Introduction
Rhododendron Linnaeus consists of a
category of species with a significant horticultural importance in family Ericaceae, also it is renowned as a world-famous alpine flower for its various ecological varieties and vibrant hues. To date, more than 1,000 species of wild
Rhododendron have been reported, mainly in the East Asia, Southeast Asia and Himalayas [
1]. In China,
Rhododendron is the largest genus of woody plants with approximately 600 species, including many species described after Flora of China was published [
2]. Possibly, southwest to central China is the origin and principal distribution area of
Rhododendron [
3]. Numerous endemic
Rhododendron species have been discovered in Guizhou Province of China so far, where wild
Rhododendron resources are abundant [
4]. Among them, the core area of the Baili-Dujuan Nature Reserve (BDNR) in Bijie City, Guizhou Province, China stretches over 125.8 square kilometers and contains 64 species of 5
Rhododendron subgenus [
5], has been by far the largest, most diverse and best-preserved primitive rhododendron forest in China, and was labeled as "
Rhododendron Kingdom" [
6]. Particularly, BDNR possesses a few locally unique species such as
R. bailiens [
5], which is an endangered species that has been just discovered herein and is on the brink of extinction [
5]. Characterization of genetic diversity and genotyping the
Rhododendron germplasms may substantially facilitate the preservation of
Rhododendron species.
With respect to the importance of rhododendron plants and the scarcity of some varieties,
in vitro culture has intensively received an increasing attention [
7].
In vitro propagation provides a technical attempt for the germplasm preservation of
Rhododendron, and the studies have been carried out on a variety of germplasms so far. Wei et al. (2018) , for example, grew
R. fortunei seedlings
in vitro and developed the most appropriate conditions for micropropagation [
7]. Nowakowska et al (2022) cultured
Rhododendron 'Kazimierz Odnowiciel'
in vitro and demonstrated the effect of cytokinins on shoot proliferation and genetic stability of plants [
8]. However, the in vitro propagation of many endangered
Rhododendron species, e.g.
R. bailiens , has not yet been established so far.
Retrotransposons are important and peculiar genetic elements derived from ancient retrovirus insertion inside plants genome, whose ability to replicate inside the genome highly contributes to the occurrence of genetic variation and the increase in genome size. Based on the sequence characters in genome, several retrotransposon-based marker systems had been established [
9], with the inter-retrotransposon amplified polymorphism (IRAP) being the most prominent. LTR retrotransposon-associated IRAP is a powerful marker because of its ability to detect insertion polymorphism through amplification of the portion of DNA fragment between double retrotransposons [
10]. Due to the simplicity, IRAP technology has been widely employed to detect genetic variation as well as to elucidate genetic diversity of plant species [
11]. However, there has not yet study on IRAP markers in rhododendron so far.
Currently, the objectives of the study was to develop a molecular marker system of rhododendron so as to detect the genetic diversity as well as the genetic fidelity/variation of germplasms. This is primarily achieved by 1) cloning and sequencing the LTR retrotransposon sequences in rhododendron, and developing IRAP markers based on these sequences; 2) evaluating the genetic diversity of 46 rhododendron accessions based on the informativeness from the combination of IRAPs and ISSRs; and 3) detecting the genetic fidelity of the in vitro cultures of R. bailiens, a very endangered species, using the above-mentioned marker system. These results highlight the availability of highly informative ISSR and IRAP markers in the genetic diversity evaluation and the genetic fidelity assessment of rhododendrons, which may facilitate the preservation and genetic breeding in rhododendron plants.
Materials and Methods
Plant Material
Totally, 46 accessions of wild rhododendrons (
Table S1) were collected from the BDNR (27.22°N, 105.84°E) from Bijie City, Guizhou Province, China, and the sampling materials were young leaves in May. Once the samples were collected, they were stored in cryopreservation tubes, placed in liquid nitrogen, transported back to the laboratory, and then stored in a refrigerator at -80 °C for long term storage.
Pods of
R. bailiens containing seeds were collected from the adult trees and brought back to laboratory for seed extraction. After the obtained seeds were first completely dried and then washed with absolute alcohol for 30 seconds. The seeds were then soaked in 10% sodium hypochlorite for approximately 15 minutes, followed by 5 washes with sterile distilled water. For germination, sterilized seeds were plated on WPM basal medium + 0.1 mM kinetin (KT). In addition, 3% (w/v) sucrose and 0.7% (w/v) agar were added to the medium and adjusted to pH 5.5, which was autoclaved at 121°C for 20 min. Seeds germinated in a culture room with a 14-h photoperiod, supplied with bright white light with a light flux density of 50 μmol m
−2 s
-1 and a room temperature of 25°C. After 45-60 days,
R. bailiens seedlings were transferred to callus induction medium (WPM + 0.2mg/L thidiazuron <TDZ> + 0.1mg/L 3-indole butyric acid <IBA>) to induce callus [
12]. Approximately 60 days later, the cultured calli (
Figure S5) were transferred to the proliferation medium (WPM + 1.0mg/L 2,4-dichlorophenoxyacetic acid <2,4-D> + 0.5mg/L 1-naphthalcetic acid <NAA>) for proliferation [
13]. Each individual callus was numbered, and the callus corresponding to each digit proliferated independently. Three groups of callus material in good growth state were selected and subcultured approximately every 40-50 days (still using the proliferation medium).
LTR retrotransposon cloning and sequencing
The reverse transcriptase (RT) domain of the LTR retrotransposon (
Copia and
Gypsy superfamily) in
Rhododendron was cloned using the leaves of
R. delavayi, and the primers were
Copia,
Copia1: 5'-ACNGCNTTYYTNCAYGG-3' and
Copia2: 5'-ARCATRTCRTCNACRTA-3' [
14];
Gypsy,
Gypsy1: 5'-AGMGRTATGTGYGTSGAYTAT-3' and
Gypsy2: 5'-CAMCCMRAAMWCACAMTT-3' [
15]. The PCR amplification volume was 50 µl, of which 25 µl 2×
Taq PCR Mastermix (Tiangen Biochemical Technology <Beijing> Co., Ltd.,containing 0.1 U/µl
Taq polymerase, 500 µM dNTP, 20 mM Tris-HCl, 100 mM KCl and 3 mM MgCl
2, pH8.3), 10-20 ng genomic DNA of
R. delavayi leaf, 0.5 µM of forward primer and reverse primer each, and ddH
2O to constitute the volume of the reaction. The PCR reaction program was pre-denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 1 min, annealing for 1 min (TY1-copia: 53°C; TY3-gypsy: 50°C), extension at 72°C for 1.5 min, and final extension at 72°C for 5 min. The PCR amplified products were electrophoresed at 150 V for 30 min in 2% agarose gel containing GelRed, and then observed and photographed in the gel imaging system Quantum CX5 Edge 18.02a. We cut and separated the target DNA bands, and then purified the resulting DNA fragments using TakaRa MiniBEST Agarose Gel DNA Extraction Kit Ver. 4.0. The PCR amplified product was ligated into the pEASY-T1 Cloning Vector (Transgene Co., Ltd.). The fusion vector was transformed into
Escherichia coli (
E. coli) strain DH5α. After culture on the plate, a single clone was picked for sequencing. The sequence generated by the sequencing was the RT domain of the LTR retrotransposon. Recombinant white clones were screened by PCR amplification (using M13F and M13R primers), after which plasmids were isolated and sequenced using the universal primers, M13F and M13R (Sangon Bioengineering (Shanghai) Co., Ltd.). Each generated sequence was the RT domain sequence of an LTR retrotransposon. The obtained sequences were performed multiple sequence alignments and calculated the evolutionary distance through the maximum likelihood method with 1000 bootstraps to construct a phylogenetic tree by employing the ClustalW in the MEGA7 software [
16] .
Design of IRAP primer
IRAP primers were designed according to the conserved sequences of the RT domains of TY1
-Copia and TY3
-Gypsy (
Table 2). Primers were synthesized by Shanghai Sangon Biotechnology Co., Ltd. Genomic DNA from leaves of
R. delavayi was selected as template DNA , and the reaction system was (10 µL): 10-50 ng of template DNA, 1 µL of primers (10 µM), 5 µL 2×
Taq PCR Mastermix and appropriate amount of ddH
2O. The PCR reaction procedure was similar to that described above see "LTR retrotransposon cloning and sequencing" section), except that the annealing temperature was set as a gradient (55-60°C) to screen the optimal annealing temperature for each IRAP primer. The PCR amplification product was electrophoresed at 150 V for 50 min in 2% agarose gel containing GelRed, and then the gel was observed and photographed in the gel imaging system.
Polymorphism detection of wild rhododendron accessions
For the dozens of rhododendron accessions for polymorphism detection (
Table S1), a total of 8 IRAP primers and 6 polymorphic ISSR primers were used (
Table 3). The reaction system for IRAP amplification and ISSR amplification was the same as described above (see "LTR retrotransposon cloning and sequencing" section), except that the annealing temperature was adjusted for each primer (
Table 1). The PCR amplification product was electrophoresed at 150V for 50 min in 2% agarose gel containing GelRed, and then the gel was observed and photographed in the gel imaging system.
