Submitted:
24 March 2023
Posted:
27 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Culture–Dependent Isolation and Identification of AAB
2.2. Optimal Fermentation Conditions for AAB
2.3. Optimal Inoculation Rate and Inoculation Order for Multiple Starters
2.4. Titratable Acidity and Growth
2.5. Microbiome Taxonomic Profiling Analysis
2.6. Volatile Compound using SPME Gas Chromatography
2.7. Statistical Analysis
3. Results and Discussion
3.1. Changes in Titratable Acidity and AAB Growth According to Fermentation Temperature, Alcohol Concentration, and Initial pH
3.2. Selecting Multiple Starter Candidates for AAF
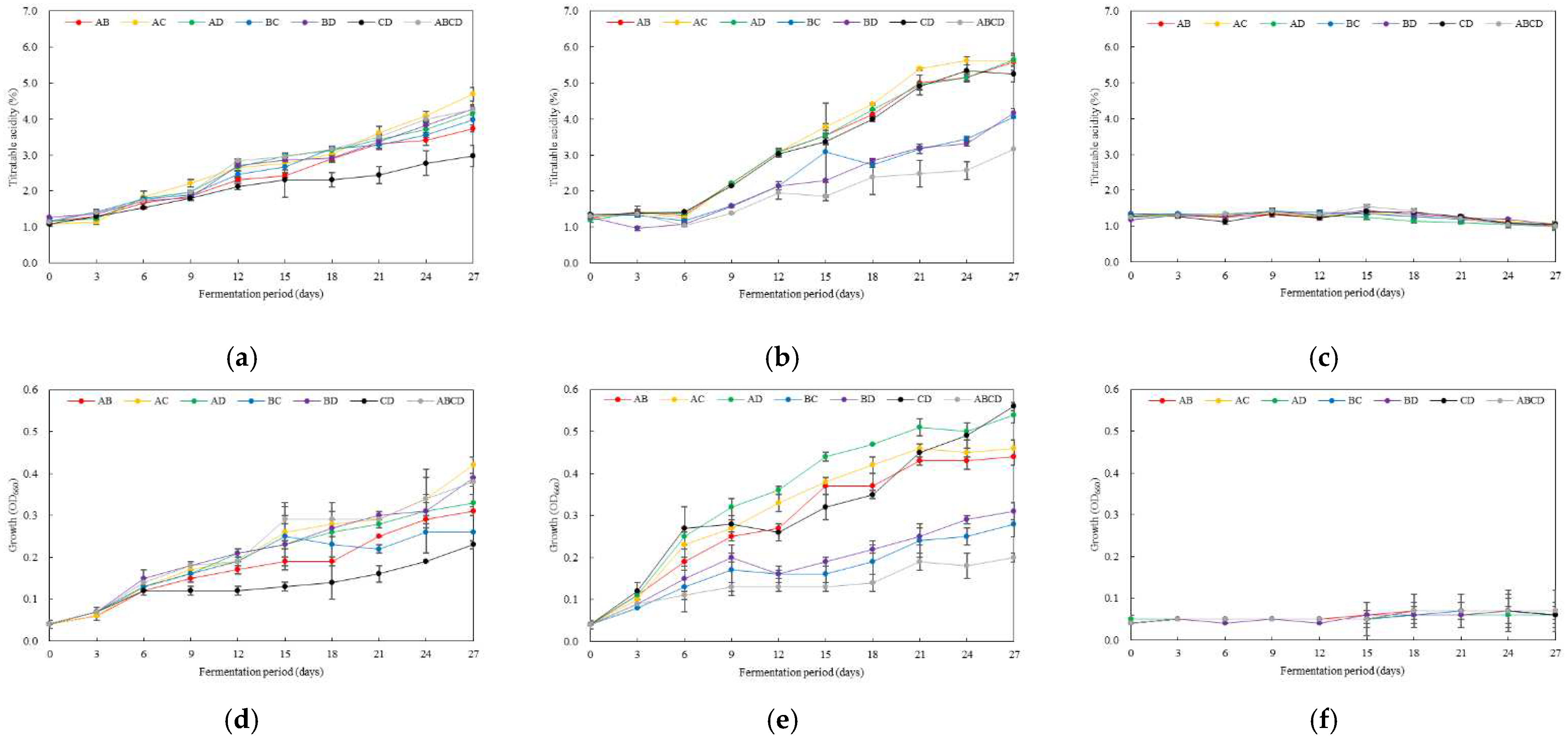
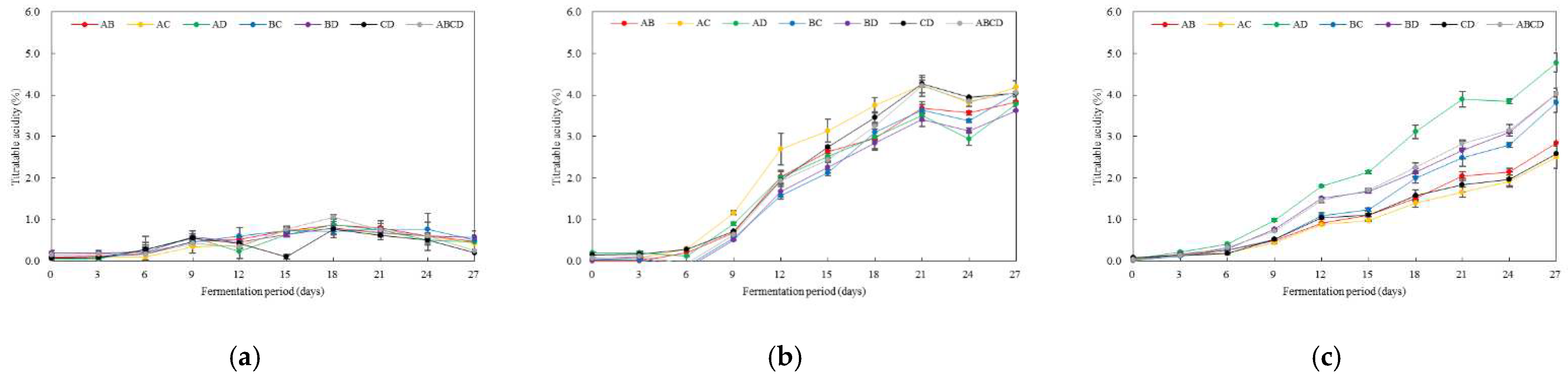
3.3. Changes in Titratable Acidity and Growth of Multiple AAB According to Fermentation Temperature and Initial pH
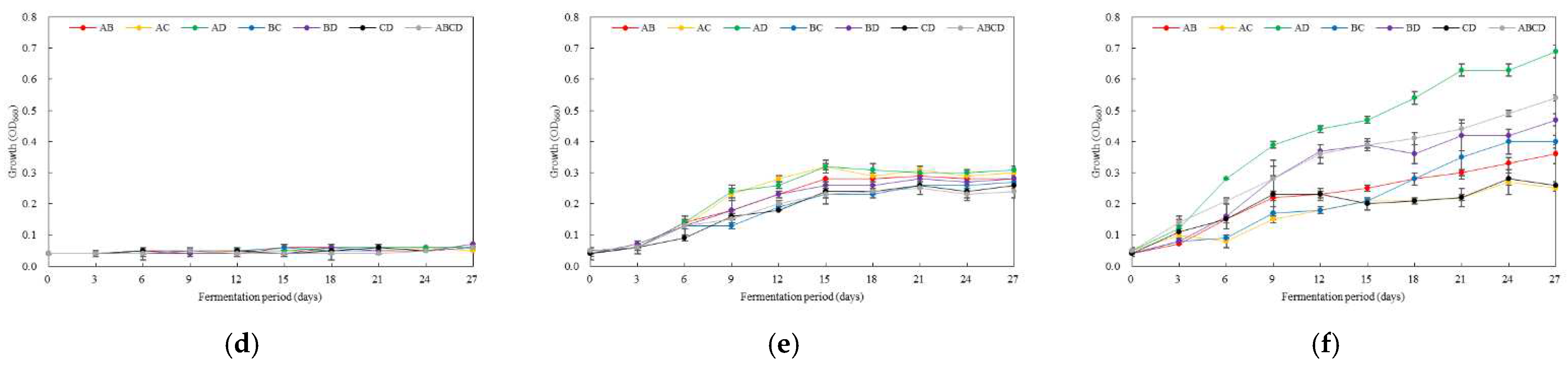
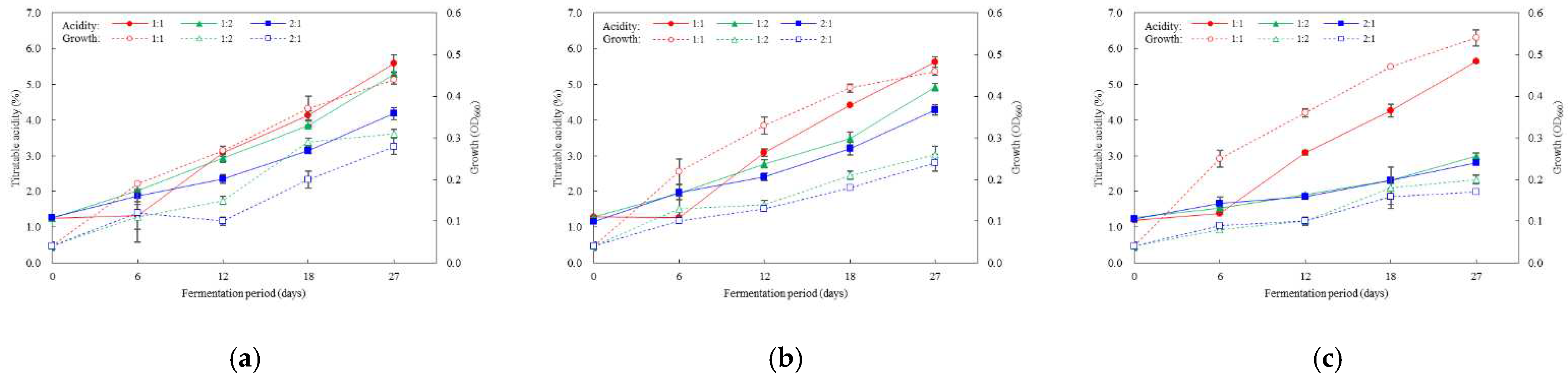
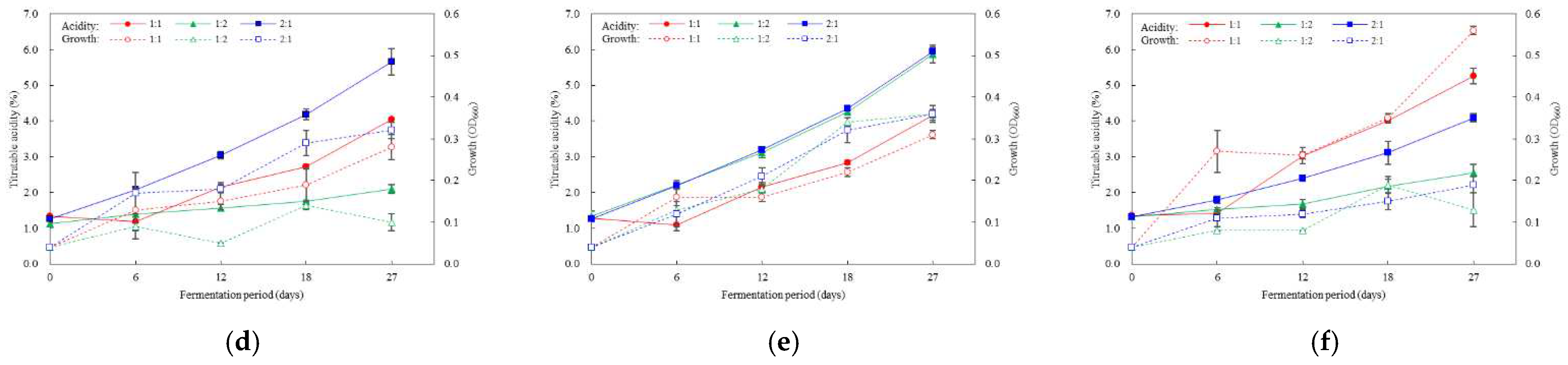
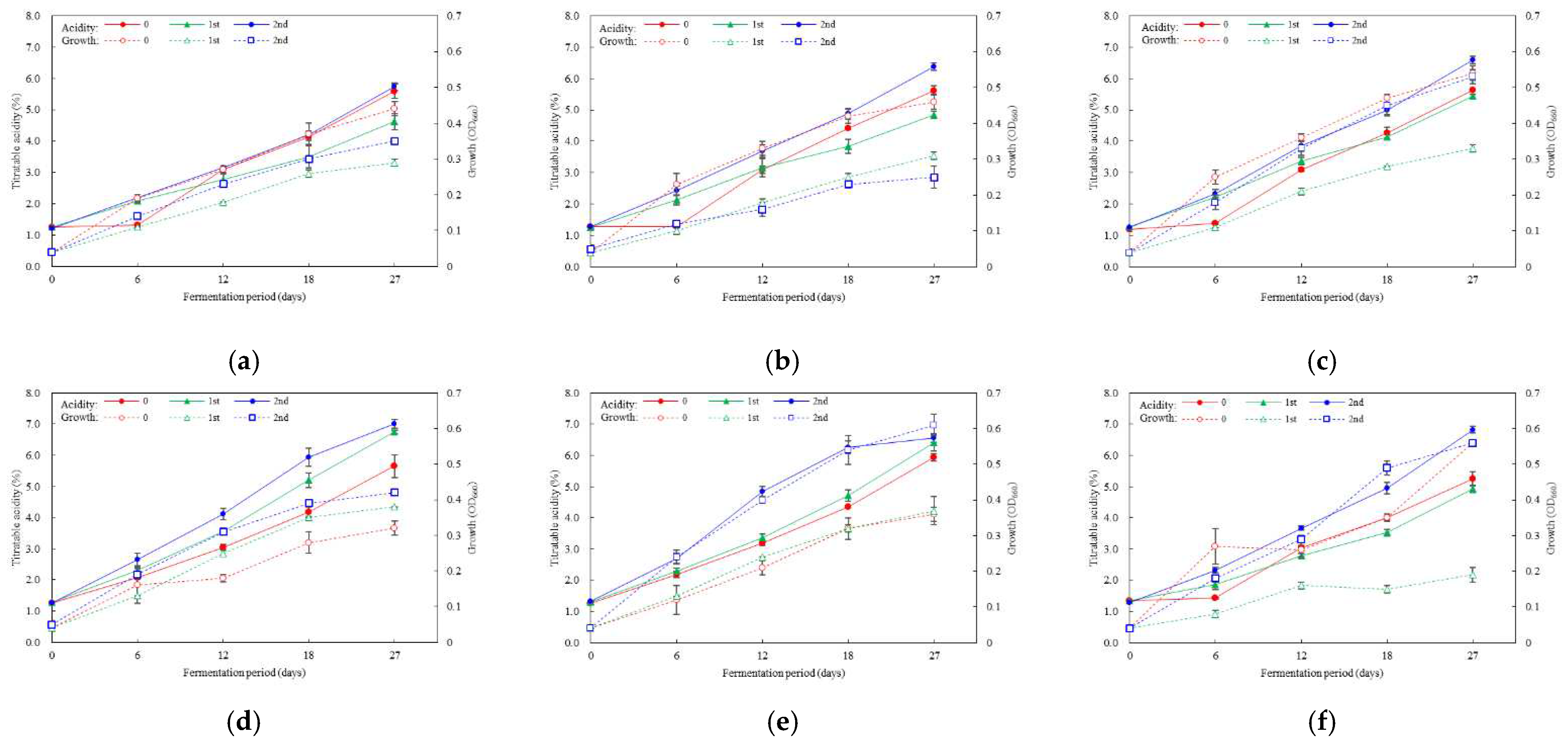
3.4. Optimal Inoculation Rate and Order of Multiple AAB
3.5. Volatile Flavor Pattern Profiling According to Fermentation Temperature and Multiple Starters
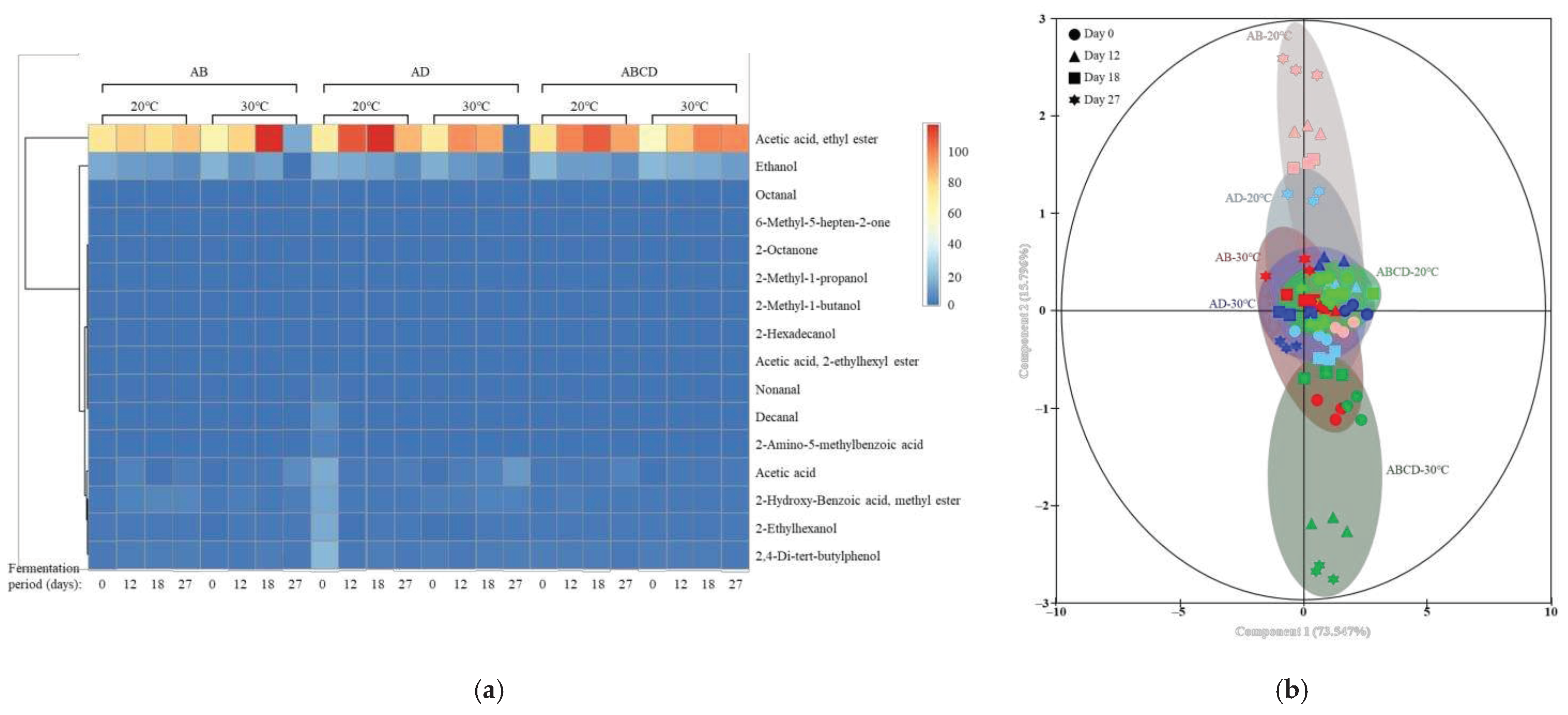
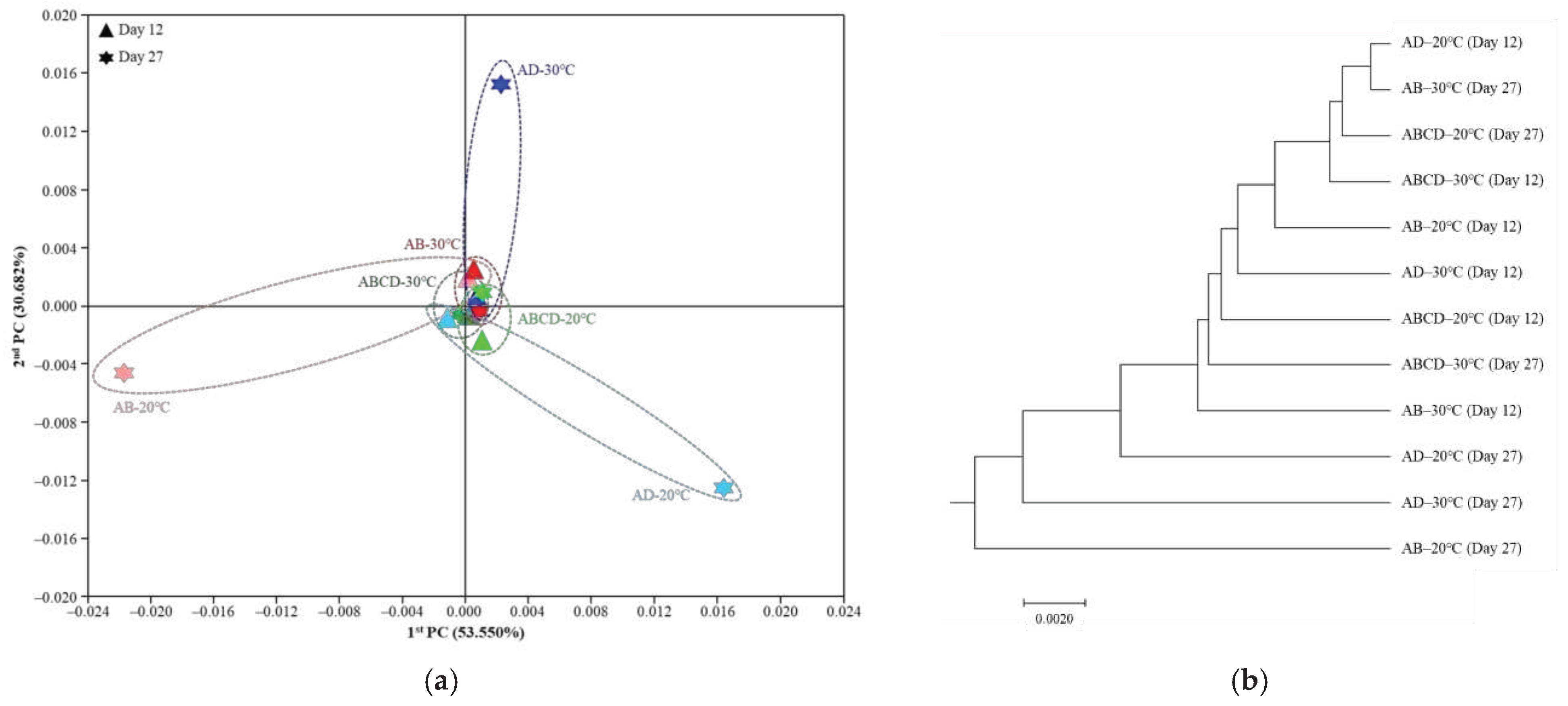
3.5. Changes in the Beta Diversity of Microbial Communities According to Fermentation Temperature and Multiple Starters
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nanda, K.; Taniguchi, M.; Ujike, S.; Ishihara, N.; Mori, H.; Ono, H.; Murooka, Y. Characterization of acetic acid bacteria in traditional acetic acid fermentation of rice vinegar (Komesu) and unpolished rice vinegar (Kurosu) produced in Japan. Appl. Environ. Microbiol. 2001, 67(2), 986–990. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.; Kim, S. H.; Jeong, W. S.; Kim, S. Y.; Yeo, S. H. Microbiome analysis of traditional grain vinegar produced under different fermentation conditions in various regions in Korea. Foods 2022, 11(22), 3573. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Chen, T.; Giudici, P.; Chen, F. Vinegar functions on health: Constituents, sources, and formation mechanisms. Compr. Rev. Food Sci. Food Saf. 2016, 15, 1124–1138. [Google Scholar] [CrossRef]
- Kim, K. O.; Kim, S. M.; Kim, S. M.; Kim, D. Y.; Jo, D.; Yeo, S. H.; Jeong, Y. J.; Kwon, J. H. Physicochemical properties of commercial fruit vinegars with different fermentation methods. J. Korean Soc. Food Sci. Nutr. 2013, 42(5), 736–742. [Google Scholar] [CrossRef]
- Kanchanarach, W.; Theeragool, G.; Inoue, T.; Yakushi, T.; Adachi, O.; Matsushita, K. Acetic acid fermentation of Acetobacter pasteurianus: relationship between acetic acid resistance and pellicle polysaccharide formation. Biosci. Biotechnol. Biochem. 2010, 74(8), 1591–1597. [Google Scholar] [CrossRef]
- Li, S.; Li, P.; Liu, X.; Luo, L.; Lin, W. Bacterial dynamics and metabolite changes in solid–state acetic acid fermentation of Shanxi aged vinegar. Appl. Microbiol. Biotechnol. 2016, 100, 4395–4411. [Google Scholar] [CrossRef]
