Submitted:
04 April 2023
Posted:
05 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Climate
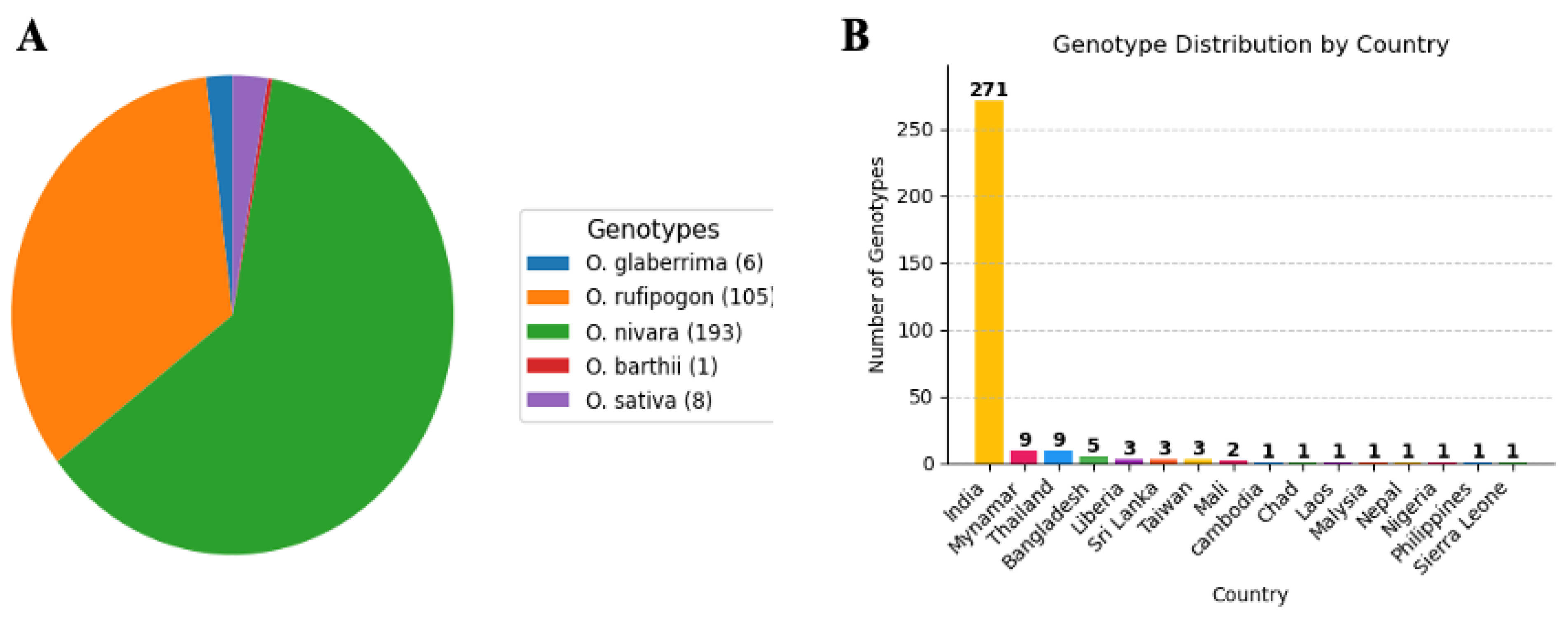
2.2. Plant Materials
2.3. Irrigation applied during experiment
2.4. Screening method for IDIC tolerance of wild germplasm
2.5. Chlorophyll measurement and leaf area index
2.6. Iron content in the leaves
2.7. Statistical analysis
3. Results
