAS-PheWAS associated with diseases with DrugBank repositories [
25]. With this approach, about 15 thousand drug-disease relationships and over 38 thousand novel drug-repurposing candidates were
in-silico identified [
25]. From this, we found five matrisome genes (
COL1A2, NOV, LPA, MMP24, PLG) with 13 distinct drug candidates for repurposing (Supplementary Table 10). Strikingly, 9 of these drugs could be used to target plasminogen (
PLG) rs783147 SNV for at least 16 disease indications (Supplementary Table 10). Thus, there is an untapped potential for repurposing drugs targeting the matrisome.
3.6. Targeting Matrisome Proteins in Rare Diseases
Unlike cancer, cardiovascular diseases, diabetes, and other highly prevalent diseases, rare and orphan diseases still lack investment in medical research, drug development, and specialist knowledge. There are around 7’000 diseases classified as “Rare and Orphan diseases”, and their number is increasing by almost 300 new diseases every year [
26] (
www.rarediseases.org). A disease qualifies as rare if less than 1 in 2000 people are affected [
27]. Approximately 80% of rare and orphan diseases are caused by genetic mutations [
28], and we sought to determine how many of those are in matrisome genes. We found 311 matrisome genes linked to 460 unique rare diseases (Supplementary Table 11).
4. Discussion
While the use of GWAS and PheWAS is an integral tool for genetic epidemiological research and elucidating potential pathways that may contribute to disease pathology, the potential for further applications can always be pursued. In the context of matrisomal genealogy, we aim to unearth the implications of ECM matrisome genotypes and SNV variants for human diseases, ultimately producing the quantified ECM diseasome. GWAS and PheWAS capabilities have also enabled the linking of diseases as co-morbidities [
29]. GWAS and PheWAS techniques also uncover a large network of potential drug targets that may be repurposed to target specific diseases that display strong redundant associations with certain polymorphisms, diversifying the array of pharmacological therapeutic targets available for exploration [
30].
Out of the 1027 total matrisome genes that were analyzed, 140 SNVs exist within this set. The most significant association was the diabetic type I neuropathy with the TNXB gene, responsible for the production of the glycoprotein tenascin XB. TNXB has previously been shown to have been moderately implicated within type I diabetes, but further analysis of this association is warranted regarding the HLA region and non-HLA class II genes [
31]. Elevated serum tenascin-C (TNC) levels have been an indicator of increased risk of cardiovascular events and death with type II diabetes, but the role of TNX in these processes must be further elucidated [
32]. TNXB has been linked to Ehlers-Danlos syndrome due to the lack of organizational structure of the collagen framework within the ECM, and the association with diabetic type I neuropathy may be apparent due to peripheral axonal stretching and pressure, increasing susceptibility to neuropathic pathology [
33].
The ECM interactome is a target of growing interest in the context of numerous diseases. ECM signaling interactions regarding movement, adhesion, and growth are implicated in breast cancer along with collagens and fibrinogen [
34]. Cell-matrix and matrisome protein-protein interactions are evident when associated with numerous diseases, as many of the diseases analyzed (270 out of 3824) were linked to 161 matrisome genes. The prevalence of the ECM interactome within disease networks must further be explored, as ECM cell, matrix, and signaling interactions have been previously described as drivers of mammalian disease [
35,
36].
The linkage of various matrisome’s SNVs to Alport syndrome [
37], Marfan syndrome [
38], and osteogenesis imperfecta [
39] is consistent with previous association studies, as these diseases are indicative of collagen irregularities or connective tissue dysfunction. Interestingly, the majority of collagen gene types (30 out of 44) are associated with disease, particularly COL2A1 and COL7A1. COL2A1 is linked to a large variation in phenotypic displays, ranging from growth and short stature disorders to retinal dysfunction. TNXB was additionally linked through PheWAS with use of insulin after 1 year of diabetic diagnosis and general use of insulin, further implicating TNXB in the pathology of diabetes and diabetic neuropathy, as previously seen. The importance of the core matrisome in disease pathology is apparent, as out of the 333 genes in the disease-matrisome, a majority are of structural nature. These structural components may be crucial in the molecular pathology of these associated diseases.
