Submitted:
24 March 2023
Posted:
27 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
Understanding Therapeutic Proteins
Reinvention Scope
Intellectual Property
Role of AI
Structure Prediction
Target identification:
- AtomNet is a convolutional neural network-based tool that applies the concepts of feature locality and hierarchical composition extracted through protein sequence, structure, and function to model bioactivity and chemical interactions of potential drug targets.[69] AtomNet’s parent AtomWise has recently enabled the rapid discovery of drugs against 27 disease targets. DeepDTA is also a deep-learning-based model that uses only sequence information of targets and drugs to predict drug-target interaction binding affinities and potential small molecules as drug candidates from given biological data.[70]
- A commercially available natural compounds database and search engine that operates using machine learning, MolPort, when used with Quantitative-Structure-Activity Relationship (QSAR), analyze the chemical structure and predicts the biological activity of potential targets in the early stages of drug discovery.[71]
- Pathway analysis also enables the identification of potential targets. Some crucial biological pathways are available on the Kegg Pathway database,[72] which provides insight into a disease mechanism. TargetNet [73] uses this pathways data and protein interaction profiles to predict potential drug targets against a specific disease.
- DeepDock is the most recent AI-driven virtual screening platform with a vast library of small molecules. For example, DeepDock virtual screen results were used to identify 15% active molecules that led to the discovery of novel compounds against the Mpro protease of SARS-CoV2.[74]
Molecular Docking
- Higher binding affinity scores from an in silico docking analysis of monoclonal antibodies (mAbs) against Alpha and Delta strains of SARS-CoV spike protein suggested that tixagevimab, regdanvimab, and cilgavimab can neutralize most Alpha strains efficiently and bamlanivimab, tixagevimab, and sotrovimab can be effective in suppressing the Delta strain[82]. Venetoclax,[83] for treating chronic lymphocytic leukemia, was designed to target the overexpressed BCL-2 protein in cancer cells by binding to its hydrophobic groove. Its development involved optimizing the binding interactions between the drug and BCL-2 through in-silico docking studies, highlighting the importance of docking in drug design.
- GOLD uses a genetic algorithm, Autodock Vina uses a grid-based energy approach with a genetic algorithm;
- ICM [84] uses multiple stochastic runs. In contrast,
- GLIDE SP [85] uses several sampling and scoring methods. Machine learning model-based pose sampling and evaluation tools like
- DeepBSP, an ML-based sampling and evaluation tool, is very useful in generating and ranking profiles close to their respective native structures.[86]