- Fleet, G. H. Microorganisms in food ecosystems. Int. J. Food Microbiol. 1999, 50(1), 101–117. [Google Scholar] [CrossRef] [PubMed]
- Gullo, M.; Giudici, P. Acetic acid bacteria in traditional balsamic vinegar: phenotypic traits relevant for starter cultures selection. Int. J. Food Microbiol 2008, 125, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, C.; Vegas, C.; Mateo, E.; Tesfaye, W.; Cerezo, A.; Callejón, R.; Poblet, M.; Guillamón, J.; Mas, A.; Torija, M. Effect of barrel design and the inoculation of Acetobacter pasteurianus in wine vinegar production. Int. J. Food Microbiol 2010, 141, 56–62. [Google Scholar] [CrossRef]
- Wu, J. J.; Ma, Y. K.; Zhang, F. F.; Chen, F. S. Biodiversity of yeasts, lactic acid bacteria and acetic acid bacteria in the fermentation of “Shanxi aged vinegar”, a traditional Chinese vinegar. Food Microbiol 2012, 30, 289–297. [Google Scholar] [CrossRef]
- Ohmori, S.; Masai, H.; Arima, K.; Beppu, T. Isolation and identification of acetic acid bacteria for submerged acetic acid fermentation at high temperature. Agric. Biol. Chem. 1980, 44(12), 2901–2906. [Google Scholar] [CrossRef]
- Astudillo–Melgar, F.; Ochoa–Leyva, A.; Utrilla, J.; Huerta–Beristain, G. Bacterial diversity and population dynamics during the fermentation of palm wine from Guerrero Mexico. Front. Microbiol. 2019, 10, 531. [Google Scholar] [CrossRef] [PubMed]
- Escalante, A. E.; Rebolleda–Gómez, M.; Benítez, M.; Travisano, M. Ecological perspectives on synthetic biology: insights from microbial population biology. Front. Microbiol. 2015, 6, 143. [Google Scholar] [CrossRef]
- Prathiviraj, R.; Rajeev, R.; Jose, C. M.; Begum, A.; Selvin, J.; Kiran, G. S. Fermentation microbiome and metabolic profiles of Indian palm wine. Gene Rep. 2022, 27, 101543. [Google Scholar] [CrossRef]
- Su, X. Elucidating the beta–diversity of the microbiome: from global alignment to local alignment. Msystems 2021, 6(4), e00363–21. [Google Scholar] [CrossRef] [PubMed]
- Baek, S. Y.; Park, H. Y.; Lee, C. H.; Yeo, S. H. Comparison of the fermented property and isolation of acetic–acid bacteria from traditional Korean vinegar. Korean J. Food Preserv. 2014, 21(6), 1738–7248. [Google Scholar] [CrossRef]
- Baek, C. H.; Baek, S. Y.; Lee, S. H.; Kang, J. E.; Choi, H. S.; Kim, J. H.; Yeo, S. H. Characterization of Acetobacter sp. strain CV1 isolated from a fermented vinegar. Microbiol. Biotechnol. Lett. 2015, 43(2), 126–133. [Google Scholar] [CrossRef]
- Gil, N. Y.; Jang, Y. J.; Gwon, H. M.; Jeong, W. S.; Yeo, S. H.; Kim, S. Y. Comparative evaluation of quality and metabolite profiles in Meju using starter cultures of Bacillus velezensis and Aspergillus oryzae. Foods 2022, 11(1), 68. [Google Scholar] [CrossRef]
- Kang, S. K.; Jang, M. J.; Kim, Y. D. Isolation and culture conditions of Acetobacter sp. for the production of citron (Citrus junos) vinegar. Korean J. Food Preserv 2006, 13(3), 357–362. [Google Scholar]
- Burbach, K.; Seifert, J.; Pieper, D. H.; Camarinha-Silva, A. Evaluation of DNA extraction kits and phylogenetic diversity of the porcine gastrointestinal tract based on Illumina sequencing of two hypervariable regions. MicrobiologyOpen 2016, 5(1), 70–82. [Google Scholar] [CrossRef]