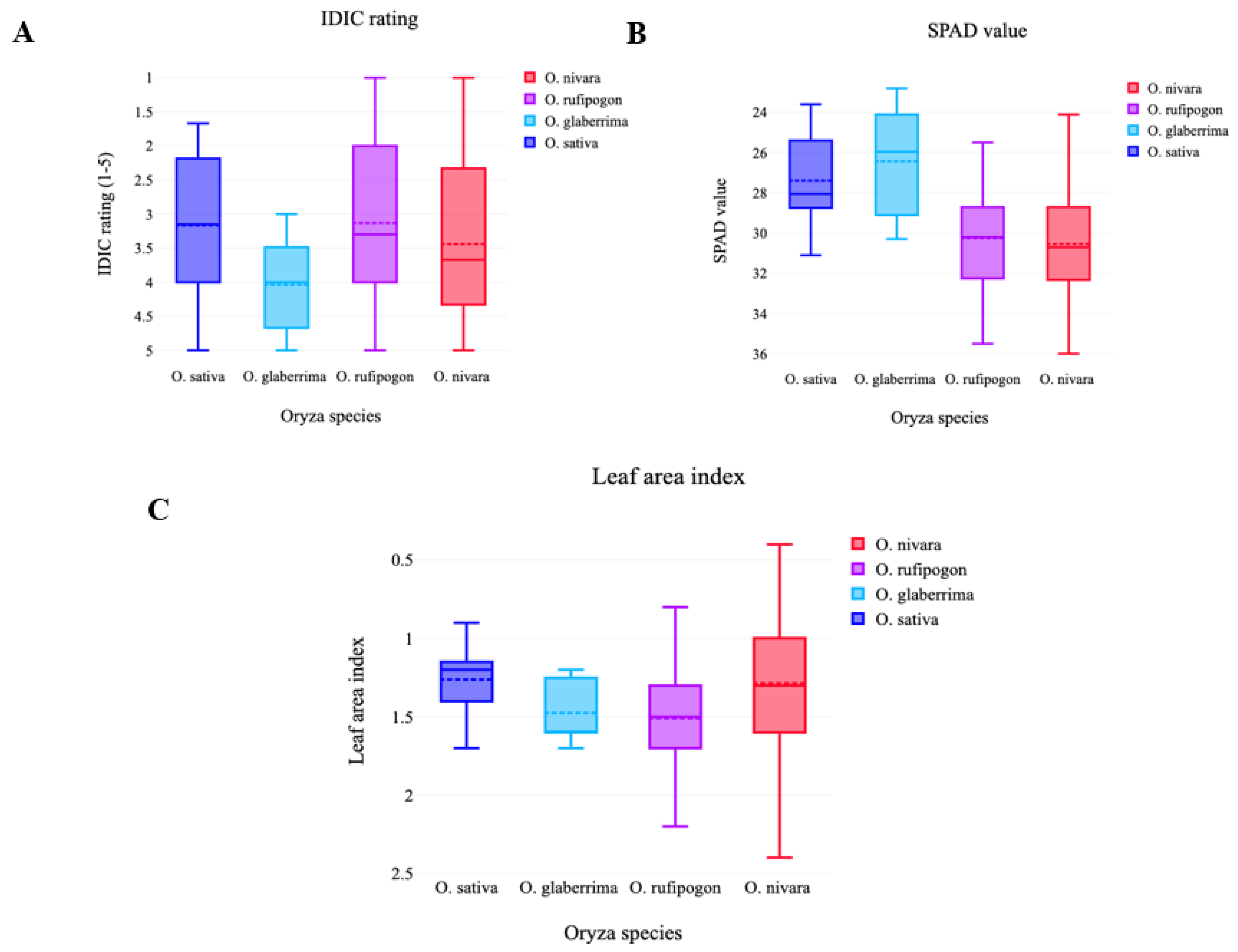
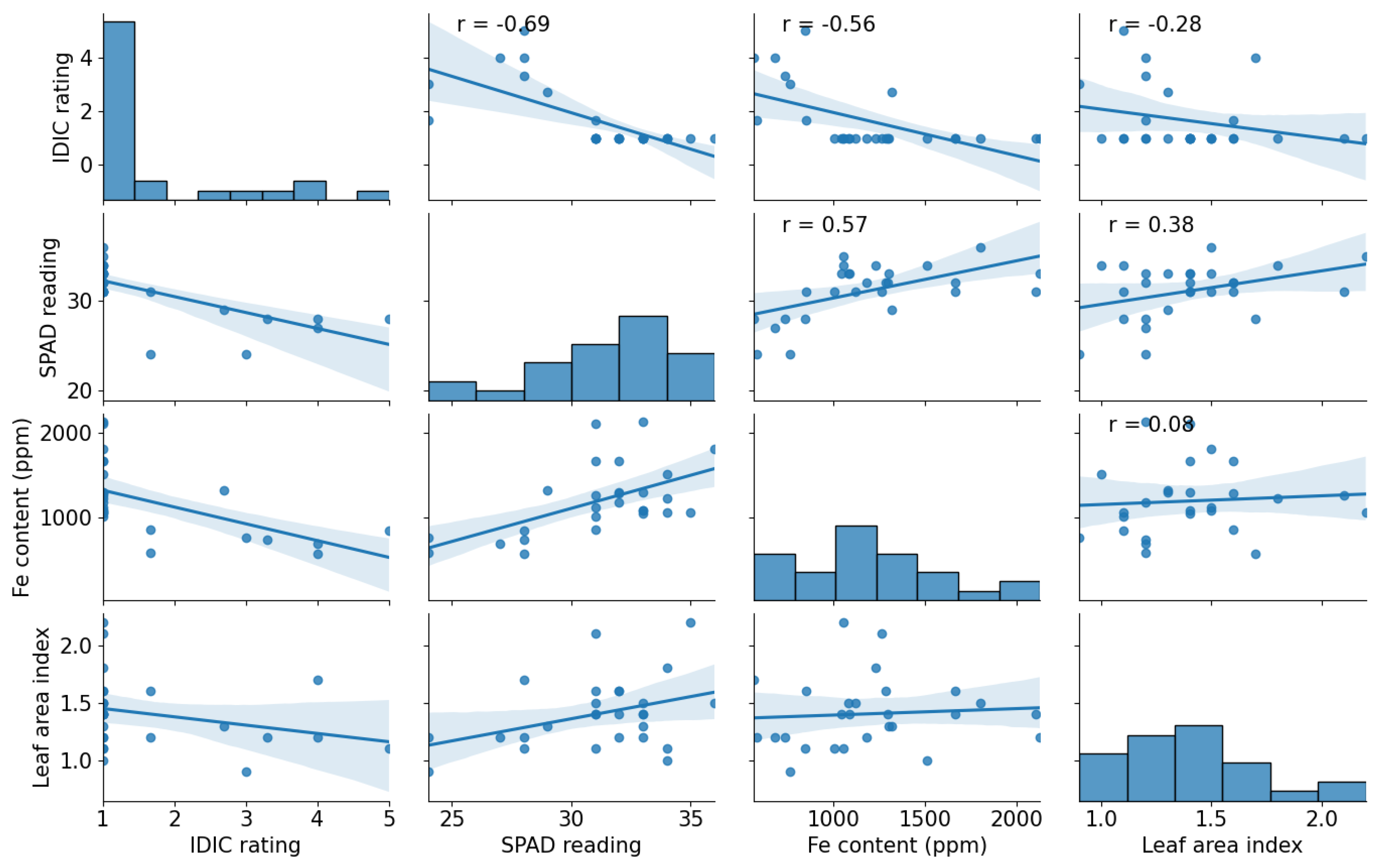
3.1. Wild Oryza germplasm exhibited significant variation for IDIC tolerance under aerobic field conditions
3.2. Wild Oryza germplasm exhibited significant variation for SPAD value
3.3. IDIC tolerant lines had more iron content in the leaves than susceptible cultivars
3.4. Wild germplasm exhibited significant variation for LAI
4. Discussion
Author Contributions
Conflict of Interest Statement
References
- Khush, G.S. Strategies for increasing the yield potential of cereals: Case of rice as an example. Plant Breed. Genet. Newsl. 2013, 67, 1–18. [Google Scholar] [CrossRef]
- FAOSTAT. FAOSTAT Database. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 30 March 2023).