- Identification of the correct view is crucial for higher binding affinity and lower steric hindrance, which can be efficiently achieved through precise AI-based tools. Structure prediction tools like AlphaFold2 and trRosetta can be integrated with other ML-based approaches to identify and optimize potential poses. One such instance is identifying transition states between the active and inactive conformations of G-Protein Coupled Receptors using multiple ML approaches.[87]
-
The effectiveness of interaction between the dynamic views and their binding partners can be weighted through the scoring systems. Scoring functions are categorized into force-filed-based, knowledge-based, and empirical scoring functions.
- ◦
- Force-field-based scoring functions utilize molecular mechanics to evaluate complex energetic affinity based on their interactions, i.e., weak Van der Waal, electrostatic forces, bond stretching, bending, and torsional angles.[88]
- ◦
- Knowledge-based scoring functions include statistical analysis of distance-dependent atom-pair potentials of protein-ligand or protein-protein complexes generated directly from experimental structures.[89,90] Empirical scoring functions like LUDI,[91] ID-Score,[92] and GlideScore[93] are based on empirical data. They correlate binding free energies to weak Van der Waal energy, electrostatic energy, desolvation, entropy, enthalpy, H-bonding, rotational and translational degrees of freedom, polar and lipophilic effects, and hydrophobicity in the form of simple equations to reproduce experimental affinity data.
- ◦
- These scores are used in combinations for better optimization, i.e., DockThor programs DockTScore[94,95] blends empirical and force-field base scoring methods, SMoG2016[96] fuses empirical and knowledge-based scoring methods, and GalaxyDock BP2 Score[97] uses all three: force-field-based knowledge-based and empirical scoring methods.[98]
- The recent integration of physics-based terms and ML in DockTScore has further enhanced binding energy prediction and conformation ranking.[99]
- GNINA docking software, based on an ensemble of convolutional neural networks as a scoring function for scoring the sample view, has outperformed AutoDock Vina,[100] once again proving that the paradigm shift from conventional methods to AI-based methods has significantly increased the impartial interpretations of scientific evidence leading to the discovery of particular targets.
Structure Modifications
Drug Conjugates
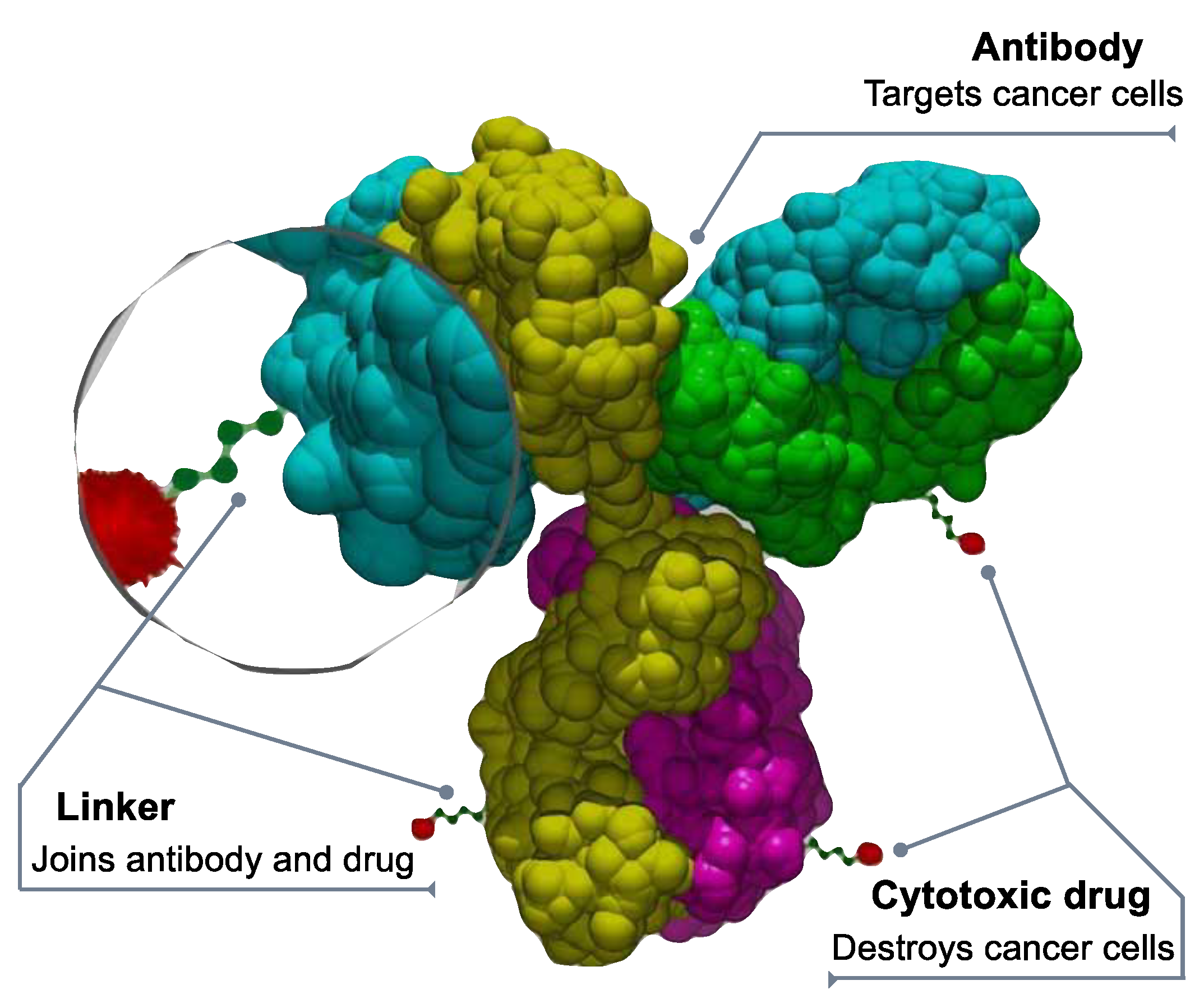
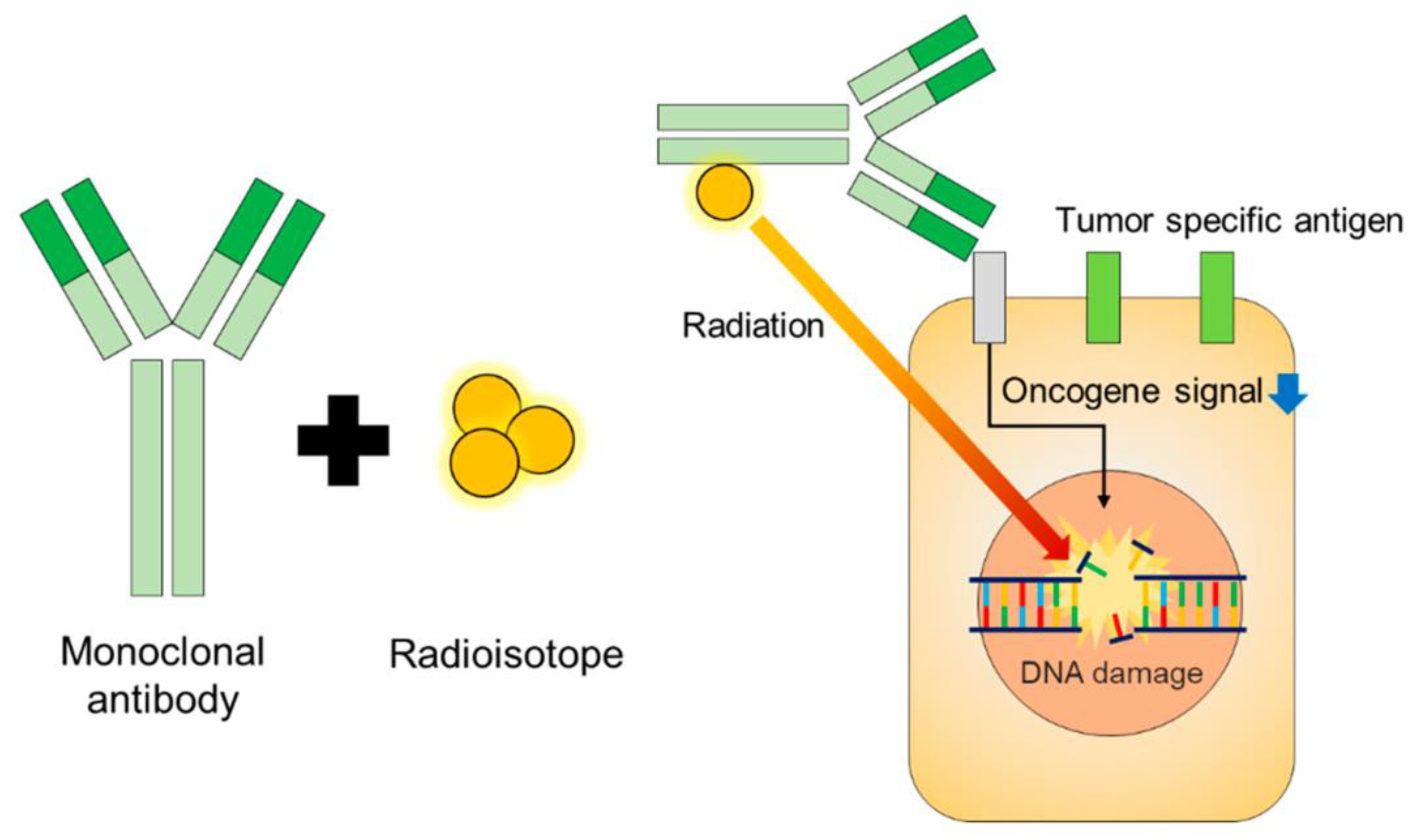
Radioimmunoconjugates (RIC)
Regulatory Perspective
Regulatory Submission
Nonclinical testing
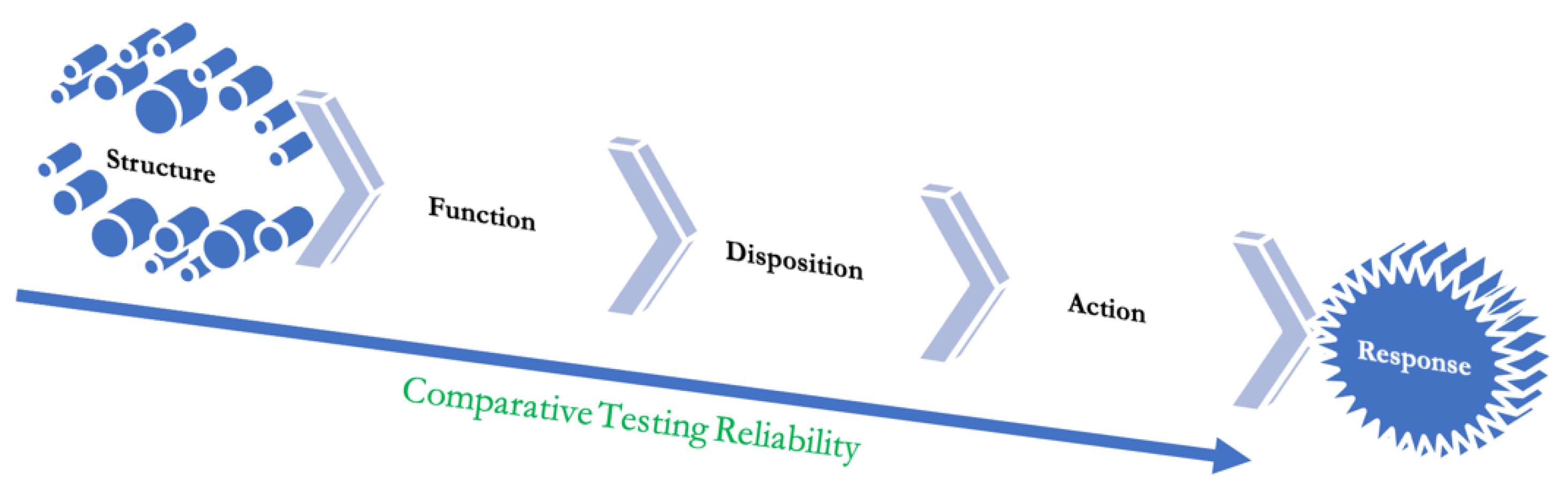
Pharmacokinetics-Pharmacodynamics
Function Testing
Immunogenic Response
Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raju, T.N. The nobel chronicles. 1988: James Whyte Black, (b 1924), Gertrude Elion (1918-99), and George H Hitchings (1905-98). Lancet 2000, 355, 022. [Google Scholar]
- Sean. The Process and Costs of Drug Development (2022). FTLOScience. Available online: https://ftloscience.com/process-costs-drug-development/ (accessed on 5 February 2023).