- Chao, A.; Lee, S. M. Estimating the number of classes via sample coverage. J. Am. Stat. Assoc. 1992, 87(417), 210–217. [Google Scholar] [CrossRef]
- Chao, A. Estimating the population size for capture–recapture data with unequal catchability. Biometrics 1987, 783–791. [Google Scholar] [CrossRef]
- Burnham, K. P.; Overton, W. S. Robust estimation of population size when capture probabilities vary among animals. Ecology 1979, 60(5), 927–936. [Google Scholar] [CrossRef]
- Magurran, A. E. Measuring biological diversity. Curr. Biol. 2021, 31(19), R1174–R1177. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.; Shen, T. J. Nonparametric estimation of Shannon’s index of diversity when there are unseen species in sample. Environ. Ecol. Stat 2003, 10, 429–443. [Google Scholar] [CrossRef]
- Faith, D. P. Conservation evaluation and phylogenetic diversity. Biol. Conserv. 1992, 61(1), 1–10. [Google Scholar] [CrossRef]
- Heck, K. L.; van Belle, G.; Simberloff, D. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 1975, 56(6), 1459–1461. [Google Scholar] [CrossRef]
- Whittaker, R. H. Dominance and diversity in land plant communities. Science 1965, 147, 250–260. [Google Scholar] [CrossRef] [PubMed]
- Lin, J. Divergence measures based on the Shannon entropy. IEEE Trans. Inf. Theory 1991, 37, 145–151. [Google Scholar] [CrossRef]
- Beals, E. W. Bray–Curtis ordination: an effective strategy for analysis of multivariate ecological data. Adv. Ecol. Res. 1984, 14, 1–55. [Google Scholar] [CrossRef]
- Chen, J.; Bittinger, K.; Charlson, E. S.; Hoffmann, C.; Lewis, J.; Wu, G. D.; Li, H. (2012). Associating microbiome composition with environmental covariates using generalized UniFrac distances. Bioinformatics 2012, 28, 2106–2113. [Google Scholar] [CrossRef]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: facilitating high–throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Doak, T. G. A parsimony approach to biological pathway reconstruction/inference for genomes and metagenomes. PLoS Comput. Biol. 2009, 5, e1000465. [Google Scholar] [CrossRef] [PubMed]
- Langille, M. G.; Zaneveld, J.; Caporaso, J. G.; McDonald, D.; Knights, D.; Reyes, J. A.; Beiko, R. G. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W. S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Kruskal, W. H.; Wallis, W. A. Use of ranks in one–criterion variance analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Lee, S. J.; Kim, S. H.; Kim, S. Y.; Yeo, S. H. Quality characteristics of Kujippong (Cudrania tricuspidata) vinegar fermented by various acetic acid bacteria. Korean J. Food Preserv. 2019, 26(7), 766–776. [Google Scholar] [CrossRef]
- Lynch, K. M.; Zannini, E.; Wilkinson, S.; Daenen, L.; Arendt, E. K. Physiology of acetic acid bacteria and their role in vinegar and fermented beverages. Compr. Rev. Food Sci. Food Saf. 2019, 18(3), 587–625. [Google Scholar] [CrossRef] [PubMed]