Genetic fidelity assessment for in vitro cultures
To detect genetic fidelity of in vitro R. bailiense, calli were collected at the primary (C0, 1 clone), the first (C1, 4 clones), and the fourth (C4, 10 clones) cycle. About 0.1 g of callus was isolated to extract DNA, which was used for IRAP and ISSR amplification to assess the genetic fidelity.
Data analysis
The GelQuest software (
https://www.sequentix.de/gelquest/) was employed for statistics, and only clear bands of ISSR or IRAP were counted as “1” (presence), conversely, absences were as “0”. Low-intensity bands (difficult to distinguish presence or absence) were not considered in this step.
We used POPGENE1.32 software [
17], as subjected to Hardy-Weinberger balance, to compute the average number of Shannon's information index (I), Nei's genetic diversity index (H) and effective number of alleles (Ne).
To compare the suitability of the IRAP and ISSR markers, the information content and level of polymorphism were assessed with several parameters including resolving power (Rp), effective multiplex ratio (EMR), marker index (MI), polymorphic information content (PIC), and mean of PIC [
18].
The calculation of PIC index for each loci comes from the following formula[
19]:
PICi = 2fi (1 - fi), where PICi represents the PIC value of loci i, and fi represents the frequency of the amplified bands. The PIC for each primer is calculated by taking the average of the PIC values for all loci.
The method of calculating MI was through the following formula [
20]:
MI = EMR × PIC, where EMR is the product of the proportion of the number of polymorphic loci an assay and the number of polymorphic loci.
The calculation of the Rp value of each primer referred to the following formula[
21]: RP=∑Ib, where Ib represents the fragment that provides information. This parameter can be calculated by the following formula: Ib = 1 - [2 × (0.5 - p)], where p represents the proportion of individuals that contain a band.
Cluster analysis
Numerical Taxonomy System (NTSYS-pc) ver. 2.10e was used to construct dendrograms and similarity matrices [
22]. The Jaccard's similarity coefficient between pair of accessions was then calculated using the binary data matrix via the SIMQUAL model [
22]. The above distance coefficients were employed to construct a dendrogram through the unweighted grouping method arithmetic mean method (UPGMA), and the Dice similarity coefficient algorithm was applied to determine the genetic diversity among accessions. To highlight the classification resolution, we performed principal coordinates analysis (PCoA) using the EIGEN module of NTSYS-pc. Once the PCoA cluster diagram was generated, Adobe illustrator CS5 (
https://www.adobe.com/products/illustrator.html) was used to embellish the image.
Results
Identification of polymorphic IRAP markers
The amplified band fragments of the reverse transcription domain (RT domain) of TY1-
Copia and TY3-
Gypsy in
R. delavayi were obtained by PCR amplification, and the sizes were approximately 250 bp and 400 bp respectively (
Figure S2). The amplified bands were purified for DNA fragment cloning. Approximately 60 single clones from each superfamilies were randomly selected for sequencing. A total of 23 independent sequences for TY1-
Copia were obtained after removing repetitions, with a length of 234-245 bp and an average GC content of 44.78% (
Table S2). The generated TY1-
Copia can be divided into 4 main clades (
Figure S3A), among which group I covers the most TY1-
Copia, reaching 15.
For TY3-
Gypsy, a total of 23 independent sequences were obtained after removing repeats, with a length of 348-503 bp and an average GC content of 43.14% (
Table S2). The sequence of TY3-
Gypsy demonstrated higher heterogeneity and might be divided into 7 clades (
Figure S3B), and the clade with the largest number of TY3-
Gypsy has only 4 TY3-
Gypsy, which were group I and group IV. According to the identified RT domain sequences of TY1-
Copia and TY3-
Gypsy, we designed IRAP primers at appropriate positions (
Table S2) for subsequent experiments.
The annealing temperature highly affects the amplification polymorphism of molecular marker, thus, it is very necessary to screen the appropriate annealing temperature of IRAP markers. The genomic DNA of
R. delavayi was selected for the annealing temperature test and polymorphism identification of IRAP markers. Many IRAP markers of TY1-
Copia showed good polymorphism (
Figure S4), such as LTR1-7 (56°C), LTR1-10 (59°C), LTR1-11 (59°C), LTR1-12 (59°C), LTR1-15 (55°C), LTR1-16 (55°C), LTR1-17 (56°C), etc., and then we obtained their optimum annealing temperature through the above screening. However, the IRAP molecular markers designed by TY3-
Gypsy generally failed to show good amplification polymorphism (
Figure S4), and only one IRAP primer was screened, LTR3-21 (57°C).
Informative identification of IRAP and ISSR markers
Hong et al. (2012) [
23] found 11 ISSR polymorphic primers suitable for
Rhododendron identification. Currently, 6 ISSR primers,
i.e. UBC826, UBC835, UBC836, UBC840, UBC890 and UBCM06, generated clear and highly polymorphic bands for the 46
Rhododendron accessions. Functional test of the selected IRAP and ISSR markers (
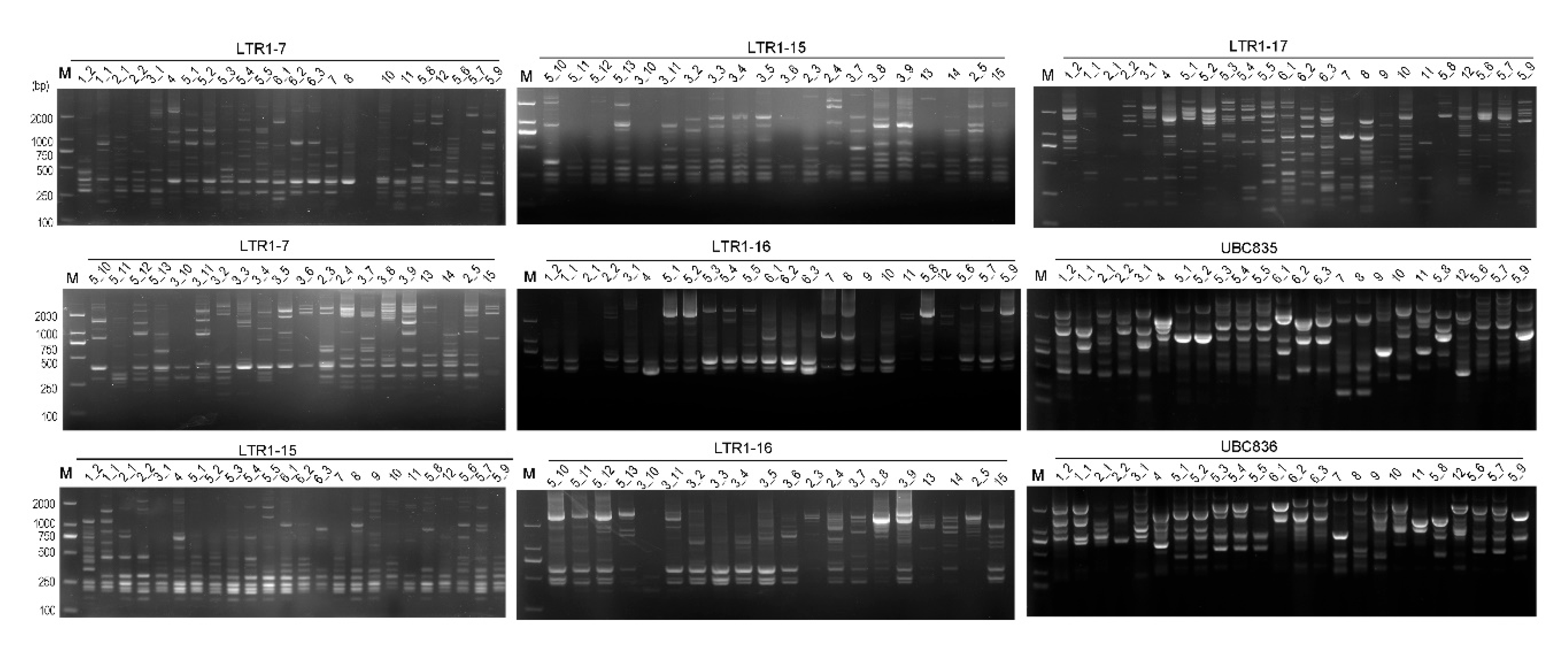
Table 1) revealed that they had good reproducibility and resolution for the 46 accessions concerned (
Figure 1). For IRAP markers, a total of 123 scorable amplified fragments (alleles) were obtained, of which 119 (96.7%) were polymorphic, with an average of 14.9 polymorphic loci generated per primer, with a size of 0.15 -2.7 kb (
Table 1). Among them, LTR3-21 produced the most polymorphic loci (17), while LTR1-7 and LTR1-11 generated the least (both 13). For ISSR markers, a total of 86 fragments were scored, 79 of which (91.9%) were polymorphic, and an average of 13.2 polymorphic loci were harvested per primer, with a size range of 0.2-2.6 kb. UBC836 and UBC840 generated the most polymorphic loci (both 16), however UBC826 generated only 10 polymorphic loci.