- Leenaars, C.H.C.; Kouwenaar, C.; Stafleu, F.R.; et al. Animal to human translation: a systematic scoping review of reported concordance rates. J Transl Med. 2019, 17, 23. [Google Scholar] [CrossRef] [PubMed]
- Papapetropoulos, A.; Szabo, C. Inventing new therapies without reinventing the wheel: the power of drug reinventing. Br J Pharmacol. 2018, 175, 65–167. [Google Scholar] [CrossRef] [PubMed]
- Pearce, R.M. Chance and the prepared mind. Science 1912, 35, 41–956. [Google Scholar] [CrossRef] [PubMed]
- Wermuth, C.G. Selective optimization of side activities: the SOSA approach. Drug Discov Today 2006, 11, 60–164. [Google Scholar] [CrossRef] [PubMed]
- Prosdocimi, M.; Zuccato, C.; Cosenza, L.C.; et al. A Rational Approach to Drug Repositioning in β-thalassemia: Induction of Fetal Hemoglobin by Established Drugs [version 3; peer review: 2 approved]. Wellcome Open Res 2022, 7, 150. [Google Scholar] [CrossRef] [PubMed]
- Bomprezzi, R. Dimethyl fumarate in the treatment of relapsing-remitting multiple sclerosis: an overview. Ther Adv Neurol Disord. 2015, 8, 20–30. [Google Scholar] [CrossRef] [PubMed]
- Blair, H.A. Dimethyl fumarate: a review in moderate to severe plaque psoriasis. Drugs 2018, 78, 23–130. [Google Scholar] [CrossRef] [PubMed]
- Santoro, M.G.; Carafoli, E. Remdesivir: from Ebola to COVID-19. Biochem Biophys Res Commun. 2021, 538, 45–150. [Google Scholar] [CrossRef]
- Beck, B.R.; Shin, B.; Choi, Y.; Park, S.; Kang, K. Predicting commercially available antiviral drugs that may act on the novel coronavirus (SARS-CoV-2) through a drug-target interaction deep learning model. Comput. Struct. Biotechnol. J. 2020, 18, 784–790. [Google Scholar] [CrossRef]
- Gilvary, C.; Elkhader, J.; Madhukar, N.; Henchcliffe, C.; Goncalves, M.D.; Elemento, O. A machine learning and network framework to discover new indications for small molecules. PLoS Comput. Biol. 2020, 16, e1008098. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Wang, M.; Xu, Y.; Wu, Z.; Wang, J.; Zhang, C.; Tang, Y. Drug repositioning by prediction of drug’s anatomical therapeutic chemical code via network-based inference approaches. Brief. Bioinform. 2021, 22, 2058–2072. [Google Scholar] [CrossRef] [PubMed]
- Cong, Y.; Shintani, M.; Imanari, F.; Osada, N.; Endo, T. A New Approach to Drug Repurposing with Two-Stage Prediction, Machine Learning, and Unsupervised Clustering of Gene Expression. OMICS 2022, 26, 339–347. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Available online: https://www.drugs.com/new-indications.html.
- Jackson, D.A.; Symons, R.H.; Berg, P. Biochemical method for inserting new genetic information into DNA of Simian Virus 40: circular SV40 DNA molecules containing lambda phage genes and the galactose operon of Escherichia coli. Proc Natl Acad Sci 1972, 69, 2904–9. [Google Scholar] [CrossRef] [PubMed]
- Berg, P.; Baltimore, D.; Boyer, H.W.; Cohen, S.N.; Davis, R.W.; Hogness, D.S.; Nathans, D.; Roblin, R.; Watson, J.D.; Weissman, S.; Zinder, N.D. Letter: Potential biohazards of recombinant DNA molecules. Science 1974, 185, 303. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, W.; Sandow, J. Recombinant Human Insulins – Clinical Efficacy and Safety in Diabetes Therapy. European Endocrinology 2016, 12, 12–17. [Google Scholar] [CrossRef]
- Usmani, S.S.; Bedi, G.; Samuel, J.S.; Singh, S.; Kalra, S.; Kumar, P.; Ahuja, A.A.; Sharma, M.; Gautam, A.; Raghava, G.P.S. THPdb: Database of FDA-approved peptide and protein therapeutics. PLoS One. 2017, 12, e0181748. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Dimitrov, D.S. Therapeutic proteins. Methods Mol. Biol. 2012, 899, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.biospace.com/article/biologics-market-size-to-hit-usd-719-94-billion-by-2030-/.
- Available online: https://www.fda.gov/media/107622/download.
- Available online: https://www.ncbi.nlm.nih.gov/books/NBK562260/#:~:text=A%20peptide%20is%20a%20short,the%20building%20block%20of%20proteins.
