Submitted:
28 March 2023
Posted:
29 March 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Methods and materials
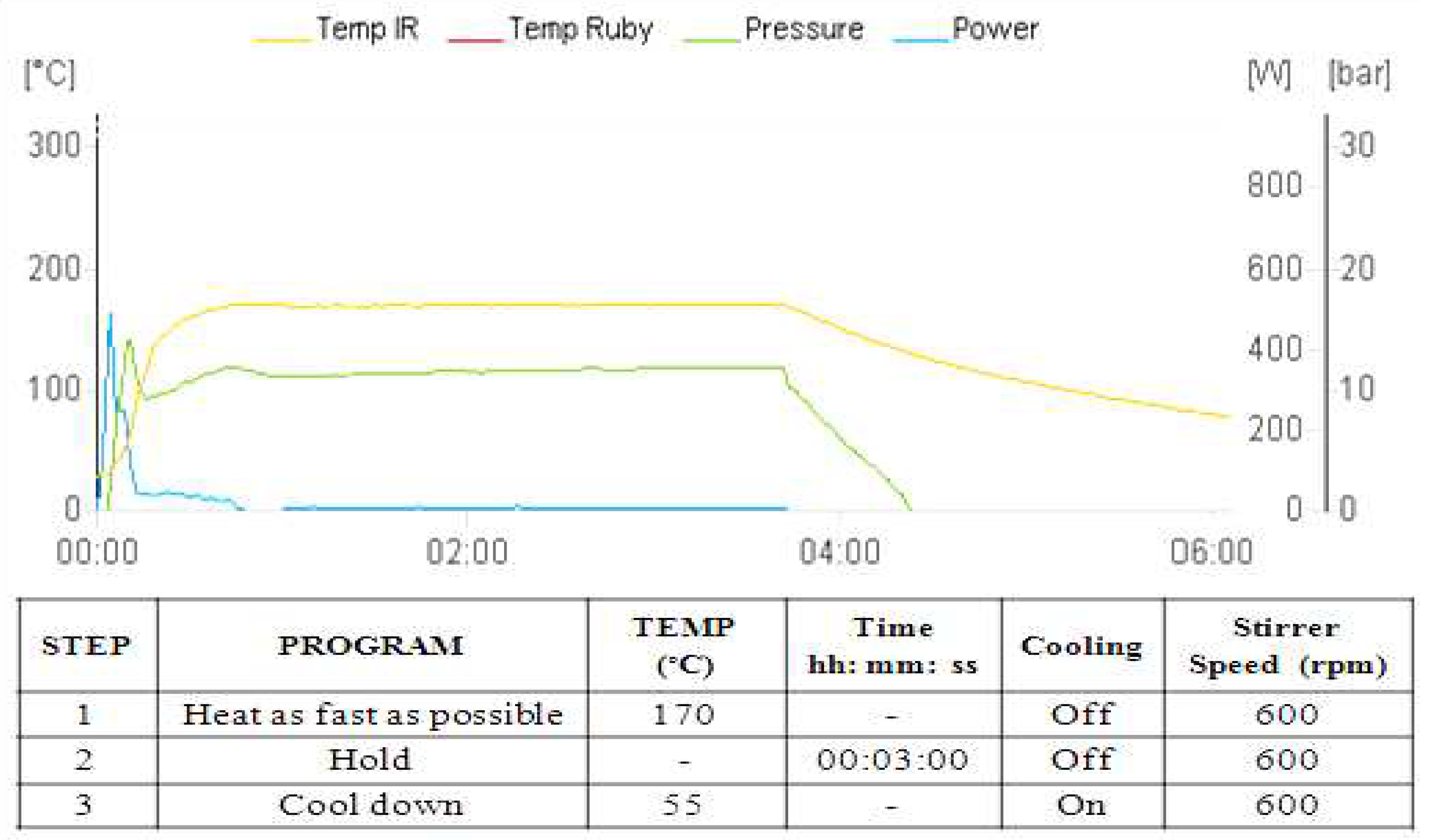
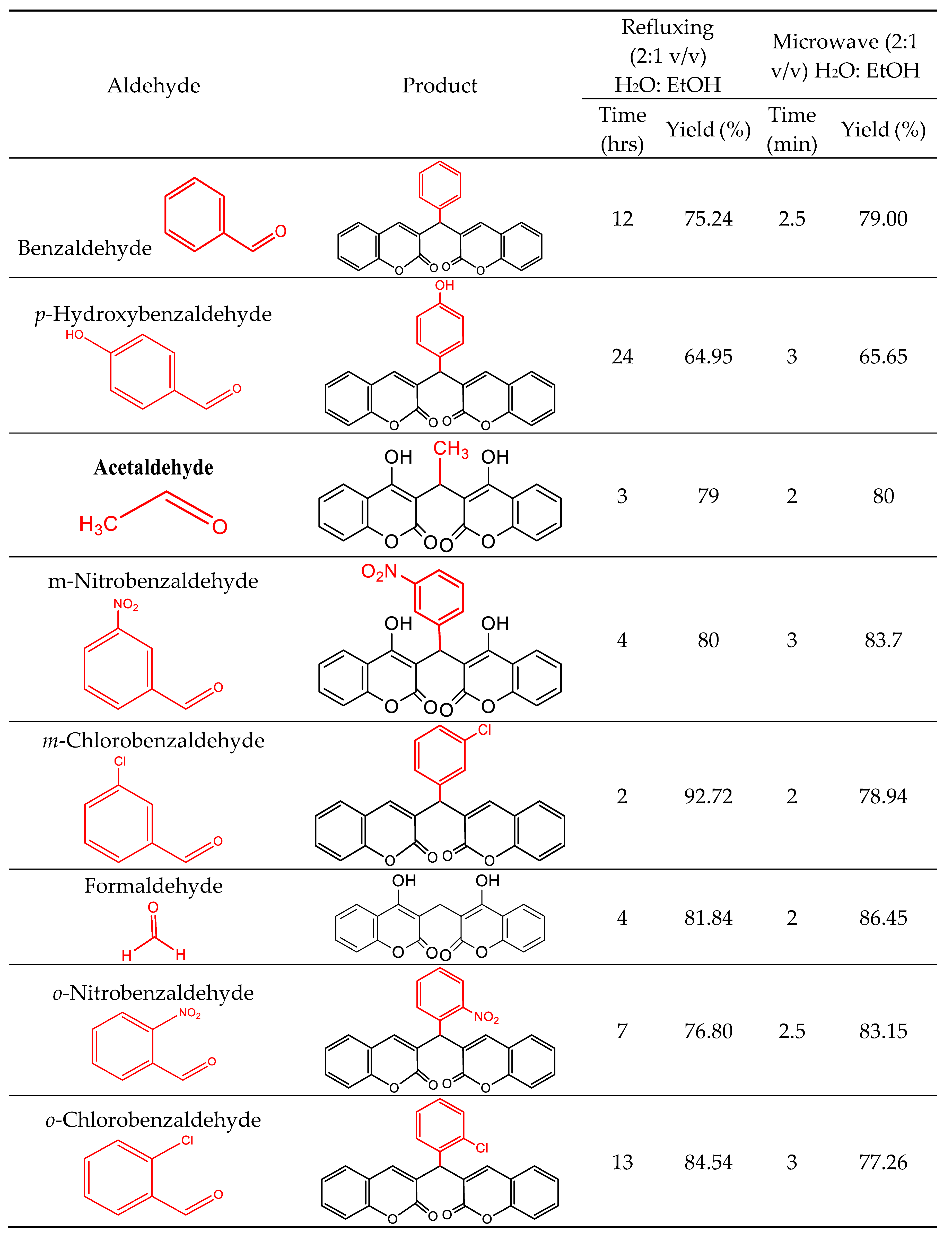
2.1. Procedure for the synthesis of Bis (4-hydroxy-2H-chromen-2-ones) a-h (refluxing conditions)
2.2. Synthesis of Bis(4-hydroxy-2H-chromen-2-ones) a-h through Method A
2.3. Synthesis of Bis(4-hydroxy-2H-chromen-2-ones) through Method B