Among IRAP primers, LTR1-17 gave the highest Ne (1.77) and I (0.61), and LTR3-21 had the highest H (0.43) (
Table 1). The average values of Ne, H and I for all IRAP primers were 1.59, 0.35 and 0.53, respectively. In terms of ISSR primers, UBC840 had the highest Ne (1.84), H (0.45) and I (0.64), and the average values of Ne, H, and I of all ISSR primers were 1.69, 0.39 and 0.57, respectively. Overall, the ISSR markers were slightly larger than the IRAP markers with respect to Ne, H, and I parameters.
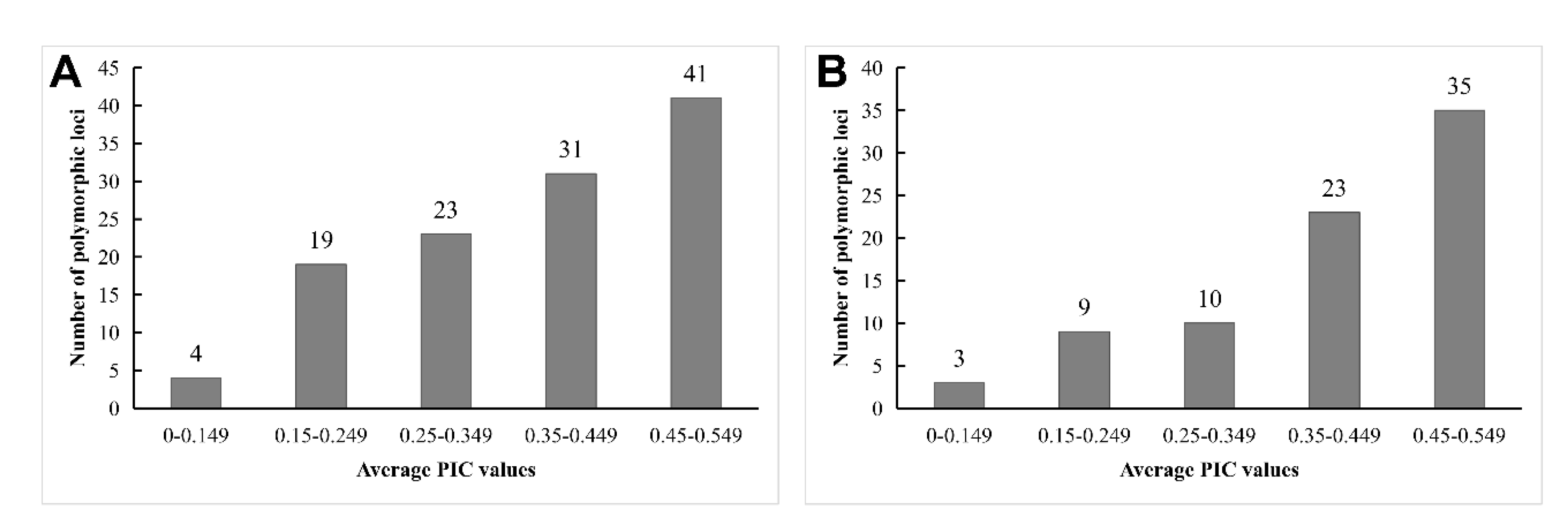
The mean PIC for IRAP markers ranged from 0.294 (LTR1-15) to 0.425 (LTR3-21), with an overall mean of 0.35 (
Table 1). Most of the polymorphic loci of IRAP markers had PIC values ranging from 0.45 to 0.549 (
Figure 2A). The mean PIC of ISSR primers was close to that of IRAP primers, 0.388, ranging from 0.326 (M06) to 0.448 (UBC840). Moreover, the PIC values of the polymorphic loci of most ISSR markers were also located at 0.45-0.549 (
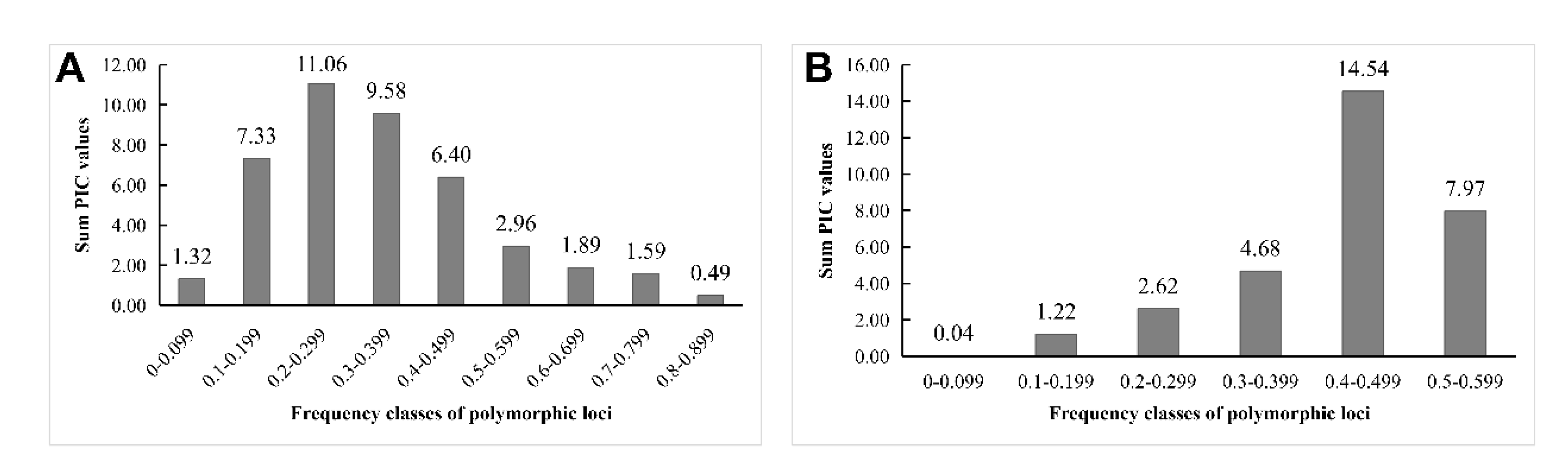
Figure 2B). To address the question of how the sum of PICs relates to the frequency of polymorphic loci, we analyzed the correlation between the two parameters (
Figure 3). For the IRAP markers, the most informative markers (total PIC value was 11.06) were located in the frequency class of polymorphic 0.200-0.299 loci, followed by the frequency category of 0.3 to 0.399. Among the ISSR loci, polymorphic loci with frequency classes 0.400-0.499 were the most informative, followed by frequency classes 0.500-0.599.
Among the IRAP markers, the highest EMR values was from LTR3-21 (17) and the lowest was from LTR1-7 (12) (
Table 1). Meanwhile, primer LTR1-17 had the highest Rp value (11.07) and MI value (7.23), while the lowest values came from LTR1-16 (6.18 and 4.11, respectively). The largest EMRs among ISSR primers were generated at UBC836 and UBC840 (both 16). And the largest Rp and MI were generated in UBC840 (12 and 7.15, respectively). Interestingly, the average Rp of ISSR markers was slightly greater than that of IRAPs, but the EMR and MI of IRAPs were greater than those of ISSRs.
Figure 1.
Electrophoresis profiles of the rhododendron accessions from BDNR as revealed by inter-retrotransposon amplified polymorphism (IRAP) and inter-simple sequence repeat (ISSR). The lanes corresponding to the accessions (
Table S1) were marked on the top of the gel map. The characters on the horizontal line were IDs of IRAP or ISSR markers. Lanes that did not exhibit bands were retested and the binary data obtained were incorporated into subsequent analyses. M, D2000 DNA marker (Beijing Tiangen Biological Company).
Figure 1.
Electrophoresis profiles of the rhododendron accessions from BDNR as revealed by inter-retrotransposon amplified polymorphism (IRAP) and inter-simple sequence repeat (ISSR). The lanes corresponding to the accessions (
Table S1) were marked on the top of the gel map. The characters on the horizontal line were IDs of IRAP or ISSR markers. Lanes that did not exhibit bands were retested and the binary data obtained were incorporated into subsequent analyses. M, D2000 DNA marker (Beijing Tiangen Biological Company).
Figure 2.
Distribution of average PICs from IRAP (A) and ISSR (B) markers among 46 rhododendron accessions.
Figure 2.
Distribution of average PICs from IRAP (A) and ISSR (B) markers among 46 rhododendron accessions.
Figure 3.
The relationship between the frequency of polymorphic loci and the total PIC value, which was amplified by IRAP (A) and ISSR (B) markers in 46 rhododendron materials.
Figure 3.
The relationship between the frequency of polymorphic loci and the total PIC value, which was amplified by IRAP (A) and ISSR (B) markers in 46 rhododendron materials.
Table 1.
Information content of IRAP and ISSR markers and other polymorphism indices from 46 wild Rhododendron accessions.
Table 1.