- Niazi, S.K. Molecular Biosimilarity—An AI-Driven Paradigm Shift. Int. J. Mol. Sci. 2022, 23, 10690. [Google Scholar] [CrossRef]
- Zwanzig, R.; Szabo, A.; Bagchi, B. Levinthal’s paradox. Proc Natl Acad Sci U S A. 1992, 89, 20–22. [Google Scholar] [CrossRef]
- Schmidt, T.; Bergner, A.; Schwede, T. Modelling three-dimensional protein structures for applications in drug design. Drug Discov Today. 2014, 19, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Tai, W.; He, L.; Zhang, X.; et al. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 2020, 17, 613–620. [Google Scholar] [CrossRef]
- Available online: https://www.cms.gov/monoclonal#:~:text=Monoclonal%20Antibodies%20to%20Treat%20Mild%2Dto%2DModerate%20COVID%2D19&text=On%20December%2023%2C%202022%2C%20the,with%20severe%20COVID%2D19%20illness.
- Available online: https://www.ema.europa.eu/en/documents/product-information/avastin-epar-product-information_en.pdf.
- Available online: https://www.ajmc.com/view/considerations-for-use-of-bevacizumab-vikg-in-wet-amd.
- Hotzel, I.; et al. A strategy for risk mitigation of antibodies with fast clearance. mAbs 2012, 4, 753–760. [Google Scholar] [CrossRef] [PubMed]
- Sharma, TW.; et al. In silico selection of therapeutic antibodies for development: viscosity, clearance, and chemical stability. Proc Natl Acad Sci 2014, 111, 18601–18606. [Google Scholar] [CrossRef]
- Daniel Lagassé, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.; Erez, T.; Reynolds, I.J.; et al. Drug reinventing from the perspective of pharmaceutical companies. Br J Pharmacol. 2018, 175, 68–180. [Google Scholar] [CrossRef] [PubMed]
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; et al. Drug reinventing approach to fight COVID-19. Pharmacol Rep. 2020, 72, 479–1508. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.; Ursu, O.; Gaulton, A.; et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017, 16, 9–34. [Google Scholar] [CrossRef]
- Available online: https://www.greyb.com/blog/biologics-patents-expiring-2022-2023-2024-2025-2026-2027/.
- Goode, R.; Chao, B. Biological patent thickets and delayed access to biosimilars, an American problem. J Law Biosci. 2022, 9, lsac022. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Moll, S.; Desmoulière, A.; Moeller, M.J.; Pache, J.C.; Badi, L.; Arcadu, F.; Richter, H.; Satz, A.; Uhles, S.; Cavalli, A.; Drawnel, F.; Scapozza, L.; Prunotto, M. DDR1 role in fibrosis and its pharmacological targeting. Biochim. Biophys. Acta (BBA) - Mol. Cell Res. 2019, 1866, 118474. [Google Scholar] [CrossRef]
- Ren, F.; Ding, X.; Zheng, M.; Korzinkin, M.; Cai, X.; Zhu, W.; Mantsyzov, A.; Aliper, A.; Aladinskiy, V.; Cao, Z.; Kong, S.; Long, X.; Man Liu, B. H.; Liu, Y.; Naumov, V.; Shneyderman, A.; Ozerov, I.V.; Wang, J.; Pun, F.W.; Zhavoronkov, A. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chemical Science 2023, 14, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www2.deloitte.com/us/en/insights/industry/life-sciences/artificial-intelligence-biopharma-intelligent-drug-discovery.html.
- Deloitte - Intelligent Drug Discovery. (n.d.). Deloitte. Available online: https://www2.deloitte.com/content/dam/Deloitte/my/Documents/risk/my-risk-sdg3-intelligent-drug-discovery.pdf (accessed on 8 March 2023).
- Dokholyan, N.V. Experimentally-Driven Protein Structure Modeling. Journal of proteomics 2020, 220, 103777. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nature protocols 2006, 1, 2876. [Google Scholar] [CrossRef] [PubMed]
- Bank, R.P.D. (n.d.). PDB Statistics: Protein-only Structures Released Per Year. Available online: https://www.rcsb.org/stats/growth/growth-protein.
- Mirdita, M.; von den Driesch, L.; Galiez, C.; Martin, M.J.; Söding, J.; Steinegger, M. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 2017, 45, D170–D176. [Google Scholar] [CrossRef]
- BFD. (n.d.). Available online: https://bfd.mmseqs.com/.
- Mitchell, A.L.; Scheremetjew, M.; Denise, H.; Potter, S.; Tarkowska, A.; Qureshi, M.; Salazar, G.A.; Pesseat, S.; Boland, M.A.; Hunter, F.M.I.; Hoopen, P.T.; Alako, B.; Amid, C.; Wilkinson, D.J.; Curtis, T.P.; Cochrane, G.; Finn, R.D. EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 2018, 46, D726–D735. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved protein structure prediction using predicted interresidue orientations. Proceedings of the National Academy of Sciences 2020, 117, 1496–1503. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Su, H.; Wang, W.; et al. The trRosetta server for fast and accurate protein structure prediction. Nat Protoc 2021, 16, 5634–5651. [Google Scholar] [CrossRef]
- Kim, D.E.; Chivian, D.; Baker, D. Protein structure prediction and analysis using the Robetta server. Nucleic Acids Res. 2004, 32, W526–W531. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; Millán, C.; Park, H.; Adams, C.; Glassman, C.R.; DeGiovanni, A.; Pereira, J.H.; Rodrigues, A.V.; Van Dijk, A.A.; Ebrecht, A.C.; Baker, D. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Costa, A.D.S.; Fazel-Zarandi, M.; Sercu, T.; Candido, S.; Rives, A. Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv. [CrossRef]
- Wu, R.; Ding, F.; Wang, R.; Shen, R.; Zhang, X.; Luo, S.; Su, C.; Wu, Z.; Xie, Q.; Berger, B.; Ma, J.; Peng, J. bioRxiv. [CrossRef]
- Database, A.P.S. (n.d.). AlphaFold Protein Structure Database. Available online: https://alphafold.ebi.ac.uk/.
- Roy, A.; Kucukural, A.; Zhang, Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols 2010, 5, 725–738. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: protein structure and function prediction. Nature methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; Lepore, R.; Schwede, T. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. Modeller: generation and refinement of homology-based protein structure models. Methods in enzymology 2003, 374, 461–491. [Google Scholar] [CrossRef]
- Deng, H.; Jia, Y.; Zhang, Y. Protein structure prediction. Int. J. Mod. Phys. B 2018, 32. [Google Scholar] [CrossRef] [PubMed]
- Segler MH, S.; Preuss, M.; Waller, M.P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 2018, 555, 604–610. [Google Scholar] [CrossRef]
- Mak, K.K.; Pichika, M.R. Artificial intelligence in drug development: present status and future prospects. Drug Discovery Today 2019, 24, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Kosugi, T.; Ohue, M. Solubility-Aware Protein Binding Peptide Design Using AlphaFold. Biomedicines 2022, 10, 1626. [Google Scholar] [CrossRef] [PubMed]
- Wong, F.; Krishnan, A.; Zheng, E.J.; et al. Benchmarking AlphaFold-enabled molecular docking predictions for antibiotic discovery. Mol Syst Biol. 2022, 18, e11081. [Google Scholar] [CrossRef]
- Available online: https://pandaomics.com/access.