- Longo, M. A.; Sanromán, M. A. Production of food aroma compounds: microbial and enzymatic methodologies. Food Technol. Biotechnol. 2006, 44(3), 335–353. [Google Scholar] [CrossRef]
- Sawamura, M.; Thi Minh Tu, N.; Onishi, Y.; Ogawa, E.; Choi, H. S. Characteristic odor components of Citrus reticulata Blanco (Ponkan) cold–pressed oil. Biosci. Biotechnol. Biochem. 2004, 68(8), 1690–1697. [Google Scholar] [CrossRef]
- Rocha, S. M.; Rodrigues, F.; Coutinho, P.; Delgadillo, I.; Coimbra, M. A. Volatile composition of Baga red wine: Assessment of the identification of the would–be impact odourants. Anal. Chim. Acta. 2004, 513(1), 257–262. [Google Scholar] [CrossRef]
- Feng, S.; Suh, J. H.; Gmitter, F. G.; Wang, Y. Differentiation between flavors of sweet orange (Citrus sinensis) and mandarin (Citrus reticulata). J. Agric. Food Chem. 2018, 66(1), 203–211. [Google Scholar] [CrossRef]
- Tandon, K. S.; Baldwin, E. A.; Shewfelt, R. L. Aroma perception of individual volatile compounds in fresh tomatoes (Lycopersicon esculentum, Mill.) as affected by the medium of evaluation. Postharvest Biol. Technol, 2000; 20, 3, 261–268. [Google Scholar] [CrossRef]
- Bett-Garber, K. L.; Bryant, R. J.; Grimm, C. C.; Chen, M. H.; Lea, J. M.; McClung, A. M. Physicochemical and sensory analysis of US rice varieties developed for the basmati and jasmine markets. Cereal Chem. 2017, 94(3), 602–610. [Google Scholar] [CrossRef]
- Su, M. S.; Chien, P. J. Aroma impact components of rabbiteye blueberry (Vaccinium ashei) vinegars. Food Chem. 2010, 119(3), 923–928. [Google Scholar] [CrossRef]
- Gullo, M.; Verzelloni, E.; Canonico, M. Aerobic submerged fermentation by acetic acid bacteria for vinegar production: Process and biotechnological aspects. Process Biochem. 2014, 49(10), 1571–1579. [Google Scholar] [CrossRef]
- Jo, Y.; Gu, S. Y.; Chung, N.; Gao, Y.; Kim, H. J.; Jeong, M. H.; Jeong, Y. J.; Kwon, J. H. Comparative analysis of sensory profiles of commercial cider vinegars from Korea, China, Japan, and US by SPME/GC–MS, E-nose, and E-tongue. Korean J. Food Sci. Technol. 2016, 48, 430–436. [Google Scholar] [CrossRef]
- Seo, H.; Lee, S.; Park, H.; Jo, S.; Kim, S.; Rahim, M. A.; Ul-Haq, A.; Barman, I.; Lee, Y.; Seo, A.; Kim, M.; Jung, I. Y.; Song, H. Y. Characteristics and microbiome profiling of Korean Gochang bokbunja vinegar by the fermentation process. Foods 2022, 11(20), 3308. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, L.; Osman, O. A.; Bertilsson, S.; Eiler, A. Microbial community composition and diversity via 16S rRNA gene amplicons: evaluating the illumina platform. PloS one 2015, 10(2), e0116955. [Google Scholar] [CrossRef] [PubMed]
- Olszewski, T. D. A unified mathematical framework for the measurement of richness and evenness within and among multiple communities. Oikos 2004, 104(2), 377–387. [Google Scholar] [CrossRef]
- Tawfik, S. A.; Azab, M. M.; Ahmed, A. A. A.; Fayyad, D. M. Illumina MiSeq sequencing for preliminary analysis of microbiome causing primary endodontic infections in Egypt. Int. J. Microbial. 2018, 2018, 2837328. [Google Scholar] [CrossRef]