To further explore these links, the division of these genes into the aforementioned categories (A, B, C, and their combinations) provided insight into how many diseases link to matrisome genes. Although age-related diseases formed the majority of the associations with matrisome genes, the most surprising results were the overwhelming number of psychiatric and neurological disorders, autism/autism spectrum disorder, schizophrenia, and intellectual disability associations. The role of the ECM in neurodevelopmental disorders has not been thoroughly stipulated, requires the use of iPSC-based methods, and prior research is greatly lacking. Certain manipulations of ECM components
in vitro have resulted in altered mechanical properties of the neocortex, but certain factors and the results of their modulation in the context of disease are still not apparent [
40].
Interestingly, certain non-neurological components of ECM physiological phenotypes are apparent in individuals with ASD, where children with ASD exhibit altered platelet functionality rather than platelet morphology compared to undiagnosed individuals due to increased collagen-ADP and collagen-epinephrine closure time, hinting to the involvement of matrisomal alterations in the disease [
41]. Ehlers-Danlos syndrome, one of the expected matrisome-related diseases and hits reconfirmed in our analysis of gene-disease networks, also shares vast phenotypic overlap with ASD and generalized hypermobility spectrum disorders (gHSDs), existing as comorbidities and co-occurrences within families of diagnosed individuals, along with other comorbidities such as intellectual learning disorders or ADHD [
42]. ASD and comorbid ADHD share a significant relation with individuals who also have generalized joint hypermobility (GJH), and the diagnosis of GJH may even serve as a biomarker for future diagnoses of ASD and comorbid ADHD [
43]. The additional roles of core matrisome genetic perturbations in proteoglycans may be another indicator of other disorders due to the high involvement of glycans in neurodevelopmental processes, warranting further exploration of glycosylation of the ECM in neurodevelopmental disease [
44].
Schizophrenia exhibited the second highest number of gene associations after ASD. The ECM of individuals afflicted with schizophrenia exhibit altered GABAergic signaling, chondroitin sulfate proteoglycans, MMPs, and ECM maintenance, resulting in neuronal abnormalities [
45,
46]. Matrisomal gene expression is also disrupted in the ECM cortical areas of individuals afflicted with schizophrenia [
47]. Neuronal migration and glial abnormalities due to dysfunctional reelin and chondroitin sulfate proteoglycans may also be additional contributors to schizophrenia pathology [
48]. Given the role of the ECM in schizophrenic pathology, numerous matrisome-related targets may be explored for disease treatment [
49].
These ECM dysfunctions are not specific only to neurodevelopmental disorders; ECM components are highly dysregulated even in age-related neurodegenerative disorders such as Alzehimer’s disease and Parkinson’s disease. AD’s primary pathological manifestation, the aggregates of amyloid beta (aB) and amyloid precursor protein (APP), are influenced by dysregulated chondroitin sulfates and heparan sulfates, which are associated with a higher protein aggregate burden along with the increased amount of matrisome components tenascin, integrin, laminin, and galectin [
50]. In PD, increases in the expression of collagen type I is evident along with other pro-inflammatory changes in the surrounding matrix [
51]. These changes may be attributed to the SNVs in matrisome genes associated with AD and PD, increasing the susceptibility of individuals to the development of neurodegenerative disorders with increasing age. Exploring the matrisome gene and neurodegenerative disease relationships within the context of dysregulated ECM mechanics may provide further targets for pharmacological therapeutic development.