- Ren, F.; Ding, X.; Zheng, M.; Korzinkin, M.; Cai, X.; Zhu, W.; Mantsyzov, A.; Aliper, A.; Aladinskiy, V.; Cao, Z.; Kong, S.; Long, X.; Man Liu, B.H.; Liu, Y.; Naumov, V.; Shneyderman, A.; Ozerov, I. V.; Wang, J.; Pun, F. W.; Zhavoronkov, A. AlphaFold accelerates artificial intelligence powered drug discovery: efficient discovery of a novel CDK20 small molecule inhibitor. Chemical Science 2023, 14, 1443–1452. [Google Scholar] [CrossRef]
- Matsuzaka, Y.; Yashiro, R. Applications of Deep Learning for Drug Discovery Systems with BigData. BioMedInformatics 2022, 2, 603–624. [Google Scholar] [CrossRef]
- Wallach, I.; Dzamba, M.; Heifets, A. AtomNet: A Deep Convolutional Neural Network for Bioactivity Prediction in Structure-based Drug Discovery. ArXiv 2015. [Google Scholar] [CrossRef]
- Öztürk, H.; Ozkirimli, E.; Özgür, A. DeepDTA: Deep Drug-Target Binding Affinity Prediction. ArXiv 2018. [Google Scholar] [CrossRef]
- Ferreira, L.T.; Borba JV, B.; Moreira-Filho, J.T.; Rimoldi, A.; Andrade, C.H.; Costa FT, M. QSAR-Based Virtual Screening of Natural Products Database for Identification of Potent Antimalarial Hits. Biomolecules 2021, 11, 459. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Nakaya, A. The KEGG databases at GenomeNet. Nucleic acids research 2002, 30, 42–46. [Google Scholar] [CrossRef]
- Yao, Z.J.; Dong, J.; Che, Y.J.; Zhu, M.F.; Wen, M.; Wang, N.N.; Wang, S.; Lu, A.P.; Cao, D.S. TargetNet: a web service for predicting potential drug-target interaction profiling via multi-target SAR models. J. Comput. -Aided Mol. Des. 2016, 30, 413–424. [Google Scholar] [CrossRef]
- Gentile, F.; Yaacoub, J.C.; Gleave, J.; Fernandez, M.; Ton, A.T.; Ban, F.; Stern, A.; Cherkasov, A. Artificial intelligence–enabled virtual screening of ultra-large chemical libraries with deep docking. Nature Protocols 2022, 17, 672–697. [Google Scholar] [CrossRef]
- Yang, C.; Chen, E.A.; Zhang, Y. Protein–Ligand Docking in the Machine-Learning Era. Molecules 2022, 27. [Google Scholar] [CrossRef]
- De Ruyck, J.; Brysbaert, G.; Blossey, R.; Lensink, M.F. Molecular docking as a popular tool in drug design, an in silico travel. Adv. Appl. Bioinform. Chem. 2016, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2. 0: New docking methods, expanded force field, and python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Venkatachalam, C.M.; Jiang, X.; Oldfield, T.; Waldman, M. LigandFit: a novel method for the shape-directed rapid docking of ligands to protein active sites. J. Mol. Graph. Model. 2003, 21, 289–307. [Google Scholar] [CrossRef] [PubMed]
- Allen, W.J.; Balius, T.E.; Mukherjee, S.; Brozell, S.R.; Moustakas, D.T.; Lang, P.T.; Rizzo, R.C. DOCK 6: Impact of new features and current docking performance. Journal of computational chemistry 2015, 36, 1132–1156. [Google Scholar] [CrossRef]
- Jones, G.; Willett, P.; Glen, R.C.; Leach, A.R.; Taylor, R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997, 267, 727–748. [Google Scholar] [CrossRef]
- Das, N.C.; Chakraborty, P.; Bayry, J.; Mukherjee, S. In Silico Analyses on the Comparative Potential of Therapeutic Human Monoclonal Antibodies Against Newly Emerged SARS-CoV-2 Variants Bearing Mutant Spike Protein. Front. Immunol. 2022, 12. [Google Scholar] [CrossRef]
- Ramos, J.; Muthukumaran, J.; Freire, F.; Paquete-Ferreira, J.; Otrelo-Cardoso, A.; Svergun, D.; Panjkovich, A.; Santos-Silva, T. Shedding Light on the Interaction of Human Anti-Apoptotic Bcl-2 Protein with Ligands through Biophysical and in Silico Studies. International Journal of Molecular Sciences 2019, 20, 860. [Google Scholar] [CrossRef]
- Neves, M.A.; Totrov, M.; Abagyan, R. Docking and scoring with ICM: the benchmarking results and strategies for improvement. Journal of computer-aided molecular design 2012, 26, 675–686. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; Shaw, D.E.; Francis, P.; Shenkin, P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef]
- Bao, J.; He, X.; Zhang, J.Z.H. DeepBSP—a Machine Learning Method for Accurate Prediction of Protein–Ligand Docking Structures. J. Chem. Inf. Model. 2021, 61, 2231–2240. [Google Scholar] [CrossRef]
- Yadav, P.; Mollaei, P.; Cao, Z.; Wang, Y.; Barati Farimani, A. Prediction of GPCR activity using machine learning. Comput. Struct. Biotechnol. J. 2022, 20, 2564–2573. [Google Scholar] [CrossRef] [PubMed]
- Vemula, D.; Jayasurya, P.; Sushmitha, V.; Kumar, Y.N.; Bhandari, V. CADD, AI and ML in drug discovery: A comprehensive review. European Journal of Pharmaceutical Sciences 2023, 181, 106324. [Google Scholar] [CrossRef]