- Dai, Y.; Tian, Z.; Meng, W.; Li, Z. Microbial diversity and physicochemical characteristics of the Maotai-flavored liquor fermentation process. J. Nanosci. Nanotechnol. 2020, 20(7), 4097–4109. [Google Scholar] [CrossRef]
- Wang, Z. M.; Lu, Z. M.; Yu, Y. J.; Li, G. Q.; Shi, J. S.; Xu, Z. H. Batch-to-batch uniformity of bacterial community succession and flavor formation in the fermentation of Zhenjiang aromatic vinegar. Food Microbiol. 2015, 50, 64–69. [Google Scholar] [CrossRef] [PubMed]
| Region (Province) | Origin | Sample Name | Species | Strain |
|---|---|---|---|---|
| Gyeongsangnam–do | Brown rice | GN_BWR | Acetobacter pasteurianus | GV–5 |
| Black rice | GN_BR | Acetobacter ascendens | GV–8 (A)1 | |
| Gyeongsangbuk–do | Five grains2 | GB_FG | Acetobacter ascendens | GV–12 (B) |
| Brown rice | GB_UR | Acetobacter pasteurianus | GV–22 (D) | |
| Chungcheongnam–do | Brown rice | CN_UR | Acetobacter pasteurianus | GV–16 |
| Acetobacter pasteurianus | GV–17 (C) |
| Temp. | Strains | Titratable acidity according to fermentation period (days) | Growth (OD660) according to fermentation period (days) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 0 | 3 | 6 | 9 | 12 | 15 | |||
| 20 °C | GV–5 | 1.25±0.001,c | 1.43±0.03b | 2.14±0.10de | 1.26±0.06d | 0.75±0.18d | 0.57±0.03d | 0.05±0.00c | 0.06±0.00cd | 0.10±0.03bc | 0.38±0.38a | 0.83±0.40a | 1.44±0.29a | |
| GV–8 | 1.30±0.01b | 1.41±0.03b | 2.01±0.02e | 1.11±0.12d | 0.73±0.20d | 0.48±0.08d | 0.06±0.00b | 0.06±0.00cd | 0.06±0.00c | 0.14±0.02b | 0.47±0.38b | 1.49±0.06a | ||
| GV–12 | 1.40±0.00a | 1.40±0.07b | 2.66±0.13bcd | 1.85±0.12d | 1.08±0.15cd | 0.44±0.02d | 0.06±0.00b | 0.06±0.00cd | 0.22±0.03a | 0.25±0.11ab | 0.79±0.20a | 1.13±0.20b | ||
| GV–16 | 1.38±0.00a | 1.40±0.06b | 2.53±0.36cde | 1.54±0.04d | 1.14±0.08cd | 0.47±0.08d | 0.07±0.01a | 0.06±0.00cd | 0.07±0.01bc | 0.22±0.07ab | 0.48±0.12b | 1.05±0.08b | ||
| GV–17 | 1.38±0.00a | 1.39±0.08b | 2.46±0.14de | 1.74±0.11d | 1.41±0.36c | 0.98±0.48c | 0.06±0.00b | 0.06±0.00cd | 0.08±0.00bc | 0.19±0.05ab | 0.45±0.11b | 1.13±0.24b | ||
| GV–22 | 1.41±0.00a | 1.41±0.12b | 2.69±0.31bcd | 1.74±0.19d | 1.07±0.23cd | 0.51±0.14d | 0.05±0.00c | 0.06±0.00cd | 0.21±0.07a | 0.20±0.08ab | 0.34±0.20bc | 1.39±0.06a | ||
| 30 °C | GV–5 | 1.27±0.03bc | 2.23±0.20a | 3.56±0.51a | 4.57±0.60ab | 5.19±0.29a | 5.64±0.16a | 0.06±0.01b | 0.06±0.02cd | 0.10±0.09bc | 0.11±0.11b | 0.13±0.07c | 0.16±0.07c | |
| GV–8 | 1.28±0.05bc | 2.19±0.14a | 3.41±0.33a | 4.77±0.72a | 5.17±0.07a | 5.33±0.19a | 0.07±0.01a | 0.06±0.02cd | 0.08±0.06bc | 0.10±0.08b | 0.10±0.05c | 0.18±0.06c | ||
| GV–12 | 1.20±0.04d | 2.14±0.14a | 3.12±0.27abc | 4.03±0.21b | 5.28±0.16a | 5.62±0.03a | 0.05±0.00c | 0.10±0.04bc | 0.11±0.07bc | 0.10±0.02b | 0.12±0.07c | 0.12±0.04c | ||
| GV–16 | 1.30±0.03b | 2.34±0.19a | 3.27±0.72ab | 4.35±1.00ab | 5.17±0.46a | 5.56±0.25a | 0.05±0.00c | 0.12±0.01ab | 0.22±0.03a | 0.22±0.03ab | 0.23±0.05bc | 0.23±0.03c | ||