Our cluster analysis of diseases with collagen variants aptly explicates the involvement of core-matrisome collagen genes in numerous diseases. Along with the previously expected associations with COL4 and COL6, a few examples which may help elucidate additional disease pathology are apparent, such as the associations of COL12 and COL18 with neurodegenerative disorders. COL6 is also important for Schwann cell differentiation in the peripheral nervous system, but the role of COL6 in the central nervous system must be further explored [
52]. COL12, associated with early onset PD, has been implicated in myopathy, but its role in age-related neurodegenerative disorders has not been elucidated [
53]. The age-related changes of the ECM, such as collagen degradation, elastase upregulation, and fibronectin upregulation, may prime the cellular environment for increased risk of disease development and accelerated aging pathology, and the crossover of certain diseases attributes from EDS and Marfan syndrome with ECM aging phenotype may provide insight into how aged ECM and certain disease states may communicate [
54]. Certain genetic associations of collagens may indicate an increased incidence of a healthier aging process, where COL1A1 rs107946 may suggest accelerated osteoporotic-related aging, but COL1A2 rs3917 suggests a reduced risk of osteoporosis [
55]. The interwoven diseases in
Figure 7 and other related associations may be crucial tethers in exploring and expounding ECM-related changes in healthy aging, particularly how certain genetic predispositions may alter the quality of life course.
When analyzing the most frequently substituted amino acid in the collagen chain, we found that glycine was most often substituted by arginine. The overwhelming display of this specific substitution cohesively strengthens the role of understanding genetic variants within the matrisome, particularly in collagen-related disorders. The substitution of glycine 661 with arginine in the COL3A1 helix results in increased intracellular retention of the protein with abnormal thermal stability, resulting in EDS [
56]. In Alport syndrome, the glycine 852 and 325 to arginine substitution is evident in the COL4A5 gene, responsible for basement membrane formation [
57,
58]. The glycine-to-arginine substitution is also evident in dominant dystrophic epidermolysis bullosa, with the variant in COL7A1 [
59]. This specific substitution has also been implicated in OI [
60], particularly in types COL1A1 and COL1A2 [
61]. As these results are consistent with the involvement of specific amino acid substitutions in typical ECM-expected disease, further analysis of the unexpected and age-related genetic variants is warranted to undercover how specific amino acid substitutions may alter aging phenotypes.
Extending the application of GWAS and PheWAS techniques to matrisome genes allows for the identification and repurposing of drugs to target specific polymorphisms, which may then ameliorate disease pathology. One of our significant target hits, the plasminogen (PLG) SNV rs783147, can be targeted for 16 disease indications. Plasminogen is a pro-fibrinolytic factor that is crucial for the removal of blood clots through its cleavage to the active enzyme plasmin [
62]. Nine drugs, alteplase, aminocaproic acid, anistreplase, aprotinin, reteplase, streptokinase, tenecteplase, tranexamic acid, and urokinase, can be used to target certain complications, which may be particularly age-related, while many of these drugs are indicated for blood clotting and myocardial infarction or other cardiovascular complications, which are some of the leading causes of death [
63]. Aging is a substantial risk factor for thrombosis and other thrombotic-related events, and with the addition of heightened interleukin 6 (IL-6) and C-reactive protein (CRP) levels, the risk of cardiovascular events is exacerbated in elderly individuals [
64]. Plasminogen activator inhibitor 1 (PAI-1), a fibrinolysis inhibitor, is implicated in many age-related metabolic disorders and cancer [
65], and PAI-1 levels significantly increase with age, predisposing elderly individuals to cardiovascular complications resulting from thrombosis and atherosclerosis [
66]. PLG and the opposing inhibition of fibrinolytic activity may be opportune targets for ameliorating cardiovascular aging through the repurposing of our identified drugs [
67]. Analyses of drug repurposing are relevant for influencing the trajectory of high throughput drug screens for ECM-related pathology [
68], and even implicate other increasingly age-related diseases to matrisome defects, such as cardiovascular disease [
69].