- Guedes, I.A.; Pereira, F.S.S.; Dardenne, L.E. Empirical Scoring Functions for Structure-Based Virtual Screening: Applications, Critical Aspects, and Challenges. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Pantsar, T.; Poso, A. Binding Affinity via Docking: Fact and Fiction. Molecules 2018, 23, 1899. [Google Scholar] [CrossRef]
- Böhm, H.J. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput. -Aided Mol. Des. 1994, 8, 243–256. [Google Scholar] [CrossRef]
- Li, H.; Leung, K.S.; Wong, M.H.; Ballester, P.J. Low-quality structural and interaction data improves binding affinity prediction via random forest. Molecules 2015, 20, 10947–10962. [Google Scholar] [CrossRef]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Shenkin, P.S. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- de Magalhães, C.S.; Almeida, D.M.; Barbosa HJ, C.; Dardenne, L.E. A dynamic niching genetic algorithm strategy for docking highly flexible ligands. Inf. Sci. 2014, 289, 206–224. [Google Scholar] [CrossRef]
- Guedes, I.A.; Barreto AM, S.; Miteva, M.A.; Dardenne, L.E. Development of empirical scoring functions for predicting protein-ligand binding affinity. Soc. Bras. Bioquim. Biol. Mol 2016, 1–174. [Google Scholar]
- Debroise, T.; Shakhnovich, E.I.; Chéron, N. A hybrid knowledge-based and empirical scoring function for protein–ligand interaction: SMoG2016. Journal of chemical information and modeling 2017, 57, 584–593. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; Shin, W.H.; Chung, H.W.; Seok, C. GalaxyDock BP2 score: a hybrid scoring function for accurate protein–ligand docking. J. Comput. -Aided Mol. Des. 2017, 31, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; Pereira FS, S.; Dardenne, L.E. Empirical Scoring Functions for Structure-Based Virtual Screening: Applications, Critical Aspects, and Challenges. Front. Pharmacol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Guedes, I.A.; Barreto, A.M.S.; Marinho, D.; et al. New machine learning and physics-based scoring functions for drug discovery. Sci Rep 2021, 11, 3198. [Google Scholar] [CrossRef] [PubMed]
- McNutt, A.T.; Francoeur, P.; Aggarwal, R.; Masuda, T.; Meli, R.; Ragoza, M.; Sunseri, J.; Koes, D.R. GNINA 1.0: molecular docking with deep learning. Journal of Cheminformatics 2021, 13. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.; Park, H.; Jung, S.; Kim, E.K.; Cho, M.L.; Min, J.K.; Moon, S.J.; Lee, S.M.; Cho, J.H.; Lee, D.H.; Nam, J.H. Therapeutic Effect of Exogenous Truncated IK Protein in Inflammatory Arthritis. Int. J. Mol. Sci. 2017, 18, 1976. [Google Scholar] [CrossRef] [PubMed]
- Rigi, G.; Kardar, G.; Hajizade, A.; Zamani, J.; Ahmadian, G. The effects of a truncated form of Staphylococcus aureus protein A (SpA) on the expression of cytokines of autoimmune patients and healthy individuals. Europe PMC (not peer-reviewed) 2022. [Google Scholar] [CrossRef]
- Xu, J.; Lloyd, D.J.; Hale, C.; Stanislaus, S.; Chen, M.; Sivits, G.; Vonderfecht, S.; Hecht, R.; Li, Y.S.; Lindberg, R.A.; Chen, J.L.; Young Jung, D.; Zhang, Z.; Ko, H.J.; Kim, J.K.; VéNiant, M.M. Fibroblast Growth Factor 21 Reverses Hepatic Steatosis, Increases Energy Expenditure, and Improves Insulin Sensitivity in Diet-Induced Obese Mice. Diabetes 2009, 58, 250–259. [Google Scholar] [CrossRef]
- Véniant, M.M.; Komorowski, R.; Chen, P.; Stanislaus, S.; Winters, K.; Hager, T.; Xu, J. Long-acting FGF21 has enhanced efficacy in diet-induced obese mice and in obese rhesus monkeys. Endocrinology 2012, 153, 4192–4203. [Google Scholar] [CrossRef]
- Charych, D.H.; Hoch, U.; Langowski, J.L.; Lee, S.R.; Addepalli, M.K.; Kirk, P.B.; Doberstein, S.K. NKTR-214, an engineered cytokine with biased IL2 receptor binding, increased tumor exposure, and marked efficacy in mouse tumor models. Clin. Cancer Res. 2016, 22, 680–690. [Google Scholar] [CrossRef]
- Peters, C.; Brown, S. Antibody-drug conjugates as novel anti-cancer chemotherapeutics. Biosci Rep. 2015, 35, e00225. Available online: https://pubmed.ncbi.nlm.nih.gov/26182432/. [CrossRef] [PubMed]
- Khongorzul, P.; Ling, C.J.; Khan, F.U.; Ihsan, A.U.; Zhang, J. Antibody-Drug Conjugates: A Comprehensive Review. Mol Cancer Res. 2020, 18, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Li, S.; Han, S.; et al. Antibody drug conjugate: the “biological missile” for targeted cancer therapy. Sig Transduct Target Ther 2022, 7, 93. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.bio-itworld.com/pressreleases/2022/11/28/fda-approved-adc-drugs-list-up-to-2022.