2.4. Antibacterial activity spectrum of biscoumarin
- Drugs, chemicals, and media
- Bacterial strains, compound dissolving, and dilution
- Determination of minimum inhibitory concentration (MIC) against Mycobacterial strains
- Determination of (MIC) against gram-positive and gram-negative bacteria
2.5. Cytotoxicity assay
2.6. Molecular docking and MD simulation studies
- Molecular Docking
- Molecular dynamics simulation (MD)
3. Results
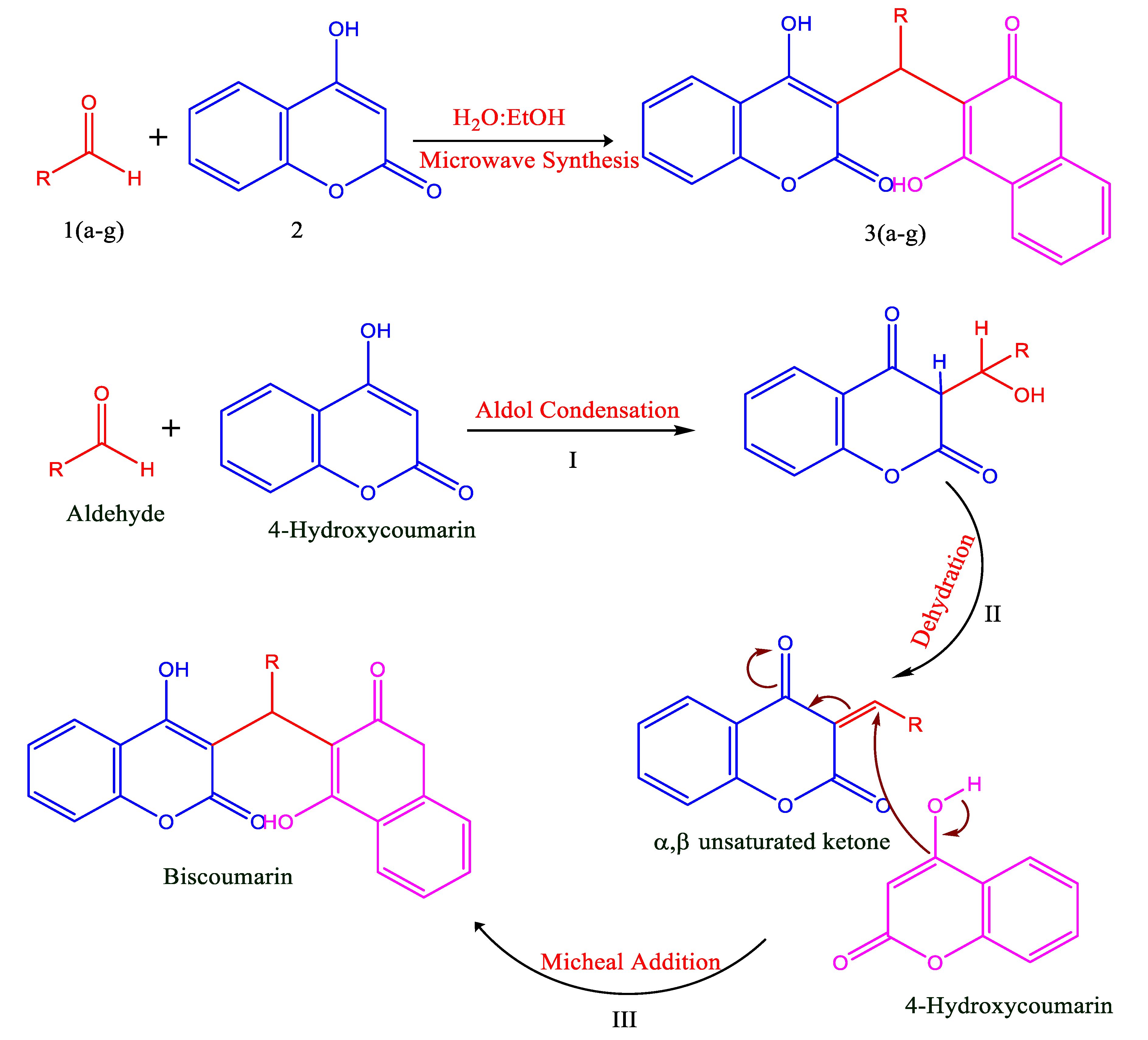
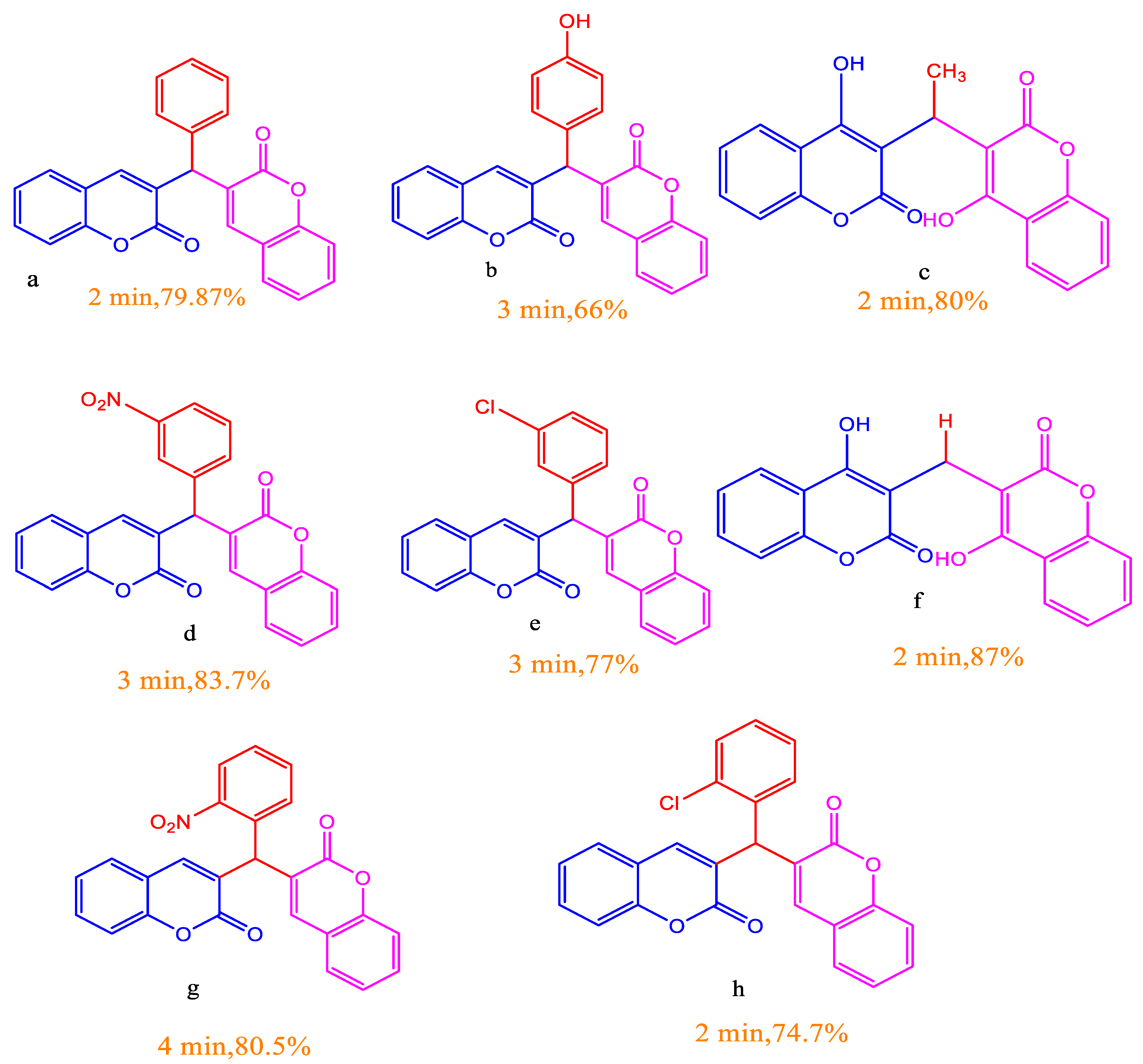
3.1. Chemistry/Synthesis
3.2. Antibacterial activity spectrum of biscoumarin
- The anti-tuberculosis activity of compounds
- Determination of MIC against gram-positive and gram-negative bacterial strains
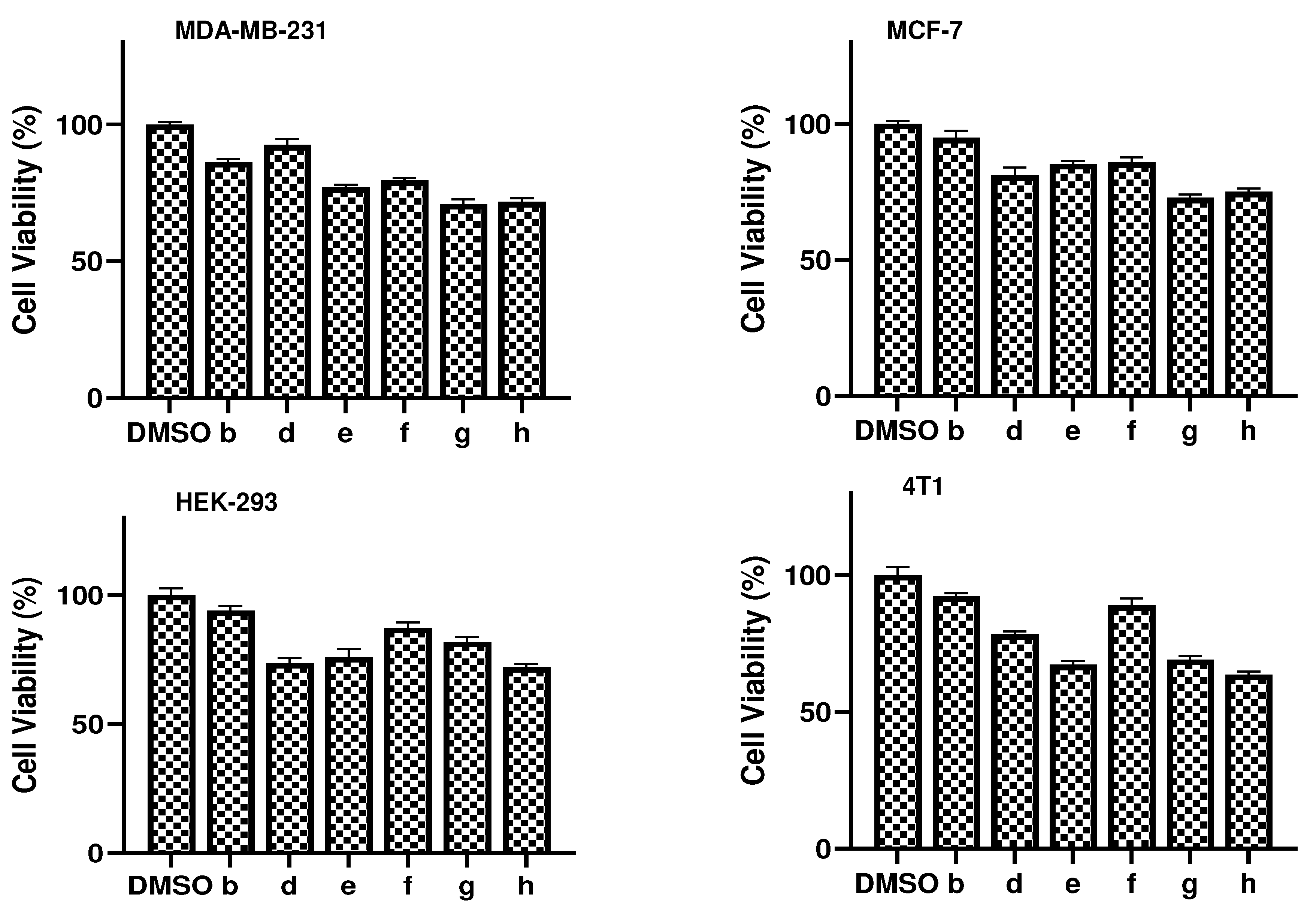
3.3. Compound cytotoxicity
3.4. Molecular Docking
4. Discussion
5. Conclusion
Author Contributions
Funding
Conflicts of Interest
References
- Khabibullina NF, Kutuzova DM, Burmistrova IA and Lyadova IV (2022) The biological and clinical aspects of a latent tuberculosis infection. Tropical Medicine and Infectious Disease 7:48.
- Sharma JB and Yadav V (2022) Tuberculosis in Pregnancy. Infections and Pregnancy, Springer, pp. 63-81.
- Dartois VA and Rubin EJ (2022) Anti-tuberculosis treatment strategies and drug development: challenges and priorities. Nature Reviews Microbiology:1-17.