In addition to the previously discussed age-related implications of the matrisome and disease networks, a majority of the genes in the matrisome-diseasome are linked to 460 rare diseases. Our findings strongly link polymorphisms located in the matrisome with many connective tissue disorders, age-related diseases, rare diseases, and neurodevelopmental disorders. The emergence of “omics” technologies in combination with association studies may serve as an essential component of further implicating the ECM in disease. As seen with squamous cell carcinoma (SqCC), integration of ECM features in aging and diseased tissue with multi-omics data of SqCC can aptly determine the risk of cancer development [
70]. The applications for precision medicine are endless, as organoid models created with patient-derived cell lines with associated SNVs and other ECM manipulations, which may be representative of certain phenotypes, can be further explored in disease pathology, particularly in other age-related diseases, inflammatory conditions [
71], osteoarthritis [
72], and processes such as tumor-stromal interactions [
73] and varying types of cancers [
74]. The matrisome is earning an increasingly prominent role in the pathology of disease (
Figure 9). The emergence of high associations of collageneous dysfunc-tion and related SNVs in disease is ever-increasing, and the implications of genetic pre-dispositions to age-related phenotypes from variations in matrisomal composition may serve a pivotal role in advancing possible therapeutic discoveries and treatments for the process of aging.
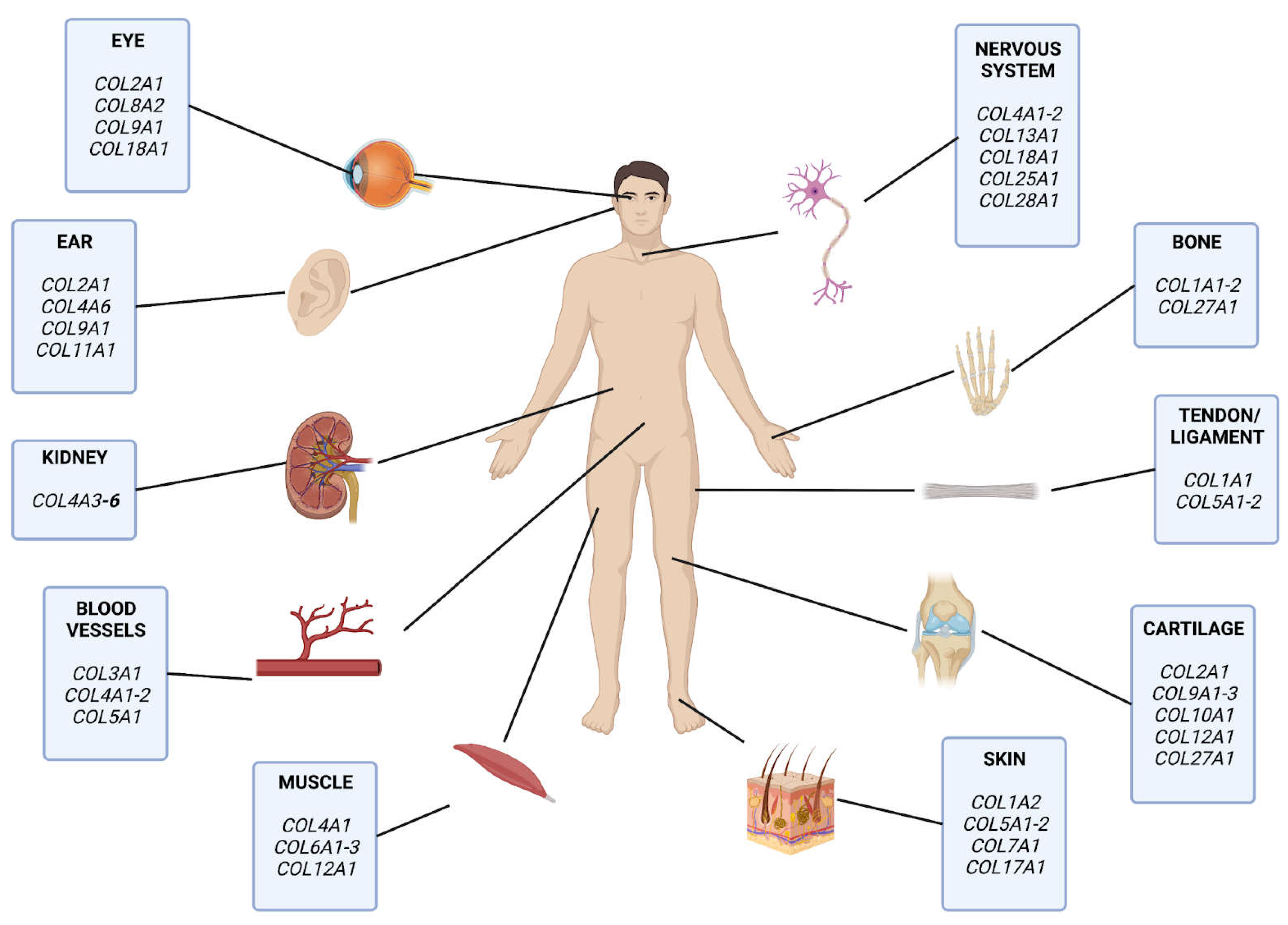
Figure 1.