| GV–17 | 1.28±0.02bc | 2.21±0.25a | 3.18±0.60ab | 4.19±0.64ab | 4.81±0.55a | 5.60±0.17a | 0.05±0.00c | 0.09±0.03bcd | 0.15±0.06ab | 0.18±0.08ab | 0.22±0.06bc | 0.22±0.04c | ||
| GV–22 | 1.27±0.03bc | 2.25±0.11a | 3.40±0.18a | 4.85±0.24a | 5.27±0.14a | 5.40±0.01a | 0.05±0.00c | 0.15±0.08a | 0.15±0.08ab | 0.16±0.07b | 0.19±0.06bc | 0.23±0.04c | ||
| 40 °C | GV–5 | 1.19±0.00d | 1.18±0.01c | 1.19±0.02f | 1.16±0.01d | 1.16±0.04cd | 1.15±0.02c | 0.05±0.00c | 0.05±0.00d | 0.05±0.00c | 0.05±0.00b | 0.05±0.00c | 0.05±0.00c | |
| GV–8 | 1.17±0.00d | 1.17±0.01c | 1.19±0.01f | 1.17±0.03d | 1.19±0.07cd | 1.16±0.04c | 0.05±0.00c | 0.05±0.00d | 0.05±0.00c | 0.05±0.00b | 0.05±0.00c | 0.05±0.00c | ||
| GV–12 | 1.25±0.05c | 1.23±0.02bc | 2.42±0.62de | 2.61±0.53c | 3.10±0.54b | 2.09±0.68b | 0.05±0.00c | 0.05±0.00d | 0.12±0.05bc | 0.09±0.03b | 0.09±0.03c | 0.09±0.03c | ||
| GV–16 | 1.30±0.00b | 1.24±0.02bc | 1.12±0.02f | 1.18±0.04d | 1.23±0.02cd | 1.20±0.08c | 0.05±0.00c | 0.05±0.00d | 0.05±0.00c | 0.05±0.00b | 0.05±0.00c | 0.05±0.00c | ||
| GV–17 | 1.26±0.01bc | 1.18±0.03c | 1.19±0.02f | 1.16±0.03d | 1.26±0.02c | 1.09±0.07c | 0.05±0.00c | 0.05±0.00d | 0.05±0.00c | 0.05±0.00b | 0.05±0.00c | 0.05±0.00c | ||
| GV–22 | 1.28±0.00bc | 1.15±0.02c | 1.15±0.02f | 1.14±0.02d | 1.20±0.02cd | 1.13±0.06c | 0.05±0.00c | 0.05±0.00d | 0.05±0.00c | 0.05±0.00b | 0.05±0.00c | 0.05±0.00c | ||
| Conc. EtOH | Strains | Titratable acidity according to fermentation period (days) | Growth (OD660) according to fermentation period (days) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 0 | 3 | 6 | 9 | 12 | 15 | |||
| 5% | GV–5 | 1.27±0.031,f | 2.23±0.20a | 3.56±0.51a | 4.57±0.40ab | 5.19±0.29abc | 5.64±0.16cd | 0.06±0.01ab | 0.06±0.02de | 0.10±0.09bcd | 0.11±0.11bcd | 0.13±0.07cdef | 0.16±0.07bcd | |
| GV–8 | 1.28±0.05ef | 2.19±0.14a | 3.41±0.33ab | 4.77±0.38a | 5.17±0.07abc | 5.33±0.19d | 0.07±0.01a | 0.06±0.02de | 0.08±0.06bcd | 0.10±0.08bcd | 0.10±0.05efg | 0.18±0.06abc | ||
| GV–12 | 1.20±0.04g | 2.14±0.14a | 3.12±0.27ab | 4.03±0.21b | 5.28±0.16ab | 5.62±0.03cd | 0.05±0.00b | 0.10±0.04bc | 0.11±0.07bcd | 0.10±0.02bcd | 0.12±0.07def | 0.12±0.04cde | ||
| GV–16 | 1.30±0.03ef | 2.34±0.19a | 3.27±0.72ab | 4.35±0.42ab | 5.17±0.46abc | 5.56±0.25cd | 0.05±0.00b | 0.12±0.01ab | 0.22±0.03a | 0.22±0.03a | 0.23±0.05a | 0.23±0.03a | ||
| GV–17 | 1.28±0.02ef | 2.21±0.25a | 3.18±0.60ab | 4.19±0.64ab | 4.81±0.55bc | 5.60±0.17cd | 0.05±0.00b | 0.09±0.03bcd | 0.15±0.06b | 0.18±0.08ab | 0.22±0.06ab | 0.22±0.04ab | ||
| GV–22 | 1.27±0.03f | 2.25±0.11a | 3.4±0.18ab | 4.85±0.24a | 5.27±0.14ab | 5.40±0.01d | 0.05±0.00b | 0.15±0.08a | 0.15±0.08b | 0.16±0.07ab | 0.19±0.06abc | 0.23±0.04a | ||
| 8% | GV–5 | 1.28±0.03ef | 1.42±0.14c | 2.05±0.25c | 3.17±0.51c | 4.13±0.30d | 5.51±0.38cd | 0.05±0.01b | 0.05±0.00de | 0.07±0.02cd | 0.07±0.02cd | 0.08±0.01fg | 0.08±0.01ef | |
| GV–8 | 1.34±0.00cdef | 1.44±0.08c | 2.06±0.24c | 3.13±0.36c | 4.09±0.28d | 5.30±0.50d | 0.06±0.01ab | 0.05±0.00de | 0.10±0.04bcd | 0.13±0.04bcd | 0.15±0.04cde | 0.16±0.02bcd | ||