- Rewari BB, Kumar A, Mandal PP and Puri AK (2021) HIV TB coinfection-perspectives from India. Expert review of respiratory medicine 15:911-930.
- Mir MA, Hamdani SS and Qadri H (2022) Significance of immunotherapy for human bacterial diseases and antibacterial drug discovery. Journal: Human Pathogenic Microbes:129-161.
- Mir MA, Albaradeh R and Agrewala JN (2013) innate–effector immune response elicitation against tuberculosis through anti-b7-1 (cd80) and anti-b7-2 (cd86) signaling in macrophages. Ijbpas,.
- da Silva Leite JM, Patriota YBG, de La Roca MF and Soares-Sobrinho JL (2022) New Perspectives in Drug Delivery Systems for the Treatment of Tuberculosis. Current Medicinal Chemistry 29:1936-1958.
- Sheikh BA, Bhat BA, Alshehri B, Mir RA, Mir WR, Parry ZA and Mir MA (2021) Nano-drug delivery systems: Possible end to the rising threats of tuberculosis. Journal of Biomedical Nanotechnology 17:2298-2318.
- Adeniji AA, Knoll KE and Loots DT (2020) Potential anti-TB investigational compounds and drugs with repurposing potential in TB therapy: a conspectus. Applied microbiology and biotechnology 104:5633-5662.
- Sheikh BA, Bhat BA, Ahmad Z and Mir MA (2022) Strategies Employed to Evade the Host Immune Response and the Mecha-nism of Drug Resistance in Mycobacterium tuberculosis: In Search of Find-ing New Targets. Current Pharmaceutical Biotechnology.
- Qadri H, Shah AH and Mir M (2021) Novel strategies to combat the emerging drug resistance in human pathogenic microbes. Current Drug Targets 22:1424-1436.
- Watroly MN, Sekar M, Fuloria S, Gan SH, Jeyabalan S, Wu YS, Subramaniyan V, Sathasivam KV, Ravi S and Rani NNIM (2021) Chemistry, Biosynthesis, Physicochemical and Biological Properties of Rubiadin: A Promising Natural Anthraquinone for New Drug Discovery and Development. Drug design, development and therapy 15:4527.
- Bhat BA, Mir WR, Sheikh BA, Rather MA and Mir MA (2022) In vitro and in silico evaluation of antimicrobial properties of Delphinium cashmerianum L., a medicinal herb growing in Kashmir, India. Journal of Ethnopharmacology 291:115046.
- Sheikh BA, Bhat BA, Mehraj U, Mir W, Hamadani S and Mir MA (2021) Development of new therapeutics to meet the current challenge of drug resistant tuberculosis. Current pharmaceutical biotechnology 22:480-500.
- Mir MA, Mir B, Kumawat M, Alkhanani M and Jan U (2022) Manipulation and exploitation of host immune system by pathogenic Mycobacterium tuberculosis for its advantage. Future Microbiology 17:1171-1198.
- Najmi A, Javed SA, Al Bratty M and Alhazmi HA (2022) Modern approaches in the discovery and development of plant-based natural products and their analogues as potential therapeutic agents. Molecules 27:349.
- Kumar M, Singh SK, Singh PP, Singh VK, Rai AC, Srivastava AK, Shukla L, Kesawat MS, Kumar Jaiswal A and Chung S-M (2021) Potential Anti-Mycobacterium tuberculosis Activity of Plant Secondary Metabolites: Insight with Molecular Docking Interactions. Antioxidants 10:1990.
- Salehian F, Nadri H, Jalili-Baleh L, Youseftabar-Miri L, Bukhari SNA, Foroumadi A, Küçükkilinç TT, Sharifzadeh M and Khoobi M (2021) A review: Biologically active 3, 4-heterocycle-fused coumarins. European Journal of Medicinal Chemistry 212:113034.
- Mir MA, Usman M, Qadri H and Aisha S (2022) Recent trends in the development of bacterial and fungal vaccines. Journal: Human Pathogenic Microbes:233-259.
- Pereira TM, Franco DP, Vitorio F and Kummerle AE (2018) Coumarin compounds in medicinal chemistry: some important examples from the last years. Current topics in medicinal chemistry 18:124-148.
- Kulasari S, Singh MF and Bhandari S (2019) Polyphenols: Phytochemistry and health benefits. J. Pharmacogn. Phytochem 8:3344-3358.
- Xu Z, Chen Q, Zhang Y and Liang C (2021) Coumarin-based derivatives with potential anti-HIV activity. Fitoterapia 150:104863.
- Pasricha S and Gahlot P (2020) Synthetic Strategies and Biological Potential of Coumarin-Chalcone Hybrids: A New Dimension to Drug Design. Current Organic Chemistry 24:402-438.
- Annunziata F, Pinna C, Dallavalle S, Tamborini L and Pinto A (2020) An overview of coumarin as a versatile and readily accessible scaffold with broad-ranging biological activities. International Journal of Molecular Sciences 21:4618.
- Banday SM, Amin A, Bashir S, Qadri RA, Khan KZ and Rizvi MA (2017) Anti-metastatic propensity of biscoumarin scaffold synthesized under catalyst free aqueous phase microwave irradiation. Croatica Chemica Acta 90:471-481.