Collagens in Human Tissues. Different types of collagen proteins experience localized expression in varying different tissue types. For example, COL2A1 is expressed in tissues with substantial components of cartilaginous fibers. .
Figure 1.
Collagens in Human Tissues. Different types of collagen proteins experience localized expression in varying different tissue types. For example, COL2A1 is expressed in tissues with substantial components of cartilaginous fibers. .
Figure 2.
PheWAS of GWAS Catalog of SNVs. The human matrisome genes are displayed on the y-axis, and the log10 P-value distribution of the associated phenotypes is shown as boxplots on the x-axis. Outliers are displayed as points, and only significantly associated phenotypes (P-value < 0.05) are shown. The ten most-significantly associated phenotypes are labeled. For more details, please see Supplementary Table 1. The phenotypes are colored according to the matrisome division the respective gene is associated with (core matrisome genes in blue, matrisome-associated genes in green).
Figure 2.
PheWAS of GWAS Catalog of SNVs. The human matrisome genes are displayed on the y-axis, and the log10 P-value distribution of the associated phenotypes is shown as boxplots on the x-axis. Outliers are displayed as points, and only significantly associated phenotypes (P-value < 0.05) are shown. The ten most-significantly associated phenotypes are labeled. For more details, please see Supplementary Table 1. The phenotypes are colored according to the matrisome division the respective gene is associated with (core matrisome genes in blue, matrisome-associated genes in green).
Figure 3.
PheWAS analysis of COL11A1, COL13A1, and COL15A1. PheWAS analysis of COL11A1, COL13A1, and COL15A1 with individual phenotypes on the x-axis and the log10 P-value on the y-axis. The most significantly enriched phenotypes (P-value < 10-20) are labeled for each collagen. The dashed line represents the significance cut-off at a P-value of 10-5. Phenotypes are colored according to their phenotype category (Supplementary Table 7).
Figure 3.
PheWAS analysis of COL11A1, COL13A1, and COL15A1. PheWAS analysis of COL11A1, COL13A1, and COL15A1 with individual phenotypes on the x-axis and the log10 P-value on the y-axis. The most significantly enriched phenotypes (P-value < 10-20) are labeled for each collagen. The dashed line represents the significance cut-off at a P-value of 10-5. Phenotypes are colored according to their phenotype category (Supplementary Table 7).
Figure 4.
Involvement of the Human Matrisome in Pathology. The composition of the human matrisome is illustrated using a circular diagram reflecting the relative abundances of the core (blue) and associated (brown) matrisome divisions in the overall matrisome (A) and in the part of the matrisome which is specific to disease (B, Supplementary Table 8). The divisions are further subdivided by the matrisome categories they contain. The subset of the human matrisome (1027 genes) responsible for the disease (333 genes) is approximately 30% and is enriched for collagens. This overrepresentation of collagens further highlights the importance of collagenopathies.