- McPherson, M.J.; Hobson, A.D. Pushing the Envelope: Advancement of ADCs Outside of Oncology In Antibody-Drug Conjugates; Tumey, L., Ed.; Methods Mol Biol. Vol. 2078; Humana: New York, NY, USA, 2020; pp. 23–36. [Google Scholar] [CrossRef] [PubMed]
- Alley, S.C.; Okeley, N.M.; Senter, P.D. Antibody-drug conjugates: targeted drug delivery for cancer. Curr Opin Chem Biol 2010, 14, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Beck, A.; et al. The next generation of antibody-drug conjugates comes of age. Discov Med 2010, 10, 329–359. [Google Scholar] [PubMed]
- Ritter, A. Antibody-drug conjugates: looking ahead to an emerging class of biotherapeutic. Pharm Tech 2012, 36, 42–47. [Google Scholar]
- Junttila, T.T.; Li, G.; Parson, K.; Phillips, G.L.; Sliwkowski, M.X. Trastuzumab-DM1 (T-DM1) retains all the mechanisms of action of trastuzumab and efficiently inhibits growth of lapatinib-insensitive breast cancer. Breast Cancer Res Treat 2011, 128, 347–356. [Google Scholar] [CrossRef]
- Schmidt, M.M.; Wittrup, K.D. A modeling analysis of the effects of molecular size and binding affinity on tumor targeting. Mol Cancer Ther 2009, 8, 2861–2871. [Google Scholar] [CrossRef]
- Francisco, J.A.; Cerveny, C.G.; Meyer, D.L.; Mixan, B.J.; Klussman, K.; Chace, D.F.; Rejniak, S.X.; Gordon, K.A.; DeBlanc, R.; Toki, B.E.; Law, C.L.; Doronina, S.O.; Siegall, C.B.; Senter, P.D.; Wahl, A.F. cAC10-vcMMAE, an anti-CD30-monomethyl auristatin E conjugate with potent and selective antitumor activity. Blood 2003, 102, 1458–65. [Google Scholar] [CrossRef] [PubMed]
- Kovtun, Y.V.; Goldmacher, V.S. Cell killing by antibody-drug conjugates. Cancer Lett. 2007, 255, 232–40. [Google Scholar] [CrossRef] [PubMed]
- Stephanie, B.; Mark, L.; Khondaker Miraz, R. Antibody–Drug Conjugates—A Tutorial Review. Molecules 2021, 26, 2943. [Google Scholar] [CrossRef]
- Tim, H.; Liu, J.; Zwaenepoel, O.; Boddin, G.; Van Leene, C.; Decoene, K.; Madder, A.; Braeckmans, K.; Gettemans, J. Nanobody click chemistry for convenient site-specific fluorescent labelling, single step immunocytochemistry and delivery into living cells by photoporation and live cell imaging. New Biotechnol. 2020, 59, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Jaffray, D.A. Image-guided radiotherapy: from current concept to future perspectives. Nat. Rev. Clin. Oncol. 2012, 9, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Nasr, D.; Kumar, P.A.; Zerdan, M.B.; Ghelani, G.; Dutta, D.; Graziano, S.; Lim, S.H. Radioimmunoconjugates in the age of modern immuno-oncology. Life Sci. 2022, 310, 121126. [Google Scholar] [CrossRef] [PubMed]
- Pouget, J.P.; Constanzo, J. Revisiting the radiobiology of targeted alpha therapy. Front. Med. 2018, 8, 692436. [Google Scholar] [CrossRef] [PubMed]
- Grillo-López, A.J. Zevalin: the first radioimmunotherapy approved for the treatment of lymphoma Expert. Rev. Anticancer. Ther. 2002, 2, 485–493. [Google Scholar] [CrossRef]
- Miranda, A.C.C.; Santos, S.N.D.; Fuscaldi, L.L.; Balieiro, L.M.; Bellini, M.H.; Guimarães, M.I.C.C.; de Araújo, E.B. Radioimmunotheranostic pair based on the anti-HER2 monoclonal antibody: influence of chelating agents and radionuclides on biological properties. Pharmaceutics 2021, 13, 971. [Google Scholar] [CrossRef] [PubMed]
- Chiu, K.; Racz, R.; Burkhart, K.; Florian, J.; Ford, K.; Iveth Garcia, M.; Geiger, R.M.; Howard, K.E.; Hyland, P.L.; Ismaiel, O.A.; Kruhlak, N.L.; Li, Z.; Matta, M.K.; Prentice, K.W.; Shah, A.; Stavitskaya, L.; Volpe, D.A.; Weaver, J.L.; Wu, W.W.; Strauss, D.G. New science, drug regulation, and emergent public health issues: The work of FDA’s division of applied regulatory science. Front. Med. 2023, 9. [Google Scholar] [CrossRef]
- U S Food and Drug Administration. Clinical Pharmacology Review for Application 214787Orig1S000 (Remdesivir). (2022). Available online at. (1 B.C.E.). Available online: https://www.accessdata.fda.gov/drugsatfda_docs/nda/2020/214787Orig1s000ClinpharmR.pdf.
- Schotland, P.; Racz, R.; Jackson, D.B.; Soldatos, T.G.; Levin, R.; Strauss, D.G.; Burkhart, K. Target Adverse Event Profiles for Predictive Safety in the Postmarket Setting. Clin. Pharmacol. &Amp; Ther. 2020, 109, 1232–1243. [Google Scholar] [CrossRef]
- Available online: https://www.fda.gov/about-fda/center-drug-evaluation-and-research-cder/division-applied-regulatory-science.
- Yan, H.; Bhagwat, B.; Sanden, D.; Willingham, A.; Tan, A.; Knapton, A.; et al. Evaluation of a TGN1412 analogue using in vitro assays and two immune humanized mouse models. Toxicol Appl Pharmacol. 2019, 372, 7–69. [Google Scholar] [CrossRef]
- Yan, H.; Semple, K.; Gonzalez, C.; Howard, K. Bone marrow-liver-thymus (BLT) immune humanized mice as a model to predict cytokine release syndrome. Transl Res. 2019, 210, 3–56. [Google Scholar] [CrossRef] [PubMed]
- Weaver, J.; Zadrozny, L.; Gabrielson, K.; Semple, K.; Shea, K. Howard KEBLT-. Immune humanized mice as a model for nivolumab-induced immune-mediated adverse events: comparison of the NOG and NOG-EXL strains. Toxicol Sci. 2019, 169, 194–208. [Google Scholar] [CrossRef] [PubMed]
- Daluwatte, C.; Schotland, P.; Strauss, D.; Burkhart, K.; Racz, R. Predicting potential adverse events using safety data from marketed drugs. BMC Bioinformatics 2020, 21, 63. [Google Scholar] [CrossRef] [PubMed]
- Schotland, P.; Racz, R.; Jackson, D.; Soldatos, T.; Levin, R.; Strauss, D.; et al. Target adverse event profiles for predictive safety in the postmarket setting. Clin Pharmacol Ther. 2021, 109, 232–43. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.pistoiaalliance.org/.
- International Council for Harmonisation of Technical Requirements for Pharmaceuticals for Human Use [ICH]. Assessment and Control of DNA Reactive (mutagenic) Impurities in Pharmaceuticals to Limit Potential Carcinogenic Risk. M7(R1). Current Step 4 Version. 2017. Available online: https://database.ich. org/sites/default/files/M7_R1_Guideline.pdf.
- Available online: https://www.youtube.com/watch?v=bNb2fEVKeEo&t=6sExternal Link Disclaimer.
- Available online: https://www.semitorr.com/specialties/particle-sentry-ai-quality-control-in-drug-product-manufacturing/.
- Available online: https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm.
- Available online: https://www.ema.europa.eu/en/medicines/what-we-publish-when/european-public-assessment-reports-background-context.
- Available online: https://www.centerforbiosimilars.com/view/opinion-a-modified-351-a-licensing-pathway-for-biosimilars.
- Wang, Y.C.; et al. Role of modeling and simulation in the development of novel and biosimilar therapeutic proteins. J Pharm Sci 2019, 108, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, S.M. Commentary on fit-for-purpose models for regulatory applications. J Pharm Sci 2019, 108, 18–20. [Google Scholar] [CrossRef] [PubMed]
- Zhu, P.; Hsu, C.H.; Liao, J.; Xu, S.; Zhang, L.; Zhou, H. Trial design and statistical considerations on the assessment of pharmacodynamic similarity. AAPS J 2019, 21, 47. [Google Scholar] [CrossRef]
- Zhu, P.; Ji, P.; Wang, Y. Using clinical PK/PD studies to support No clinically meaningful differences between a proposed biosimilar and the reference product. AAPS J 2018, 20, 89. [Google Scholar] [CrossRef]
- US Food and Drug Administration. FDA Guidance: Bioanalytical Method Validation. 2018. Available online: https://www.fda.gov/media/70858/download (accessed on 1 April 2022).