- Hussain A, Rather MA, Bhat ZS, Majeed A, Maqbool M, Shah AM, Aga MA, Shah A, Mushtaq S and Sangwan PL (2019) In vitro evaluation of dinactin, a potent microbial metabolite against Mycobacterium tuberculosis. International journal of antimicrobial agents 53:49-53.
- Moradipoodeh B, Jamalan M, Zeinali M, Fereidoonnezhad M and Mohammadzadeh G (2019) In vitro and in silico anticancer activity of amygdalin on the SK-BR-3 human breast cancer cell line. Molecular Biology Reports 46:6361-6370.
- Abraham CS, Muthu S, Prasana JC, Armaković SJ, Armaković S and As BG (2018) Spectroscopic profiling (FT-IR, FT-Raman, NMR and UV-Vis), autoxidation mechanism (H-BDE) and molecular docking investigation of 3-(4-chlorophenyl)-N, N-dimethyl-3-pyridin-2-ylpropan-1-amine by DFT/TD-DFT and molecular dynamics: A potential SSRI drug. Computational Biology and Chemistry 77:131-145.
- Shree P, Mishra P, Selvaraj C, Singh SK, Chaube R, Garg N and Tripathi YB (2022) Targeting COVID-19 (SARS-CoV-2) main protease through active phytochemicals of ayurvedic medicinal plants–Withania somnifera (Ashwagandha), Tinospora cordifolia (Giloy) and Ocimum sanctum (Tulsi)–a molecular docking study. Journal of Biomolecular Structure and Dynamics 40:190-203.
- Zhang C-H, Stone EA, Deshmukh M, Ippolito JA, Ghahremanpour MM, Tirado-Rives J, Spasov KA, Zhang S, Takeo Y and Kudalkar SN (2021) Potent noncovalent inhibitors of the main protease of SARS-CoV-2 from molecular sculpting of the drug perampanel guided by free energy perturbation calculations. ACS central science 7:467-475.
- Bhadresha K, Upadhyay V, Kumar SP, Pandya P, Jain N and Rawal RM (2021) Computational investigation of ginkgetin and theaflavin as potential inhibitors of heat shock protein 90 (Hsp90). Journal of Biomolecular Structure and Dynamics:1-7.
- Al-Otaibi JS, Mary YS, Mary S, Trivedi R, Chakraborty B, Yadav R, Celik I and Soman S (2022) DFT and MD investigations of the biomolecules of phenothiazine derivatives: interactions with gold and water molecules and investigations in search of effective drug for SARS-CoV-2. Journal of Biomolecular Structure and Dynamics:1-12.
- Mukne A, Momin M, Betkar P, Rane T and Valecha S (2021) Cell-Based Assays in Natural Product-Based Drug Discovery. Evidence Based Validation of Traditional Medicines, Springer, pp. 211-248.
- Ferraz CAN, Tintino SR, Teixeira AMR, Bandeira PN, Santos HS, Cruz BG, Nogueira CES, Moura TF, Pereira RLS and Sena Jr DM (2020) Potentiation of antibiotic activity by chalcone (E)-1-(4′-aminophenyl)-3-(furan-2-yl)-prop-2-en-1-one against gram-positive and gram-negative MDR strains. Microbial Pathogenesis 148:104453.
- Alsayed SSR, Lun S, Payne A, Bishai WR and Gunosewoyo H (2021) Design, synthesis and antimycobacterial evaluation of novel adamantane and adamantanol analogues effective against drug-resistant tuberculosis. Bioorganic chemistry 106:104486.
- Mir MA (2022) Human Pathogenic Microbes: Diseases and Concerns. Elsevier.
- Parameshwarappa G, Lingamani J, Patil SB and Goudgaon NM (2009) Synthesis and anti-mirobial activity of thiazole substituted coumarins. Heterocyclic Communications 15:343-348.
- Patil SB (2022) Medicinal significance of novel coumarin analogs: Recent studies. Results in Chemistry 4:100313.
- Phutdhawong W, Chuenchid A, Taechowisan T, Sirirak J and Phutdhawong WS (2021) Synthesis and biological activity evaluation of coumarin-3-carboxamide derivatives. Molecules 26:1653.
- Coronado L, Zhang X-Q, Dorta D, Escala N, Pineda LM, Ng MG, Del Olmo E, Wang C-Y, Gu Y-C and Shao C-L (2021) Semisynthesis, antiplasmodial activity, and mechanism of action studies of isocoumarin derivatives. Journal of Natural Products 84:1434-1441.
- Sahar A, Khan ZA, Ahmad M, Zahoor AF, Mansha A and Iqbal A (2017) Synthesis and antioxidant potential of some biscoumarin derivatives. Tropical Journal of Pharmaceutical Research 16:203-210.
- Joy MN, Bakulev VA, Bodke YD and Telkar S (2020) Synthesis of coumarins coupled with benzamides as potent antimicrobial agents. Pharmaceutical Chemistry Journal 54:604-621.
- Mk BKBJG (2019) Singh JV Gulati HK Singh A. Kaur K. Kaur G. Sharma S. Rana K. Singh H. Sharma S. Singh Bedi PM ACS Omega 4:8720-8730.
- Godge R and Kunkulol R (2018) Synthesis of Coumarin heterocyclic derivatives with In-Vitro antituberculer activity. Journal of Drug Delivery and Therapeutics 8:217-223.