| GV–12 | 1.29±0.05ef | 1.76±0.12b | 2.89±0.29b | 4.16±0.64ab | 5.7±0.60a | 7.27±0.46a | 0.06±0.01ab | 0.07±0.01cde | 0.09±0.02bcd | 0.11±0.07bcd | 0.12±0.01def | 0.11±0.06de | ||
| GV–16 | 1.30±0.03ef | 1.77±0.06b | 2.86±0.21b | 3.99±0.42b | 4.70±0.17c | 6.10±0.34c | 0.06±0.01ab | 0.06±0.02de | 0.13±0.00bc | 0.14±0.02abc | 0.17±0.03abcd | 0.21±0.09ab | ||
| GV–17 | 1.29±0.06ef | 1.88±0.23b | 3.30±0.67ab | 4.64±1.12ab | 5.55±0.92a | 6.95±1.14ab | 0.05±0.00b | 0.07±0.02cde | 0.14±0.07bc | 0.16±0.05ab | 0.17±0.05abcd | 0.20±0.04ab | ||
| GV–22 | 1.30±0.02ef | 1.47±0.12c | 1.96±0.06cd | 3.35±0.23c | 4.69±0.22c | 6.64±0.50b | 0.05±0.00b | 0.05±0.00de | 0.08±0.04bcd | 0.10±0.10bcd | 0.16±0.01bcde | 0.22±0.02ab | ||
| 11% | GV–5 | 1.38±0.00abcd | 1.48±0.01c | 1.48±0.14de | 1.34±0.11d | 1.37±0.09e | 1.37±0.08e | 0.05±0.00b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | |
| GV–8 | 1.40±0.01abc | 1.45±0.10c | 1.42±0.04e | 1.36±0.16d | 1.32±0.03e | 1.31±0.02e | 0.05±0.00b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–12 | 1.41±0.05ab | 1.55±0.08c | 1.36±0.03e | 1.28±0.01d | 1.28±0.07e | 1.28±0.03e | 0.06±0.01ab | 0.04±0.00e | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–16 | 1.35±0.04bcde | 1.46±0.02c | 1.37±0.04e | 1.28±0.03d | 1.21±0.05e | 1.28±0.09e | 0.05±0.01b | 0.04±0.00e | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–17 | 1.40±0.00abc | 1.45±0.05c | 1.40±0.04e | 1.27±0.02d | 1.21±0.03e | 1.28±0.04e | 0.05±0.00b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–22 | 1.42±0.00a | 1.44±0.07c | 1.35±0.07e | 1.25±0.03d | 1.18±0.01e | 1.26±0.04e | 0.05±0.00b | 0.04±0.00e | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| 14% | GV–5 | 1.30±0.05ef | 1.47±0.02c | 1.40±0.08e | 1.28±0.01d | 1.38±0.05e | 1.28±0.03e | 0.05±0.00b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | |
| GV–8 | 1.27±0.02f | 1.42±0.06c | 1.31±0.02e | 1.35±0.16d | 1.28±0.03e | 1.39±0.08e | 0.05±0.00b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–12 | 1.27±0.03f | 1.38±0.05c | 1.35±0.08e | 1.27±0.05d | 1.32±0.06e | 1.20±0.18e | 0.06±0.01ab | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–16 | 1.28±0.00ef | 1.38±0.02c | 1.32±0.01e | 1.28±0.01d | 1.38±0.02e | 1.44±0.11e | 0.05±0.00b | 0.04±0.00e | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–17 | 1.29±0.10ef | 1.40±0.04c | 1.41±0.12e | 1.33±0.03d | 1.34±0.01e | 1.30±0.03e | 0.05±0.00b | 0.04±0.00e | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| GV–22 | 1.32±0.00def | 1.44±0.08c | 1.30±0.01e | 1.28±0.02d | 1.30±0.03e | 1.32±0.02e | 0.05±0.01b | 0.05±0.00de | 0.04±0.00d | 0.04±0.00d | 0.04±0.00g | 0.04±0.00f | ||
| Initial pH1 | Strains | Titratable acidity according to fermentation period (days) | Growth (OD660) according to fermentation period (days) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 0 | 3 | 6 | 9 | 12 | 15 | |||
| pH 2.0 | GV–5 | 0.02±0.032,bc | 0.03±0.01e | 0.09±0.10g | 0.06±0.04h | 0.13±0.14h | 0.09±0.04h | 0.06±0.00a | 0.06±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.00g | 0.05±0.00e | |