Information content of IRAP and ISSR markers and other polymorphism indices from 46 wild Rhododendron accessions.
| |
Ne |
H |
I |
Number of polymorphic alleles |
Range of alleles (kb) |
PIC |
EMR |
Rp |
MI |
Annealing temperature (℃) |
| IRAP |
|
|
|
|
|
|
|
|
|
|
| LTR1-7 |
1.62 |
0.37 |
0.55 |
13 |
0.2-2.3 |
0.37 |
12.07 |
6.98 |
4.41 |
56 |
| LTR1-10 |
1.54 |
0.33 |
0.51 |
15 |
0.3-2.7 |
0.33 |
15.00 |
7.20 |
5.01 |
59 |
| LTR1-11 |
1.64 |
0.37 |
0.56 |
13 |
0.4-2.4 |
0.37 |
14.00 |
7.07 |
5.25 |
59 |
| LTR1-12 |
1.47 |
0.30 |
0.47 |
16 |
0.3-2.9 |
0.30 |
16.00 |
6.38 |
4.83 |
59 |
| LTR1-15 |
1.51 |
0.32 |
0.49 |
15 |
0.2-2.4 |
0.32 |
13.24 |
6.70 |
4.20 |
55 |
| LTR1-16 |
1.51 |
0.31 |
0.48 |
14 |
0.15-2.6 |
0.31 |
13.07 |
6.18 |
4.11 |
55 |
| LTR1-17 |
1.77 |
0.42 |
0.61 |
16 |
0.2-2.5 |
0.42 |
16.00 |
11.07 |
6.78 |
56 |
| LTR3-21 |
1.76 |
0.43 |
0.61 |
17 |
0.2-2.5 |
0.43 |
17.00 |
11.00 |
7.23 |
57 |
| Mean |
1.60 |
0.36 |
0.53 |
14.88 |
- |
0.36 |
14.55 |
7.82 |
5.23 |
57 |
| |
|
|
|
|
|
|
|
|
|
|
| ISSR |
|
|
|
|
|
|
|
|
|
|
| UBC826 |
1.79 |
0.43 |
0.62 |
10 |
0.3-2.0 |
0.43 |
8.33 |
7.16 |
3.61 |
59 |
| UBC835 |
1.72 |
0.41 |
0.60 |
15 |
0.3-2.4 |
0.41 |
15.00 |
9.14 |
6.20 |
59 |
| UBC836 |
1.69 |
0.39 |
0.58 |
16 |
0.2-2.6 |
0.38 |
16.00 |
9.54 |
6.05 |
59 |
| UBC840 |
1.83 |
0.45 |
0.64 |
16 |
0.2-2.3 |
0.45 |
16.00 |
12.00 |
7.15 |
59 |
| UBC890 |
1.59 |
0.34 |
0.51 |
11 |
0.3-2.3 |
0.34 |
9.31 |
6.18 |
3.18 |
59 |
| UBCM06 |
1.48 |
0.30 |
0.46 |
11 |
0.2-2.2 |
0.30 |
8.64 |
4.75 |
2.55 |
59 |
| Mean |
1.69 |
0.39 |
0.57 |
13.17 |
- |
0.38 |
12.21 |
8.13 |
4.79 |
59 |
Detection of genetic diversity among rhododendron accessions
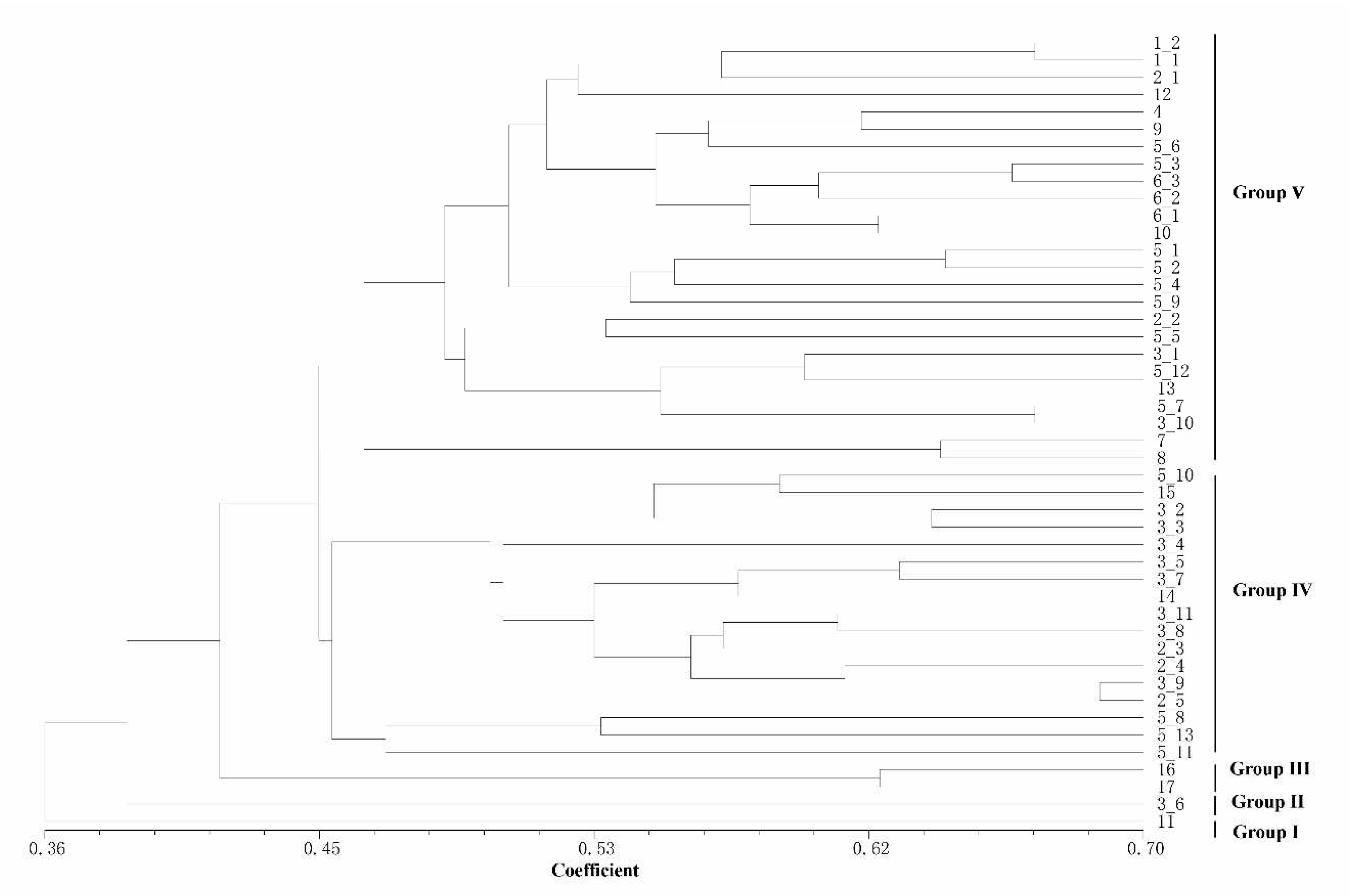
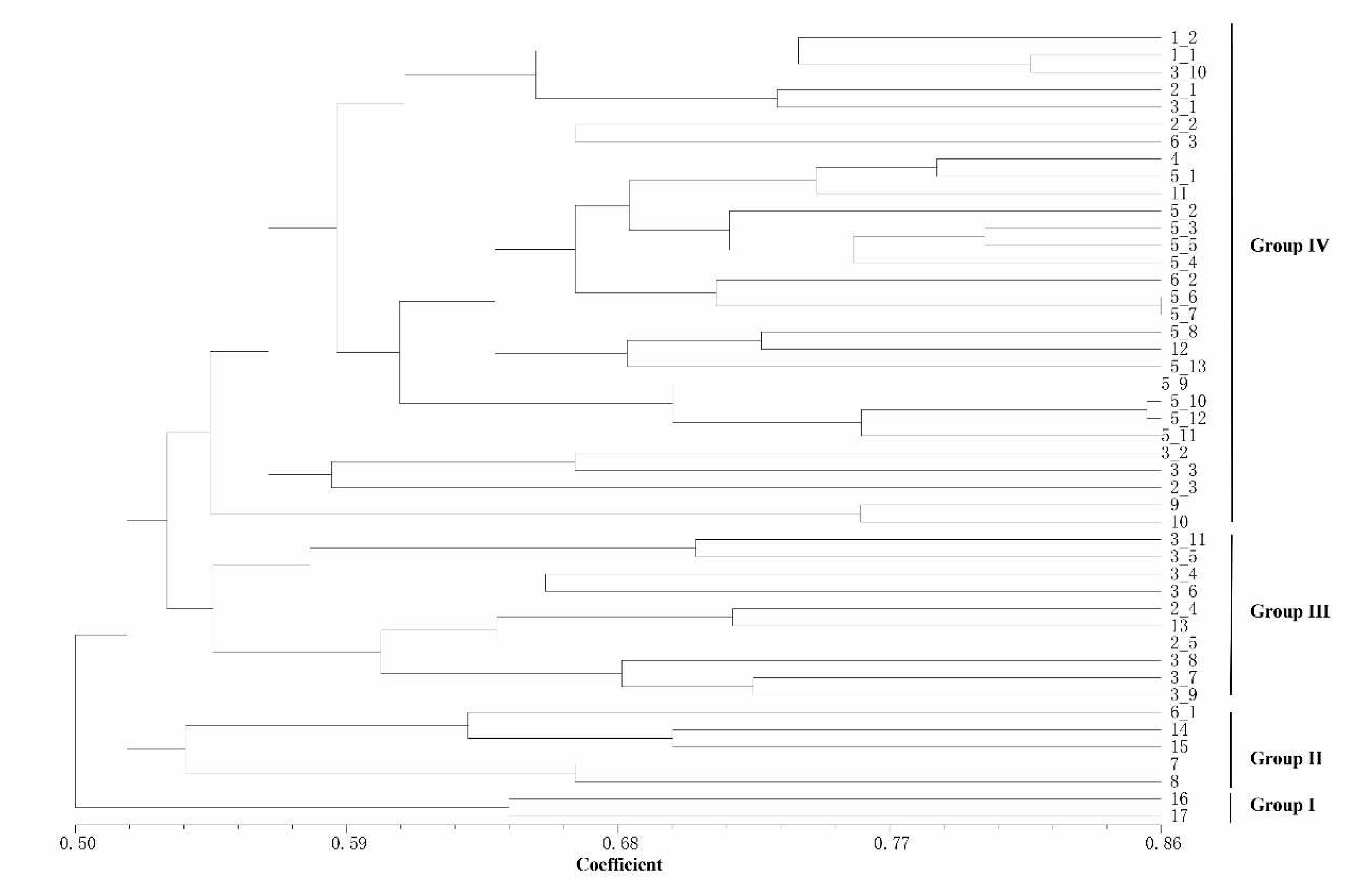
Based on the detection results of IRAP and ISSR markers, the UPGMA clustering algorithm was used to cluster 17
Rhododendron species (46 accessions), which can be divided into 5 and 4 clusters, respectively (
Figure 4 and
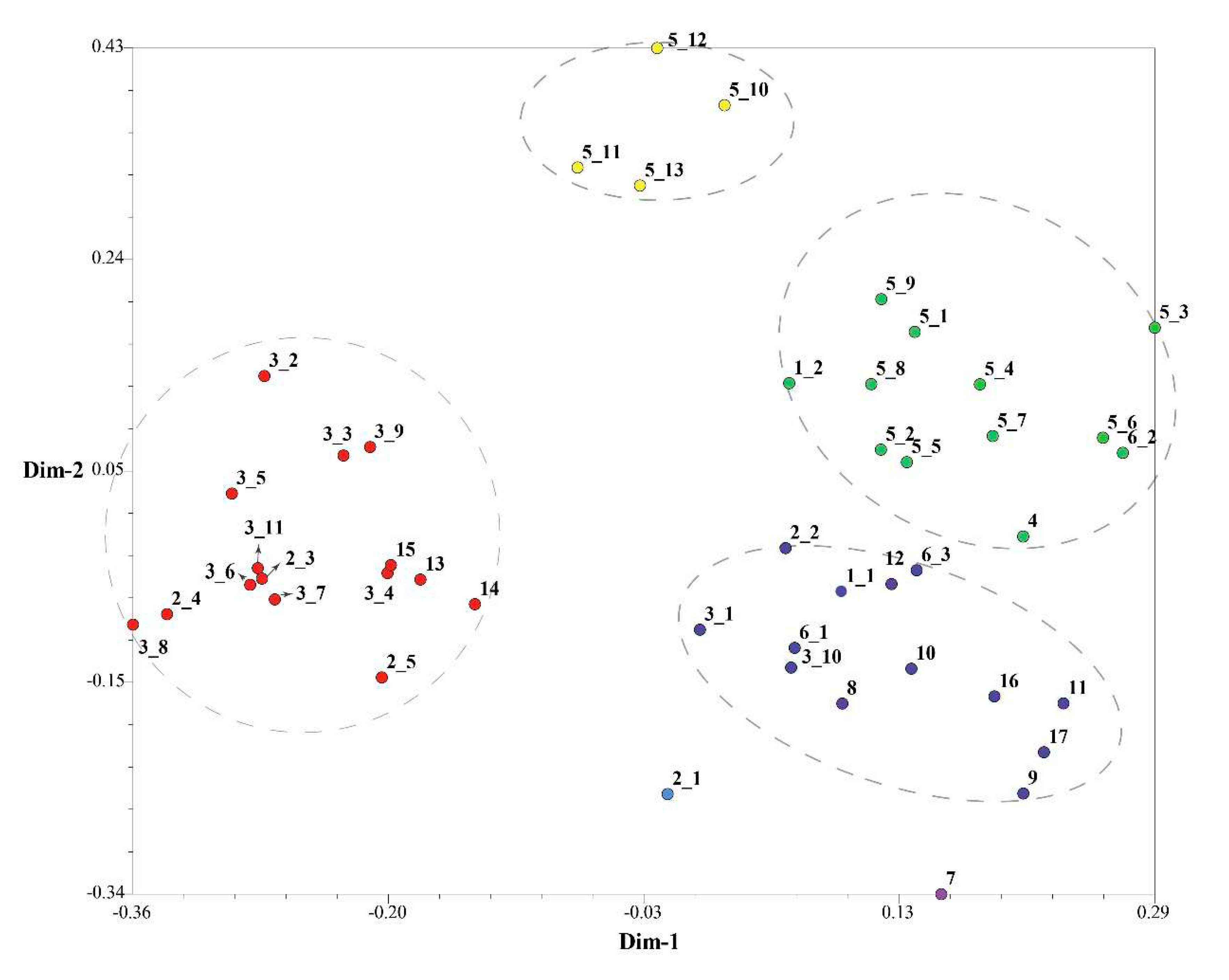