- KhanYusufzai S, Osman H, Khan MS, Mohamad S, Sulaiman O, Parumasivam T, Gansau JA and Johansah N (2017) Design, characterization, in vitro antibacterial, anti-tubercular evaluation and structure–activity relationships of new hydrazinyl thiazolyl coumarin derivatives. Medicinal Chemistry Research 26:1139-1148.
- Pollo LAE, Martin EF, Machado VR, Cantillon D, Wildner LM, Bazzo ML, Waddell SJ, Biavatti MW and Sandjo LP (2021) Search for antimicrobial activity among fifty-two natural and synthetic compounds identifies anthraquinone and polyacetylene classes that inhibit Mycobacterium tuberculosis. Frontiers in Microbiology:3619.
- Rather MA, Lone AM, Teli B, Bhat ZS, Singh P, Maqbool M, Shairgojray BA, Dar MJ, Amin S and Yousuf SK (2017) The synthesis, biological evaluation and structure–activity relationship of 2-phenylaminomethylene-cyclohexane-1, 3-diones as specific anti-tuberculosis agents. MedChemComm 8:2133-2141.
- Acharya P, Ramana MMV, Upadhyay M and Pavale G (2021) Unveiling the Anti-tubercular Properties of Biscoumarins, through Biological Evaluation and Docking Studies. Letters in Drug Design & Discovery 18:57-66.
- Qasaymeh RM, Rotondo D, Oosthuizen CB, Lall N and Seidel V (2019) Predictive binding affinity of plant-derived natural products towards the protein kinase g enzyme of mycobacterium tuberculosis (mtpkng). Plants 8:477.