| GV–8 | 0.04±0.03bc | 0.04±0.03e | 0.03±0.05g | 0.16±0.20h | 0.22±0.35h | 0.16±0.20h | 0.06±0.00a | 0.06±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.00g | 0.05±0.01e | ||
| GV–12 | 0.09±0.10abc | 0.06±0.03e | 0.07±0.09g | 0.04±0.08h | 0.07±0.09h | 0.11±0.07h | 0.05±0.01b | 0.06±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.01g | 0.05±0.00e | ||
| GV–16 | 0.12±0.11ab | 0.07±0.03e | 0.06±0.03g | 0.20±0.22h | 0.17±0.08h | 0.40±0.36h | 0.06±0.00a | 0.05±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.00g | 0.05±0.00e | ||
| GV–17 | 0.04±0.02bc | 0.05±0.03e | 0.03±0.03g | 0.05±0.03h | 0.06±0.05h | 0.13±0.02h | 0.05±0.00b | 0.06±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.00g | 0.05±0.00e | ||
| GV–22 | 0.03±0.02bc | 0.07±0.03e | 0.15±0.06g | 0.19±0.10h | 0.25±0.16h | 0.31±0.16h | 0.05±0.00b | 0.07±0.00e | 0.05±0.00g | 0.05±0.00e | 0.05±0.00g | 0.05±0.00e | ||
| pH 3.0 | GV–5 | 0.08±0.05abc | 0.09±0.06e | 0.32±0.06g | 2.06±0.47g | 2.19±0.47g | 2.04±0.50g | 0.05±0.00b | 0.06±0.00e | 0.09±0.00g | 0.34±0.05cd | 0.40±0.06ef | 0.43±0.09d | |
| GV–8 | 0.04±0.05bc | 0.11±0.03e | 4.30±0.16c | 5.13±0.24d | 4.90±0.32d | 4.39±0.15de | 0.05±0.00b | 0.06±0.01e | 0.23±0.05def | 0.29±0.02cde | 0.70±0.35bc | 0.88±0.09ab | ||
| GV–12 | 0.06±0.07abc | 0.08±0.03e | 3.41±0.70d | 4.35±0.77e | 4.12±0.43e | 3.78±0.38ef | 0.06±0.00a | 0.07±0.00e | 0.29±0.02d | 0.28±0.06cde | 0.27±0.17fg | 0.48±0.05d | ||
| GV–16 | 0.15±0.09a | 0.10±0.03e | 2.69±0.66de | 3.79±0.47ef | 3.99±0.03ef | 3.81±0.66ef | 0.06±0.01a | 0.06±0.00e | 0.17±0.03f | 0.16±0.00de | 0.46±0.26cdef | 0.44±0.03d | ||
| GV–17 | 0.05±0.09abc | 0.10±0.03e | 2.04±0.77ef | 3.57±0.75f | 3.55±0.22f | 3.44±0.96f | 0.05±0.00b | 0.06±0.00e | 0.10±0.05g | 0.15±0.01de | 0.15±0.01g | 0.17±0.08e | ||
| GV–22 | 0.08±0.06abc | 0.10±0.06e | 1.49±0.23f | 2.57±0.09g | 2.47±0.23g | 2.07±0.78g | 0.06±0.01a | 0.06±0.01e | 0.19±0.01df | 0.32±0.12cde | 0.41±0.09def | 0.49±0.16d | ||
| pH 4.0 | GV–5 | 0.03±0.02bc | 1.60±0.04c | 6.78±0.07a | 7.05±0.02c | 6.99±0.24b | 6.79±0.07a | 0.05±0.00b | 0.22±0.09c | 0.35±0.04c | 0.87±0.04a | 0.94±0.12a | 1.07±0.02a | |
| GV–8 | 0.01±0.02c | 2.99±0.19a | 6.88±1.44a | 8.04±0.19a | 7.55±0.40a | 5.81±0.31b | 0.06±0.00a | 0.53±0.06a | 0.62±0.03a | 0.91±0.00a | 0.90±0.13ab | 1.03±0.23a | ||
| GV–12 | 0.01±0.01c | 2.19±0.10b | 7.16±0.27a | 7.69±0.45ab | 7.51±0.51a | 7.21±0.54a | 0.05±0.00b | 0.28±0.02b | 0.41±0.10b | 0.49±0.13bc | 0.65±0.18cd | 0.78±0.12bc | ||
| GV–16 | 0.02±0.01bc | 1.12±0.11d | 4.60±0.68c | 5.64±0.18d | 5.62±0.15c | 5.41±0.12bc | 0.06±0.01a | 0.15±0.05d | 0.25±0.05de | 0.35±0.07cd | 0.54±0.05cde | 0.61±0.29cd | ||
| GV–17 | 0.02±0.01bc | 1.06±0.11d | 4.54±0.19c | 5.69±0.29d | 5.56±0.13c | 4.80±0.26cd | 0.05±0.00b | 0.11±0.04de | 0.23±0.03def | 0.31±0.22cde | 0.51±0.05cdef | 0.52±0.08d | ||
| GV–22 | 0.03±0.01bc | 1.19±0.21d | 5.71±0.15b | 7.40±0.10bc | 7.13±0.23ab | 7.04±0.36a | 0.05±0.00b | 0.10±0.03de | 0.24±0.01de | 0.66±0.53ab | 0.70±0.15bc | 0.75±0.13bc | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