Figure 5). The dendrogram plotted by the IRAP markers showed that accessions 11 and 3_6 fell into two distinct clusters (respectively Group I and Group II). Accessions 16 and 17 were also split into a separate group (Group III). The other accessions were grouped into two further subclusters (Group IV and Group V). In the dendrogram plotted by ISSR markers, Group IV was the most (29), followed by Group III (10), Group II (5) and Group I (2). Because ISSRs and IRAPs represent different genomic components (repeated sequences and retrotransposons, respectively), we merged the two sets of data, and finally 6 groups after superclustering were obtained. This joint analysis resulted in the separation of the two accessions 2_1 and 7 separately (
Figure 6).
The accessions 2_1 and 7 are far apart from another group of
Rhododendron decorum (5_10-5_13).
R. decorum and other ones (5_1-5_9) did not gather together, indicating that their genomes might have mutated. Interestingly, the two separated
R. decorum clusters also belonged to two populations (
Table S1), suggesting that geographical distances may have contributed to intraspecies differences. Furthermore, accessions 3_1 and 3_10 (
R. delavayi) were not found to cluster with any other
R. delavayi (accessions 3_2 and 3_3, etc.), which also showed that their genomes were different from other
R. delavayi accessions. The three groups with the largest number of accessions (the red dots, the purple dots and the green dots) clustered 15, 13 and 12
Rhododendron accessions, respectively. In summary, the combinatorial clustering format with the combination of the two marker systems was expected to be more discriminative than the single procedure.
For the three populations involved in this study (
R. delavayi,
R. decorum and
R. agastum),
R. decorum demonstrated the highest Ne (1.38), H (0.25) and I (0.39) (
Table 2), which may indicate that the population diversity of
R. decorum should be the most abundant in this area, partially reflecting the better adaptation of
R. decorum to the environment in BDNR of Guizhou Province, China.
Table 2.
Genetic variation information of three Rhododendron populations.
Table 2.
Genetic variation information of three Rhododendron populations.