- Gleick, P.H. Water use. Annu. Rev. Environ. Resour. 2003, 28, 275–314. [Google Scholar] [CrossRef]
- Tuong, T.P.; Bouman, B.A. Rice production in water-scarce environments. Water productivity in agriculture: Limits and opportunities for improvement 2003, 1, 13–42. [Google Scholar]
- Kumar, N.; Chhokar, R.S.; Meena, R.P.; Kharub, A.S.; Gill, S.C.; Tripathi, S.C.; Singh, G.P. Challenges and opportunities in productivity and sustainability of rice cultivation system: a critical review in Indian perspective. Cereal Res. Commun. 2021, 1–29. [Google Scholar] [CrossRef]
- Bouman, B.A.; Humphreys, E.; Tuong, T.P.; Barker, R. Rice and water. Adv. Agron. 2007, 92, 187–237. [Google Scholar]
- Fukai, S.; Mitchell, J. Factors determining water use efficiency in aerobic rice. Crop Environ. 2022, Mar 28.
- Bouman, B. Water-efficient management strategies in rice production. Int. Rice Res. Notes 2001, 16, 17–22. [Google Scholar]
- Farooq, M.K.H.M.; Siddique, K.H.; Rehman, H.; Aziz, T.; Lee, D.J.; Wahid, A. Rice direct seeding: experiences, challenges and opportunities. Soil Tillage Res. 2011, 111, 87–98. [Google Scholar] [CrossRef]
- Sagare, D.B.; Abbai, R.; Jain, A.; Jayadevappa, P.K.; Dixit, S.; Singh, A.K.; Kumar, A. More and more of less and less: Is genomics-based breeding of dry direct-seeded rice (DDSR) varieties the need of hour? Plant Biotechnol. J. 2020, 18, 2173–2186. [Google Scholar] [CrossRef]
- Fan, X.; Karim, M.R.; Chen, X.; Zhang, Y.; Gao, X.; Zhang, F.; Zou, C. Growth and iron uptake of lowland and aerobic rice genotypes under flooded and aerobic cultivation. Commun. Soil Sci. Plant Anal. 2012, 43, 1811–1822. [Google Scholar] [CrossRef]
- Shi, R.L.; Hao, H.M.; Fan, X.Y.; Karim, M.R.; Zhang, F.S.; Zou, C.Q. Responses of aerobic rice (Oryza sativa L.) to iron deficiency. J. Integr. Agric. 2012, 11, 938–945. [Google Scholar] [CrossRef]
- Nogiya, M.; Pandey, R.N.; Singh, B. Physiological basis of iron chlorosis tolerance in rice (Oryza sativa) in relation to the root exudation capacity. J. Plant Nutr. 2016, 39, 1536–1546. [Google Scholar] [CrossRef]
- Nogiya, M.; Pandey, R.N.; Singh, B.; Singh, G.; Meena, M.C.; Datta, S.C.; Meena, A.L. Responses of aerobically grown iron chlorosis tolerant and susceptible rice (Oryza sativa L.) genotypes to soil iron management in an Inceptisol. Arch. Agron. Soil Sci. 2019.