- Lim, S.H.; Kim, K.; Choi, C.I. Pharmacogenomics of Monoclonal Antibodies for the Treatment of Rheumatoid Arthritis. J Pers Med. 2022, 12, 1265. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Niazi, S. Volume of distribution as a function of time. J Pharm Sci. 1976, 65, 452–4. [Google Scholar] [CrossRef] [PubMed]
- Wesolowski, C.A.; Wesolowski, M.J.; Babyn, P.S.; Wanasundara, S.N. Time Varying Apparent Volume of Distribution and Drug Half-Lives Following Intravenous Bolus Injections. PLoS One. 2016, 11, e0158798. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Gadkar, K.; et al. Mathematical PKPD and safety model of bispecific TfR/BACE1 antibodies for the optimization of antibody uptake in brain. Eur J Pharm Biopharm 2016, 101, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Wittrup, K.D.; et al. Practical theoretic guidance for the design of tumor-targeting agents. Methods Enzymol 2012, 503, 255–268. [Google Scholar] [PubMed]
- Yeung, Y.A.; et al. Engineering human IgG1 affinity to human neonatal Fc receptor: impact of affinity improvement on pharmacokinetics in primates. J Immunol 2009, 182, 7663–7671. [Google Scholar] [CrossRef] [PubMed]
- Deng, R.; et al. Pharmacokinetics of humanized monoclonal anti-tumor necrosis factor-alpha antibody and its neonatal Fc receptor variants in mice and cynomolgus monkeys. Drug Metab Dispos 2010, 38, 600–605. [Google Scholar] [CrossRef]
- Robbie, G.J. A novel investigational Fc-modified humanized monoclonal antibody, motavizumab-YTE, has an extended half-life in healthy adults. Antimicrob Agents Chemother 2013, 57, 6147–6153. [Google Scholar] [CrossRef]
- Kamath, A.V. Translational pharmacokinetics and pharmacodynamics of monoclonal antibodies. Drug Discov Today Technol. 2016, 21, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Castelli, M.S.; McGonigle, P.; Hornby, P.J. The pharmacology and therapeutic applications of monoclonal antibodies. Pharmacol Res Perspect. 2019, 7, e00535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Sweeney, G.D. Variability in the human drug response. Thromb Res Suppl. 1983, 4, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Marchant, B. Pharmacokinetic factors influencing variability in human drug response. Scand J Rheumatol Suppl. 1981, 39, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Babin, V.; Taran, F.; Audisio, D. Late-Stage Carbon-14 Labeling and Isotope Exchange: Emerging Opportunities and Future Challenges. JACS Au. 2022, 2, 1234–1251. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Holford, N.H.; Sheiner, L.B. Kinetics of pharmacologic response. Pharmacol. Ther. 1982, 16, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Keutzer, L.; You, H.; Farnoud, A.; Nyberg, J.; Wicha, S.G.; Maher-Edwards, G.; Vlasakakis, G.; Moghaddam, G.K.; Svensson, E.M.; Menden, M.P.; Simonsson US, H. Machine Learning and Pharmacometrics for Prediction of Pharmacokinetic Data: Differences, Similarities and Challenges Illustrated with Rifampicin. Pharmaceutics 2022, 14, 1530. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Leil, T.A.; Gibiansky, L.; Krishna, M.; Zhang, H.; Gu, H.; Sun, H.; Throup, J.; Banerjee, S.; Girgis, I. Modeling and Simulation of the Pharmacokinetics and Target Engagement of an Antagonist Monoclonal Antibody to Interferon-γ–Induced Protein 10, BMS-986184, in Healthy Participants to Guide Therapeutic Dosing. Clin. Pharmacol. Drug Dev. 2020, 9, 689–698. [Google Scholar] [CrossRef] [PubMed]
- McClellan, J.E.; et al. The ’totality-of-the-evidence’ approach in the development of PF-06438179/GP1111, an infliximab biosimilar, and in support of its use in all indications of the reference product. Therap Adv Gastroenterol. 2019, 12, 756284819852535. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Ryding, J.; Stahl, M.; Ullmann, M. Demonstrating biosimilar and originator antidrug antibody binding comparability in antidrug antibody assays: a practical approach. Bioanalysis 2017, 9, 1395–1406. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, Z.; Luo, W.; Xia, N.; Zhao, Q. Molecular and functional analysis of monoclonal antibodies in support of biologics development. Protein Cell. 2018, 9, 74–85. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Todoroki, K.; Yamada, T.; Mizuno, H.; Toyo’oka, T. Current Mass Spectrometric Tools for the Bioanalyses of Therapeutic Monoclonal Antibodies and Antibody-Drug Conjugates. Anal Sci. 2018, 34, 397–406. [Google Scholar] [CrossRef] [PubMed]
- Láng, J.A.; Balogh, Z.C.; Nyitrai, M.F.; Juhász, C.; Gilicze, A.K.B.; Iliás, A.; Zólyomi, Z.; Bodor, C.; Rábai, E. In vitro functional characterization of biosimilar therapeutic antibodies. Drug Discov Today Technol. 2020, 37, 1–50. [Google Scholar] [CrossRef] [PubMed]
- Cymera, F.; Becka, H.; Rohde, A.; Reusch, D. Therapeutic monoclonal antibody N-glycosylation—structure, function and therapeutic potential. Biologicals 2018, 52, 1–11. [Google Scholar] [CrossRef]
- Prior, S.; et al. International standards for monoclonal antibodies to support pre- and post-marketing product consistency: evaluation of a candidate international standard for the bioactivities of rituximab. MAbs 2018, 10, 129–42. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, H.P.; et al. Characterization and non-clinical assessment of the proposed etanercept biosimilar GP2015 with originator etanercept (Enbrel(®)). Expert Opin Biol Ther. 2016, 16, 1185–95. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/clinical-immunogenicity-considerations-biosimilar-and-interchangeable-insulin-products.
- Zhou, F.; Zhong, Z. Drug Design and Discovery: Principles and Applications. Mol. A J. Synth. Chem. Nat. Prod. Chem. 2017, 22. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://michaelschlander.com/publications-since-2020.html?file=files/downloads/publications/2018/Schlander-et-al-Cost-Drug-Development-2021-PharmacoEconomics.pdf&cid=5702#:~:text=Results%20Estimates%20of%20total%20average,%244.54%20billion%20(2019%20US%24).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