Figure 4.
Involvement of the Human Matrisome in Pathology. The composition of the human matrisome is illustrated using a circular diagram reflecting the relative abundances of the core (blue) and associated (brown) matrisome divisions in the overall matrisome (A) and in the part of the matrisome which is specific to disease (B, Supplementary Table 8). The divisions are further subdivided by the matrisome categories they contain. The subset of the human matrisome (1027 genes) responsible for the disease (333 genes) is approximately 30% and is enriched for collagens. This overrepresentation of collagens further highlights the importance of collagenopathies.
Figure 5.
Total Matrisome Disease-Gene-Associations. Cluster analysis of matrisome disease-gene-associations, composed primarily of age-related diseases and overlapping categories (Supplementary Table 9). .
Figure 5.
Total Matrisome Disease-Gene-Associations. Cluster analysis of matrisome disease-gene-associations, composed primarily of age-related diseases and overlapping categories (Supplementary Table 9). .
Figure 6.
Matrisome Gene and Disease Associations. The numerous diseases on the x-axis that display association with a certain number of matrisome genes on the y-axis (A). For example, autism spectrum disorder, schizophrenia, and intellectual disability display the largest number of associated matrisome genes and belong to the group B of diseases that would not particularly be expected to correlate strongly with matrisome related SNVs. The various matrisome genes on the x-axis (B) are associated with more than one displayed disease state (Supplementary Table 9). .
Figure 6.
Matrisome Gene and Disease Associations. The numerous diseases on the x-axis that display association with a certain number of matrisome genes on the y-axis (A). For example, autism spectrum disorder, schizophrenia, and intellectual disability display the largest number of associated matrisome genes and belong to the group B of diseases that would not particularly be expected to correlate strongly with matrisome related SNVs. The various matrisome genes on the x-axis (B) are associated with more than one displayed disease state (Supplementary Table 9). .
Figure 7.
Disease-Collagen-Gene Associations. Numerous disease states are associated with varying mutations in different collagen genes. Groups of diseases within the collagen diseasome form a network in this cluster analysis due to relatedness to a mutation in specific collagens. Multiple diseases, as seen in beige, share numerous mutations in different types of collagen genes, surrounding the beige centroid (Supplementary Table 9). .
Figure 7.
Disease-Collagen-Gene Associations. Numerous disease states are associated with varying mutations in different collagen genes. Groups of diseases within the collagen diseasome form a network in this cluster analysis due to relatedness to a mutation in specific collagens. Multiple diseases, as seen in beige, share numerous mutations in different types of collagen genes, surrounding the beige centroid (Supplementary Table 9). .
Figure 8.
Predominant Mutations in the Collagen Helix. Heatmaps displaying the (A) frequency of amino acid substitution, where glycine is the most substituted amino acid, and (B) most frequent mutation substitution of glycine for arginine. Highest values indicate predominant amino acid mutation (Supplementary Table 9). .
Figure 8.
Predominant Mutations in the Collagen Helix. Heatmaps displaying the (A) frequency of amino acid substitution, where glycine is the most substituted amino acid, and (B) most frequent mutation substitution of glycine for arginine. Highest values indicate predominant amino acid mutation (Supplementary Table 9). .