| Populations |
Sample Size |
Ne |
H |
I |
| R. delavayi |
11 |
1.32 |
0.21 |
0.33 |
| R. decorum |
13 |
1.38 |
0.25 |
0.39 |
| R. agastum |
5 |
1.35 |
0.22 |
0.33 |
Genetic fidelity assessment of in vitro cultures of the endangered R. bailiens
Based on the IRAPs and ISSRs profiles from the 46
Rhododendron accessions, IRAP and ISSR markers were employed to evaluate genetic variation of callus subcultured different cycles of
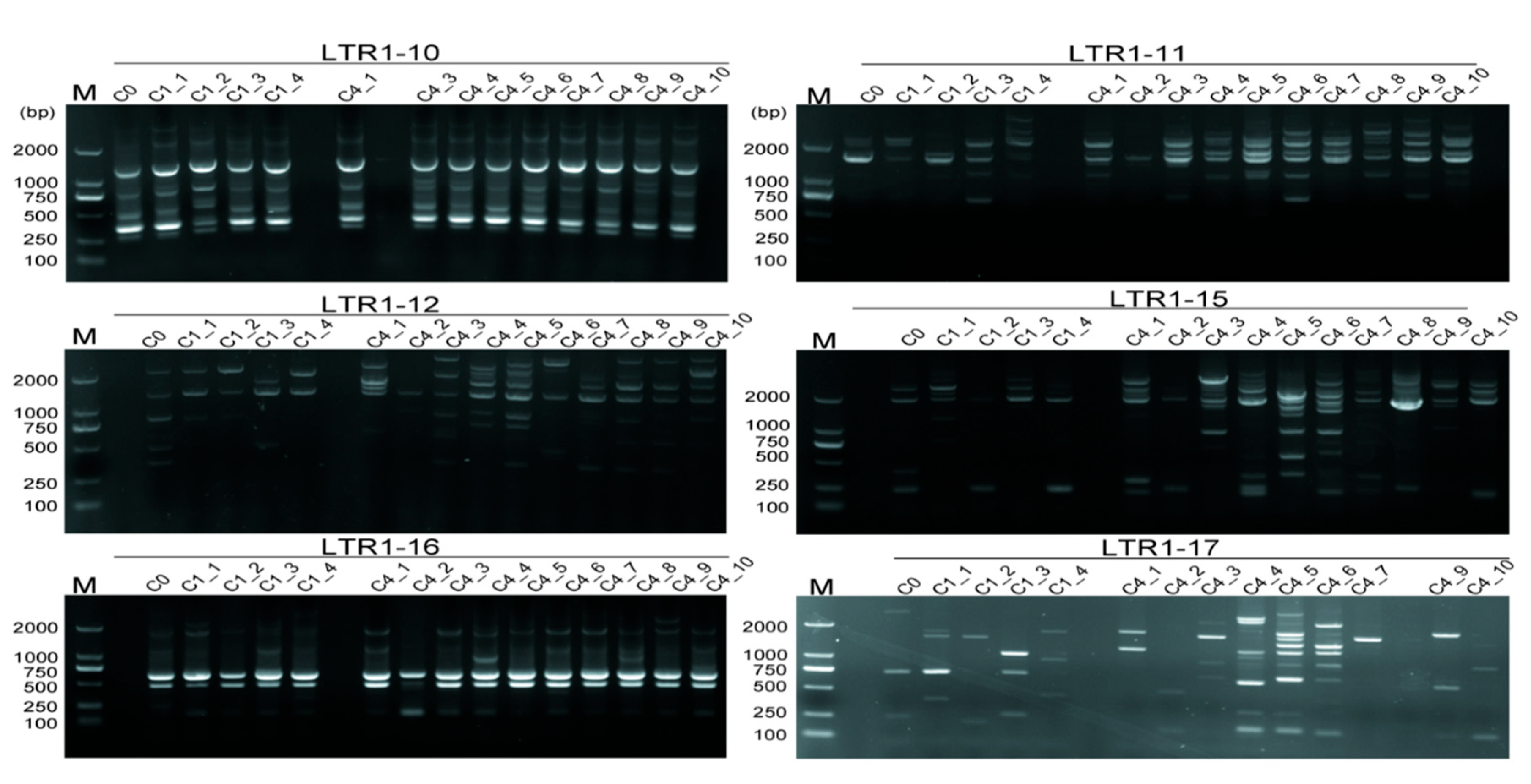
R. bailiens, an endangered species only discovered in BDNR. The fingerprints yielded from 6 IRAP primers and 6 ISSR primers with the most polymorphic loci demonstrated that they were effective for somaclonal variation detection (
Table 3). In
R. bailiens, IRAPs were effective in detecting genetic fidelity from
in vitro cultures (
Figure 7). Among them, some primers,
e.g. LTR1-15 and LTR1-17, produced more polymorphic bands. In terms of ISSRs, the 6 ISSR primers were also effective in detecting the genetic fidelity of
R. bailiens during subculture (
Figure S6), however, ISSRs were obviously less effective than IRAPs in somaclonal variation assessment since the former detected far fewer polymorphic fragments (
Figure 7 and
Figure S6,
Table 3).
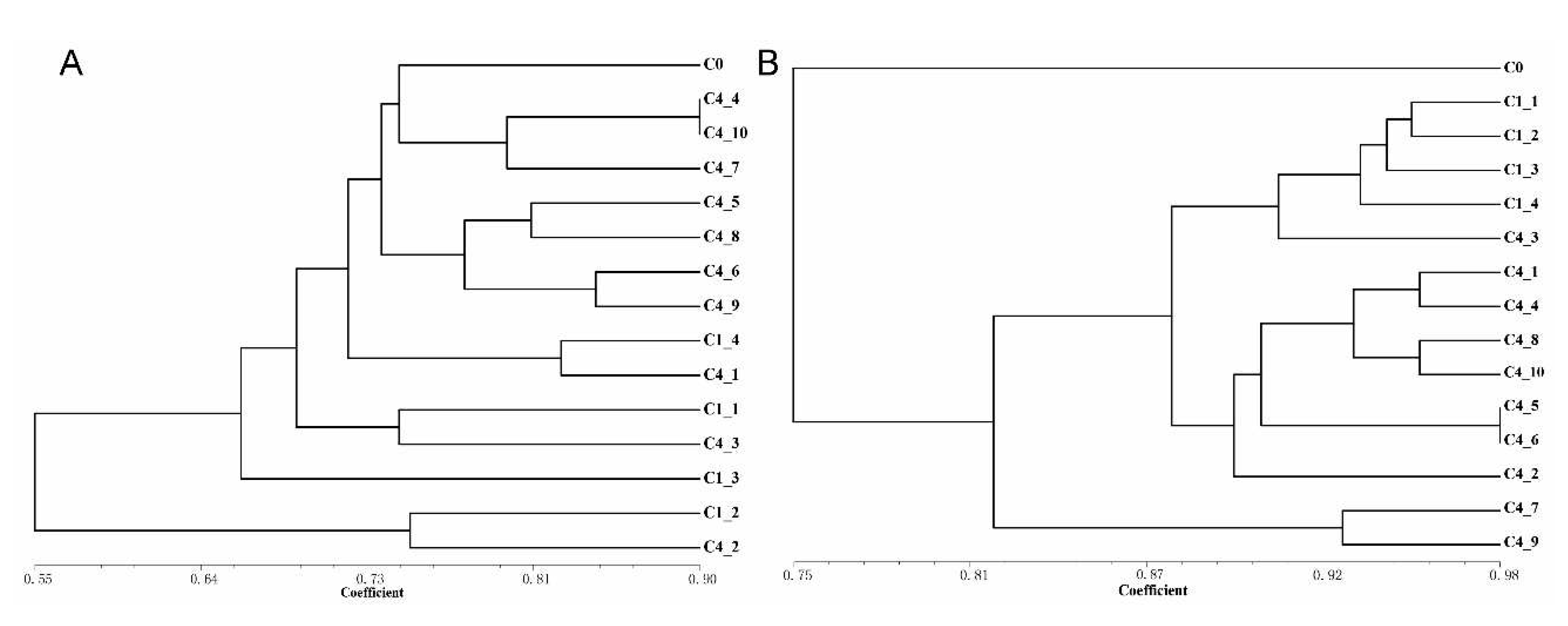
The Jaccard similarity coefficients between the callus of different subculture cycles were calculated based on the results of marker detection, and the UPGMA cluster analysis was carried out (
Figure 8). Generally, the most similarity in IRAPs was investigated among the subclones within same cycle, and the higher genetic abberation was observed beyond the cycles (
Figure 8A). Within the ISSRs detection, the 4 subclones of C1 and C4_3 were clustered into the same group (
Figure 8B), and the remaining C4 subclones gathered into another one. The similarity coefficients generated by ISSRs were generally larger than those generated by IRAPs, indicating that IRAPs are more effective in somaclonal variation detection, which at least partially reflected that the cause of somaclonal variation of
R. bailiens was ascribed to the activation of retrotransposons.
Table 3.
IRAPs and ISSRs information selected to detect the genetic fidelity of R. bailiens callus.
Table 3.
IRAPs and ISSRs information selected to detect the genetic fidelity of R. bailiens callus.
| Primer ID |
Primer sequence (5'-3') |
Number of scorable bands |
Number of polymorphic band |
| IRAP |
|
|
|
| LTR1-10 |
ATGGTCTTAAGCAGTCACCTC |
8 |
2 |
| LTR1-11 |
GGGCTGAAACAGTCTCCAAGA |
7 |
6 |
| LTR1-12 |
CTCCCAGACAGTGGTACAGAA |
8 |
6 |
| LTR1-15 |
CCCAGACAGTGGTACAGAAAG |
7 |
7 |
| LTR1-16 |
ACAGGCTCCCAGACAGTGGTA |
6 |
2 |
| LTR1-17 |
GAGTCTGTATGGTTTGAAACA |
9 |
9 |
| |
|
|
|
| ISSR |
|
|
|
| UBC826 |
ACACACACACACACACC |
9 |
7 |
| UBC835 |
AgAgAgAgAgAgAgAgTC |
7 |
4 |
| UBC836 |
AgAgAgAgAgAgAgAgYA |
5 |
3 |
| UBC840 |
gAgAgAgAgAgAgAgAYT |
5 |
2 |
| UBC890 |
VHVgTgTgTgTgTgTgT |
7 |
0 |
| UBCM06 |
AgCAgCAgCAgCY |
9 |
5 |
Discussion
Transposons make up a large proportion of plant genomes. In
Rhododendron, repetitive elements account for over a half (57.00%) of the genome, and LTR retrotransposons consist of the largest part, reaching 24.76% [
1]. The difference between
Copia and
Gypsy is not only reflected in the gene structure, but also in the copy number, insertion time, characteristics and influence on the genome [
24]. In the current case, we gained 7 IRAP primers designed with
Copia sequences and one IRAP primer designed with
Gypsy sequence, and their average PIC value was 0.36. The 6 highly informative ISSR primers tested herein yielded an average of 13.2 polymorphic loci per primer (
Table 1). The combination of two marker systems were justified to be effective for genetic diversity evaluation and somaclonal variation detection in
Rhododendron germplasm.