- Aljahdali MO, Molla MHR and Ahammad F (2022) Immunoinformatics and Computer-Aided Drug Design as New Approaches against Emerging and Re-Emerging Infectious Diseases.
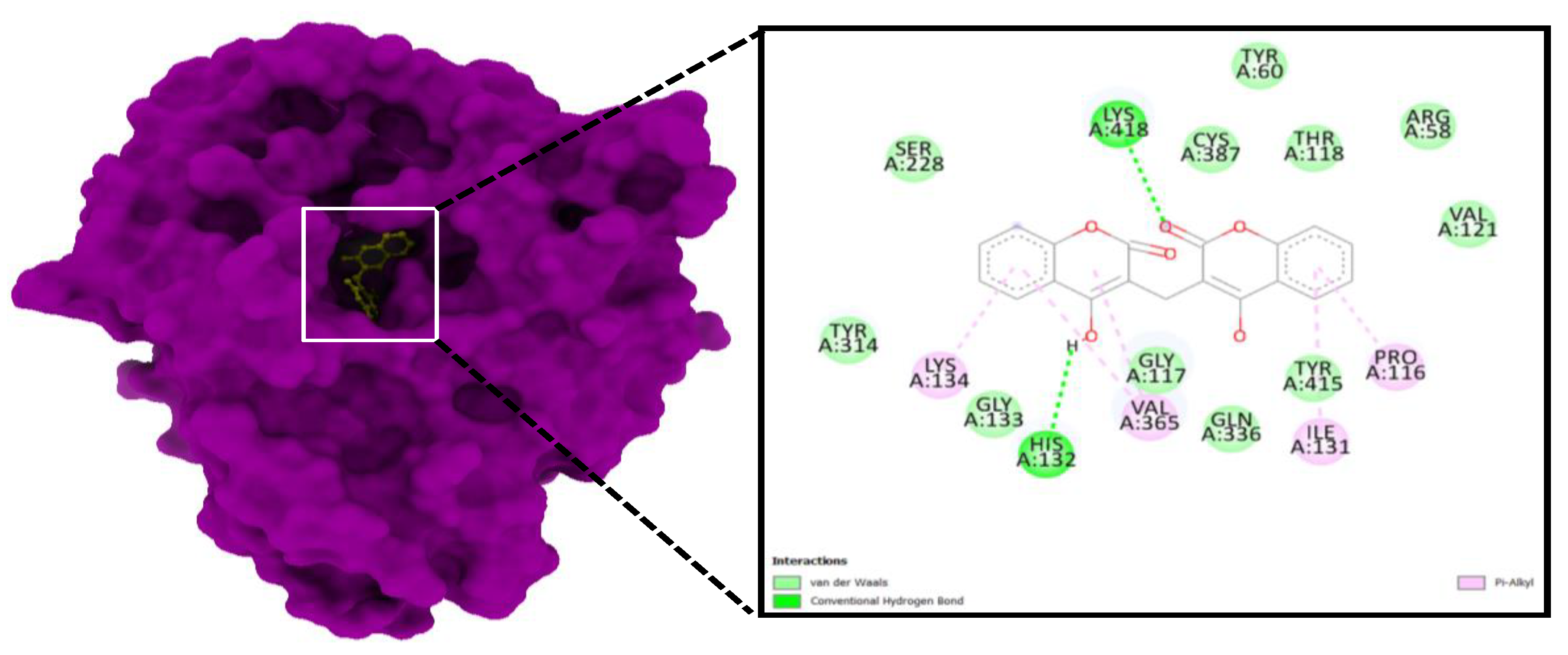
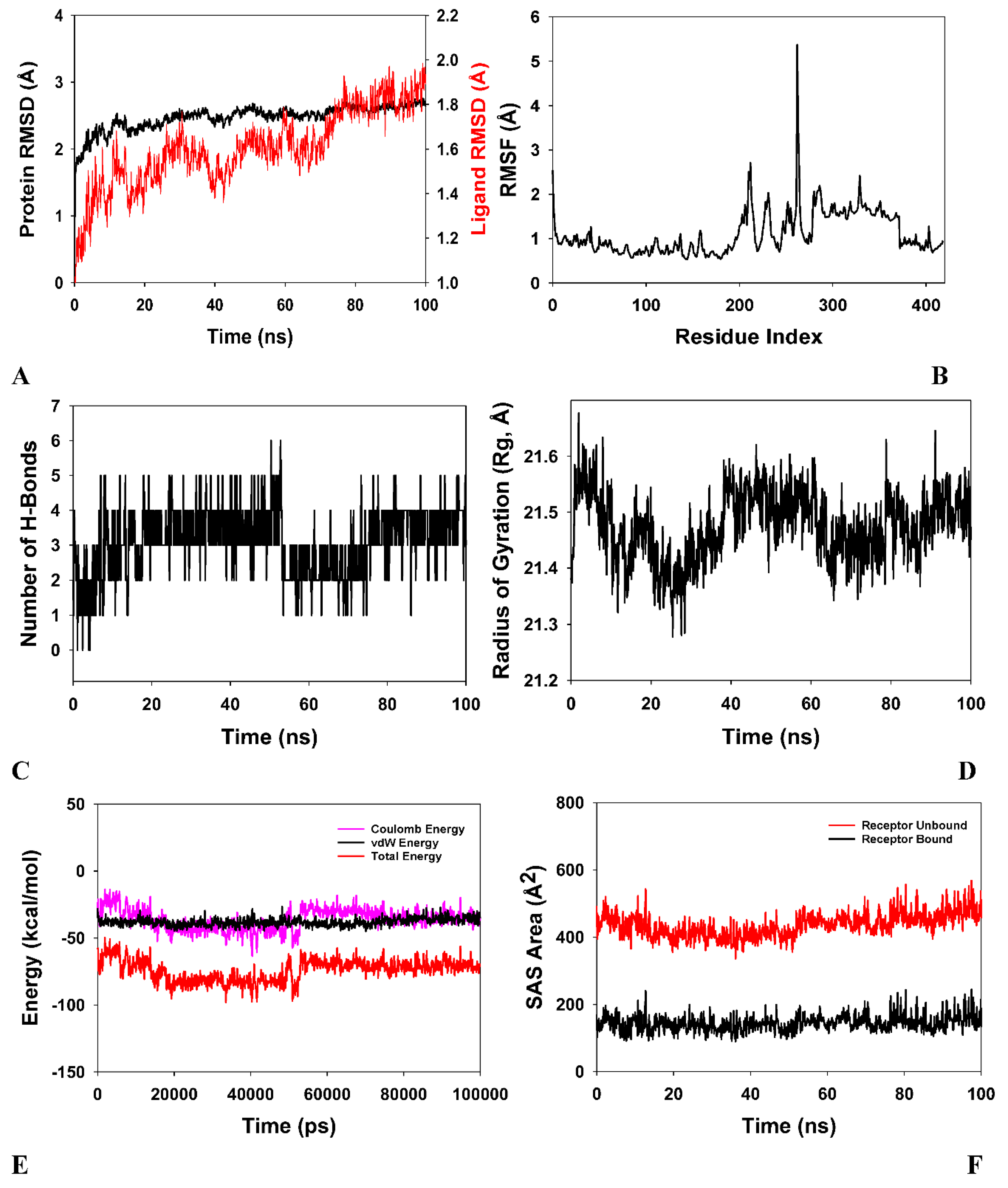
| S. No | Compound | E. coli | E. faecalis | B. subtilis | P. aeruginosa | Mtb H37 Rv | Mtb H37Ra | M. smegmatis |
| 1 | b | 128 | 128 | 64 | 128 | 64 | 64 | 128 |
| 2 | d | 128 | 128 | 64 | 128 | 32 | 32 | 128 |
| 3 | e | 128 | 128 | 64 | 128 | 64 | 64 | 128 |
| 4 | f | 128 | 64 | 64 | 128 | 16 | 16 | 128 |
| 5 | g | 128 | 128 | 64 | 128 | 64 | 64 | 128 |
| 6 | h | 128 | 128 | 64 | 128 | 64 | 64 | 128 |
| INH | NA | NA | NA | NA | 0.325 | 0.325 | 1.25 | |
| ETB | NA | NA | NA | NA | 1.25 | 1.25 | 0.039 | |
| LVX | 0.031 | 0.156 | 0.125 | 0.078 | 0.15 | 0.15 | 0.039 |
| Compound | MCF-7 | HEK-293 | MDA-MB-231 | 4T1 |
| b | 5.15 | 6.02 | 13.72 | 7.76 |
| d | 18.91 | 26.36 | 7.456 | 21.73 |
| e | 14.69 | 24.07 | 22.97 | 32.80 |
| f | 14.13 | 12.75 | 20.46 | 11.13 |
| g | 27.20 | 18.13 | 29.08 | 30.93 |
| h | 24.95 | 27.92 | 28.33 | 36.48 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