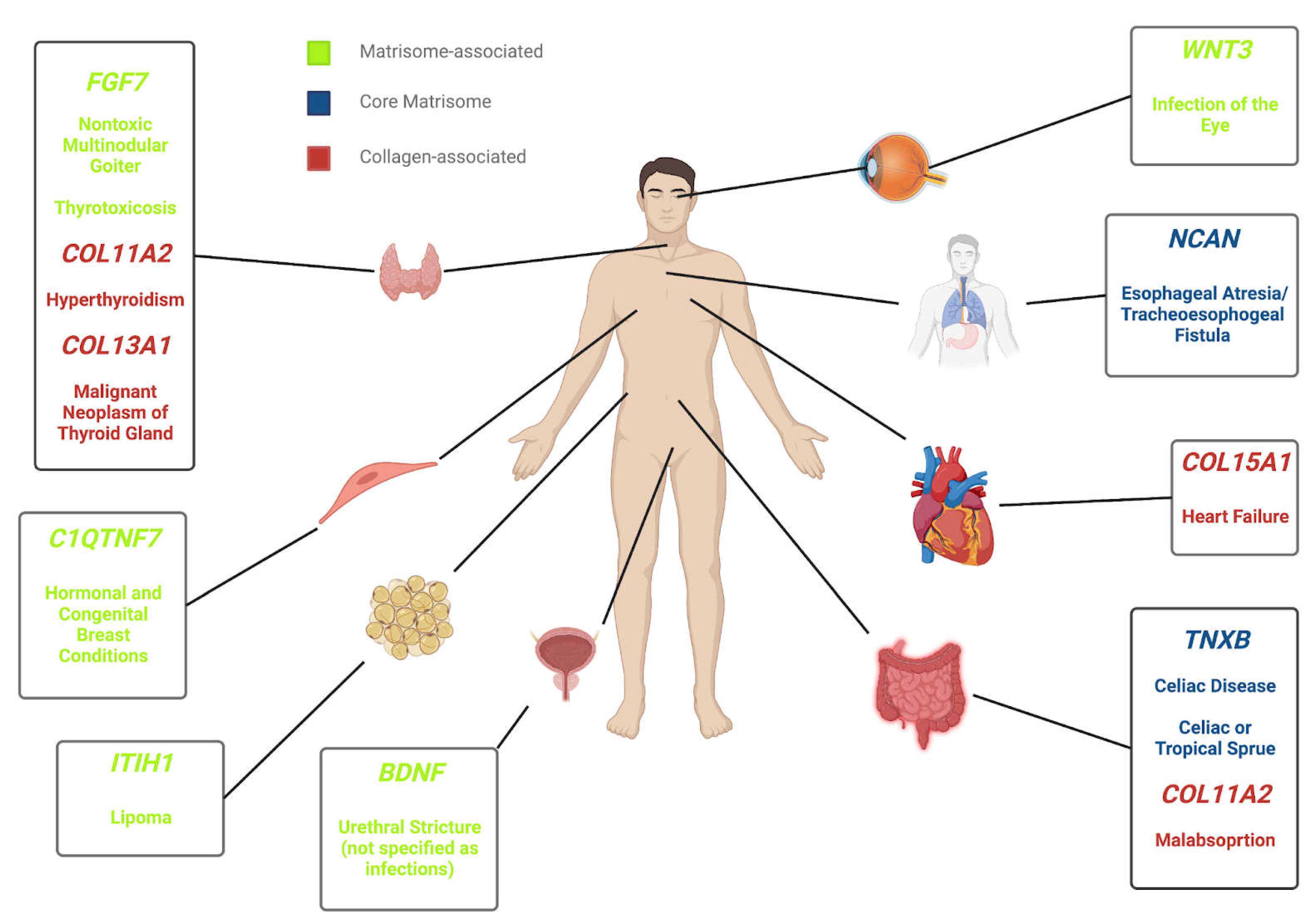
Figure 9.
The ECM-Diseasome Body Map. An example of a proposed body map of matrisome genes that are associated with numerous disease states. Certain matrisome genes localize to various tissues in the body and may be linked to more than one disease state, implicating therapeutic repurposing. This body map can be expanded and various other body maps can be created through other gene-disease associations which may be explored in the future.
Figure 9.
The ECM-Diseasome Body Map. An example of a proposed body map of matrisome genes that are associated with numerous disease states. Certain matrisome genes localize to various tissues in the body and may be linked to more than one disease state, implicating therapeutic repurposing. This body map can be expanded and various other body maps can be created through other gene-disease associations which may be explored in the future.