IRAPs and its effectiveness in genetic divergence detection for Rhododendron germplasm
Previously, the ISSR marker system was justified to be suitable for the identification of genetic diversity in rhododendron as well as the detection of genetic fidelity in somatic clone reproduction [
23]. In the present study, IRAP marker was firstly development in
Rhododendron plants based on the sequences of retrotransposons. To evaluate the effectiveness of the two marker systems, currently, A comparison between ISSR and IRAP markers was carried out since they represent different genome components.
The IRAP markers stand for LTR retrotransposons and the ISSR markers represent repeat sequence. Although the areas they cover may overlap, there exists a remarkable difference. The two markers based on different sequence features may be an intriguing and worthy direction to explore for the characteristic parameters they exhibit. The PICs of IRAP (0.36) and ISSR (0.38) were not much different, and the number of polymorphic bands was also close, being IRAP (14.9) and ISSR (13.2), respectively. The IRAP markers showed a high polymorphism in
Rhododendron plants, as high as 96.7%, which are close to those of 94% of polymorphisms in
Bletilla striata [
25], and 98% of polymorphisms in
Phyllostachys species [
26] as calculated by IRAPs, respectively. Previously, ISSRs were demonstrated to be polymorphic for genetic variation detection [
8], currently, IRAP marker was justified to be more effective in polymorphism assessment for rhododendrons. Likewise, EMR is the number of markers generated per primer assay [
22], and the EMR value for IRAPs (14.55) was greater than that for ISSRs (12.21). Sheik et al (2019) analyzed the polymorphism of 24 rhododendron accessions using 14 microsatellite markers, and scored an average of 2.92 alleles per primer [
27]. The marker index (MI), which evaluates the efficiency of markers to detect polymorphisms, was also about 10% higher for IRAPs (5.23) than for ISSRs (4.79). Markers with higher MI values are the preferred markers in plant species identification and plant DNA fingerprinting [
22]. Therefore, IRAP marker system developed herein is considerably superior in genetic diversity detection to the highly informative ISSRs as identified previously.
As in sugarcane, IRAPs and ISSRs exhibited different characteristics in the species clustering analysis [
22], similar tendency was also investigated in the rhododendrons (
Figure 4 and
Figure 5). For example, the coefficient of the IRAPs is 0.36-0.7, while that of ISSRs is 0.5-0.86, mirroring that IRAPs was somehow more sensitive in detecting genetic differences of rhododendrons. The IRAPs could divide the 46 rhododendron accessions into 5 groups, whereas the ISSRs divided them into 4 groups. Interestingly, some accessions from different population of same species were clustered into separate groups,
e.g., accessions 3_1, 3_6 and 3_10 of
R. delavayi, as revealed by IRAP markers. In ISSR-marker-associated clustering, 7 accessions of
R. delavayi were classified in Group III and another 4 were clustered in Group IV. The results just-mentioned above also reflected that IRAPs was more effective in genetic difference detection in rhododendrons. Additionally, a comparatively high polymorphism was investigated by IRAPs among the accessions from different populations of
R. delavayi distributed within 2 km in the Nature Reserve, indicating the high divergences may exist among the different populations of this species, possibly being ascribe to the occurrence of retrotransposons, finally leading to the better adaptation to the environments in BDNR. The above results also indicate that IRAP markers can easily identify genomic variation within
Rhododendron species.
A combination of IRAPs and ISSRs shows more effectiveness in genetic diversity assessment in rhododendrons
Considering the differences of IRAP and ISSR markers, we carried out the combination of IRAPs and ISSRs so as to evaluate the clustering effect among the rhododendron accessions. IRAP marker system of rhododendrons was developed herein based on retrotransposon sequences, and ISSRs were from the repeat sequences. Interestingly, the clustering efficiency of combining two markers is better than that using a single one, and the accessions from R. delavayi were more likely to cluster together in comparison with those as identified by one marker system.
In exception of the seven accessions from R. delavayi, two accessions 3_1 and 3_10 were clustered into another category, actually, the two accessions were geographically apart from the others by about one kilometer, possibly leading to subtle genetic differences from the others, which could be more effectively evaluated by the combination of two marker systems. Also in R. decorum, four accessions, i.e. 5_10-5_13, were grouped separately. Geographically, the four accessions were separated from the other population (accessions 5_1-5_9) by approximately two kilometers, where the topographical characters were somewhat different, leading to the differences in rainfall and temperature, which might promote transposition events so as to enhance the adaptive evolution. Most interestingly, rhododendrons that located closer were more likely to cluster together as revealed by the combination of two marker systems, even though they were different species. For example, the accessions 13, 14, and 15 demonstrated very closely genetic relationship. This may be due to the proximity in geographic location, which can easily lead to cross pollination between species, resulting in the mutual merging of genomes between offspring. Overall, the combination of the IRAP markers and the ISSR markers appears be able to better reveal the synteny and divergence among species.
Somaclonal variation as detected by IRAPs and ISSRs
The use of ISSRs to detect genetic fidelity of somaclonal clones has been reported in
Rhododendron [
8]. In this study, IRAPs and ISSRs were employed to detect the somaclonal variation among the 15 individuals of
R. bailiens callus which were subcultured for different cycles. Both could effectively distinguish some somaclonal variation in
R. bailiens genome (
Figure 7,
Figure S6). However, the two markers showed different detection properties. This is primarily due to certain IRAP markers that can facilitate the discovery of variants in the somatic clones of
Rhododendron, such as LTR1-11, LTR1-12 and LTR1-15 (
Figure 7). Interestingly, some IRAP primers used herein,
e.g. LTR1-11, LTR1-12 and LTR1-15, detected a large number of polymorphic bands during
in vitro culture, which might be ascribed to the occurrences in transposon events activated by
in vitro conditions. The similar results had been also documented in rice [
28] and sugarcane [
29], in which the transposon was strongly activated under tissue culture conditions. Meanwhile, the above findings also suggested that retrotransposons from
Copia,
e.g. TY1-30, TY1-33 and TY1-42, which correspond to IRAP primers LTR1-11, LTR1-12, and LTR1-15, might be the active transposons in
R. bailiens, which are easily activated by the external factors to trigger mutations in the genome.
Compared with IRAPs, the variation detected by ISSRs was significantly less (
Figures 7, 8 and S6), which was reflected by the higher similarity coefficient values of tissue-cultured
R. bailiens callus as displayed by ISSR markers. It is noteworthy that the rate of genetic variation detected by ISSR markers was low (less than 5%) in some species of
Rhododendron, such as
R. 'Kazimierz Odnowiciel' [
8] and
R. mucronulatum [
30], which might also be due to the difference in
in vitro materials (seedlings and callus) as well as in medium formula. The callus used herein should be more prone to variation during subculture in comparison to the seedling [
31]. Therefore, although IRAPs and ISSRs have similar PIC values, the former are obviously better suited to detect somatic variation in somatoclonal replication.
It is noteworthy that most of the transposons are also associated with repeat sequences [
32], which may lead to the genetic variation as detected by ISSRs due to the activity of transposons. Evidences in sugarcane also showed that polymorphic fragments amplified by the ISSR markers might be transposable element sequences generated by the transposon activity [
22]. Therefore, somaclonal variation detected by ISSR markers may also be related to transposon events.
Conclusively, IRAP marker developed in the present study provides a novel methodology to detect somaclonal variation in Rhododendron tissue culture. The in vitro cultures used herein was callus, which is generally prone to genetic unstability, thus demonstrated high frequency in somaclonal variation. Our preliminary investigation in plantlets of Rhododendron species also showed that IRAPs can also detect genetic variation, and a systematic study should be further carried out for in vitro plantlet detection so as to facilitate the preservation of Rhododendron germplasms.
Conclusions
In conclusion, we developed the IRAP marker system in Rhododendron, and examined both IRAPs and ISSRs as possible candidates with the goal of establishing a reliable system for identifying genetic fidelity and diversity. They were found to have complementary effects when testing the genetic diversity of multiple Rhododendron species and the genetic fidelity of R. bailiens somaclonal clones. Particularly, IRAPs were superior to ISSRs in the number of polymorphic bands, EMR, MI, and were more suitable for detecting the somatic variations of R. bailiens.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org.
Author Contributions
G.Q conceived and supervised the project. G.Q. and S.L.W designed the experiments. S.L.W, H.Z and M.Y.Z. performed the experiments. S.L.W analyzed the data and wrote the article. G.Q and X.H.S provided research suggestions and assisted in reviewing the manuscript. G.Q. acquired the funding. All the authors read and approved the final version for publication.
Funding
This project is supported by grants from the National Natural Science Foundation of China (32160700), the Special Program (2022-2023) for Seed Engineering from Guizhou Department of Agriculture and Rural Area, as well as the Innovation Talent Program of Guizhou Province (2016-4010), China.
Acknowledgements
We are extremely indebted to Mr. Jiayong Huang of the Baili Rhododendron Reserve, Bijie, Guizhou, China, for helping to collect the rhododendron materials.
Conflict Of Interest: The authors declare that they have no conflicts of interests associated with this work.
References
- Ma, H.; Liu, Y.; Liu, D.; Sun, W.; Liu, X.; Wan, Y.; Zhang, X.; Zhang, R.; Yun, Q.; Wang, J.; et al. Chromosome-level genome assembly and population genetic analysis of a critically endangered rhododendron provide insights into its conservation. The Plant Journal 2021, 107, 1533–1545. [Google Scholar] [CrossRef]
- Tian, X.L.; Chang, Y.H.; Neilsen, J.; Wang, S.H.; Ma, Y.P. A new species of Rhododendron (Ericaceae) from northeastern Yunnan, China. Phytotaxa 2019, 395, 66–70. [Google Scholar] [CrossRef]
- Yang, B.; Huang, M.; Wang, L.; Wu, X.; Xiaoyong, D. Fruit morphology of Ericaceaeand seed characteristics of 21 species of Rhododendron in Guizhou. Guizhou Forestry Science and Technology 2020, 48, 8–14. [Google Scholar]
- Chen, X.; Huang, J.; Xie, H.; chen, X. A new species and a new variety of Rhododendron (Ericaceae) in Guizhou. Seed 2010, 29, 65–67. [Google Scholar]
- Ma, Y.; Chamberlain, D.; Sun, W.; Zhang, C. A new species of Rhododendron (Ericaceae) from Baili Rhododendron nature reserve, NW Guizhou, China. Phytotaxa 2015, 195. [Google Scholar] [CrossRef]
- Li, D.; Xia, G.; Zhang, M.; Huang, C. Research on the priority conservation order of rhododendron in baili rhododendron provincial nature reserve, Guizhou. Xiang Cun Ke ji 2022, 13, 81–85. [Google Scholar]
- Wei, X.Y.; Chen, J.J.; Zhang, C.Y.; Wang, Z.H. In vitro shoot culture of Rhododendron fortunei: An important plant for bioactive phytochemicals. Industrial Crops and Products 2018, 126, 459–465. [Google Scholar] [CrossRef]
- Nowakowska, K.; Pinkowska, A.; Siedlecka, E.; Pacholczak, A. The effect of cytokinins on shoot proliferation, biochemical changes and genetic stability of Rhododendron 'Kazimierz Odnowiciel' in the in vitro cultures. Plant Cell Tissue and Organ Culture 2022, 149, 675–684. [Google Scholar] [CrossRef]
- Lee, S.I.; Kim, J.H.; Park, K.C.; Kim, N.S. LTR-retrotransposons and inter-retrotransposon amplified polymorphism (IRAP) analysis in Lilium species. Genetica 2015, 143, 343–352. [Google Scholar] [CrossRef]
- Biswas, M.K.; Xu, Q.; Deng, X.X. Utility of RAPD, ISSR, IRAP and REMAP markers for the genetic analysis of Citrus spp. Scientia Horticulturae 2010, 124, 254–261. [Google Scholar] [CrossRef]
- Kalendar, R. Flavell; A, J.; Ellis; Hn, T.; Sjakste; T; Moisy; C. Analysis of plant diversity with retrotransposon-based molecular markers. Heredity 2011. [Google Scholar] [CrossRef] [PubMed]
- Zaytseva, Y.G.; Skaptsov, M.V.; Kutsev, M.G.; Novikova, T.I. In vitro establishment and TDZ-induced regeneration from shoot and leaf explants of Rhododendron sichotense Pojark. Turczaninowia 2020, 23, 106–111. [Google Scholar] [CrossRef]
- Vejsadova, H.; Pretova, A. Somatic embryogenesis in rhododendron catawbiense 'grandiflorum'. In Proceedings of the 1st international symposium on acclimatization and establishment of micropropagated plants, Sani-Halkidiki, Greece, 2001, Sep 19-22; pp. 467–470.
- Kumar, A.; Pearce, S.R.; McLean, K.; Harrison, G.; Heslop-Harrison, J.S.; Waugh, R.; Flavell, A.J. The Ty1-copia group of retrotransposons in plants: genomic organisation, evolution, and use as molecular markers. Genetica 1997, 100, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Kumekawa, N.; Ohtsubo, E.; Ohtsubo, H. Identification and phylogenetic analysis of Gypsy-type retrotransposons in the plant kingdom. Genes & Genetic Systems 1999, 74, 299–307. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology And Evolution 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Yeh, F.; Yang, R.; Boyle, T. POPGENE Version 1.32 Microsoft Windows-based freeware for populations genetic analysis. University of Alberta, Edmonton 1999. [Google Scholar]
- Adhikari, S.; Saha, S.; Bandyopadhyay, T.K.; Ghosh, P. Efficiency of ISSR marker for characterization of Cymbopogon germplasms and their suitability in molecular barcoding. Plant Systematics and Evolution 2014, 301, 439–450. [Google Scholar] [CrossRef]
- Roldan-Ruiz, I.; Dendauw, J.; Van Bockstaele, E.; Depicker, A.; De Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Molecular Breeding 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Varshney, R.K.; Chabane, K.; Hendre, P.S.; Aggarwal, R.K.; Graner, A. Comparative assessment of EST-SSR, EST-SNP and AFLP markers for evaluation of genetic diversity and conservation of genetic resources using wild, cultivated and elite barleys. Plant Science 2007, 173, 638–649. [Google Scholar] [CrossRef]
- Prevost, A.; Wilkinson, M.J. A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theoretical And Applied Genetics 1999, 98, 107–112. [Google Scholar] [CrossRef]
- Shingote, P.R.; Mithra, S.V.A.; Sharma, P.; Devanna, N.B.; Arora, K.; Holkar, S.K.; Khan, S.; Singh, J.; Kumar, S.; Sharma, T.R.; et al. LTR retrotransposons and highly informative ISSRs in combination are potential markers for genetic fidelity testing of tissue culture-raised plants in sugarcane. Molecular Breeding 2019, 39. [Google Scholar] [CrossRef]
- Hong, Y.; Wen, X. Establishment and Optimization of in vitro Micropropagation of Rhododendron delavayi Franch. Journal of Southwest University. Natural Science Edition 2012, 34, 61–66. [Google Scholar]
- He, G.-Q.; Jin, H.-Y.; Cheng, Y.-Z.; Yu, Y.-H.; Guo, D.-L. Characterization of genome-wide long terminal repeat retrotransposons provide insights into trait evolution of four grapevine species. Journal of Systematics and Evolution 2022, n/a. [Google Scholar] [CrossRef]
- Guo, Y.; Zhai, L.N.; Long, H.; Chen, N.P.; Gao, C.X.; Ding, Z.S.; Jin, B. Genetic diversity of Bletilla striata assessed by SCoT and IRAP markers. Hereditas 2018, 155. [Google Scholar] [CrossRef]
- Li, S.; Ramakrishnan, M.; Vinod, K.K.; Kalendar, R.; Yrjala, K.; Zhou, M. Development and deployment of high-throughput retrotransposon-based markers reveal genetic diversity and population structure of asian bamboo. Forests 2020, 11. [Google Scholar] [CrossRef]
- Sheik, M.L.; LaBounty, K.L.; Mitchell, E.; Gillespie, E.L. Fourteen polymorphic microsatellite markers for the widespread Labrador tea (Rhododendron groenlandicum). Appl Plant Sci 2019, 7, e11306. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Han, F.P.; Tan, M.; Shan, X.H.; Dong, Y.Z.; Wang, X.Z.; Fedak, G.; Hao, S.; Liu, B. Activation of a rice endogenous retrotransposon Tos17 in tissue culture is accompanied by cytosine demethylation and causes heritable alteration in methylation pattern of flanking genomic regions. Theoretical And Applied Genetics 2004, 109, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Araujo, P.G.; Rossi, M.; de Jesus, E.M.; Saccaro, N.L.; Kajihara, D.; Massa, R.; de Felix, J.M.; Drummond, R.D.; Falco, M.C.; Chabregas, S.M.; et al. Transcriptionally active transposable elements in recent hybrid sugarcane. The Plant Journal 2005, 44, 707–717. [Google Scholar] [CrossRef]
- Novikova, T.I.; Asbaganov, S.V.; Ambros, E.V.; Zaytseva, Y.G. TDZ-induced axillary shoot proliferation of Rhododendron mucronulatum Turcz and assessment of clonal fidelity using DNA-based markers and flow cytometry. In Vitro Cellular & Developmental Biology-Plant 2020, 56, 307–317. [Google Scholar] [CrossRef]
- Mehbub, H.; Akter, A.; Akter, M.A.; Mandal, M.S.H.; Hoque, M.A.; Tuleja, M.; Mehraj, H. Tissue culture in ornamentals: Cultivation factors, propagation techniques, and its application. Plants-Basel 2022, 11. [Google Scholar] [CrossRef]
- Dhillon, B.; Gill, N.; Hamelin, R.C.; Goodwin, S.B. The landscape of transposable elements in the finished genome of the fungal wheat pathogen Mycosphaerella graminicola. Bmc Genomics 2014, 15. [Google Scholar] [CrossRef] [PubMed]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).