- Sperotto, R.A.; Ricachenevsky, F.K.; Waldow, V.A.; Fett, J.P. Iron biofortification in rice: it’s a long way to the top. Plant Sci. 2012, 190, 24–39. [Google Scholar] [CrossRef] [PubMed]
- Cakmak, I. Enrichment of cereal grains with zinc: agronomic or genetic biofortification? Plant Soil 2008, 302, 1–17. [Google Scholar] [CrossRef]
- Brar, D.S.; Khush, G.S. Alien introgression in rice. In Oryza: from Molecule to Plant; Khush, G.S., Ed.; Springer: Dordrecht, 1997; pp. 35–47. [Google Scholar]
- Tanksley, S.D.; McCouch, S.R. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 1997, 277, 1063–1066. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, D.A.; Lu, B.R.; Tomooka, N. The evolving story of rice evolution. Plant Sci. 2003, 164, 394–397. [Google Scholar] [CrossRef]
- Wang, J.; McLean, P.E.; Lee, R.; Goos, R.J.; Helms, T. Association mapping of iron deficiency chlorosis loci in soybean (Glycine max L. Merr.) advanced breeding lines. Theor. Appl. Genet. 2008, 116, 777–787. [Google Scholar] [CrossRef]
- Isaac, R.A.; Kerber, J.D. Atomic absorption and flame photometry: techniques and uses in soil, plant and water analysis. In Instrumental methods for analysis of soils and plant tissue; Walsh, L.M., Ed.; Soil Soc Am: Madison, WI, USA, 1971; pp. 18–37. [Google Scholar]
- He, K.; Lu, H.; Zheng, H.; Yang, Q.; Sun, G.; Zheng, Y.; Huan, X. Role of dynamic environmental change in sustaining the protracted process of rice domestication in the lower Yangtze River. Quaternary Sci. Rev. 2020, 242, 106456. [Google Scholar] [CrossRef]
- Razzaq, A.; Wani, S.H.; Saleem, F.; Yu, M.; Zhou, M.; Shabala, S. Rewilding crops for climate resilience: economic analysis and de novo domestication strategies. J. Exp. Bot. 2021, 72, 6123–6139. [Google Scholar] [CrossRef]
- Terry, N. Physiology of trace element toxicity and its relation to iron stress. J. Plant Nutr 1981, 3, 561–578. [Google Scholar] [CrossRef]
- Briat, J.F.; Lobréaux, S. Iron transport and storage in plants. Trends Plant Sci. 1997, 2, 187–193. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Suzuki, M.; Tsukamoto, T.; Suzuki, K.; Nakazono, M.; Kobayashi, T.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 2006, 45, 335–346. [Google Scholar] [CrossRef]
- Marschner, H.; Römheld, V.; Kissel, M. Different strategies in higher plants in mobilization and uptake of iron. J. Plant Nutr. 1986, 9, 695–713. [Google Scholar] [CrossRef]
- Zhang, F.; Cui, F.; Zhang, L.; Wen, X.; Luo, X.; Zhou, Y.; Xie, J. Development and identification of a introgression line with strong drought resistance at seedling stage derived from Oryza sativa L. mating with Oryza rufipogon Griff. Euphytica 2014, 200, 1–7. [Google Scholar] [CrossRef]
- Huang, F.; He, N.; Yu, M.; Li, D.; Yang, D. Identification and fine mapping of a new bacterial blight resistance gene, Xa43 (t), in Zhangpu wild rice (Oryza rufipogon). Plant Biol. 2023. [CrossRef] [PubMed]
- Yuan, R.; Zhao, N.; Usman, B.; Luo, L.; Liao, S.; Qin, Y.; Li, R. Development of chromosome segment substitution lines (CSSLs) derived from Guangxi wild rice (Oryza rufipogon Griff.) under rice (Oryza sativa L.) background and the identification of QTLs for plant architecture, agronomic traits and cold tolerance. Genes 2020, 11, 980. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Li, W.; Lin, Y.L.; Gao, L.Z. Genome-wide analysis of WRKY genes and their response to salt stress in the wild progenitor of Asian cultivated rice, Oryza rufipogon. Front. Genet. 2020, 11, 359. [Google Scholar] [CrossRef]
- Kumar, R.; Sahi, G.K.; Kaur, R.; Khanna, R.; Choudhary, O.P.; Mangat, G.S.; Singh, K. Tolerance response of wild and cultivated Oryza species under iron deficiency condition. J. Crop Improv. 2013, 40, 168–172. [Google Scholar]

| Species | First year | Second year | ||||
|---|---|---|---|---|---|---|
| IDIC rating* | SPAD reading* | Fe content (ppm)* |
LAI* | IDIC rating* | SPAD Reading* |
|
| O. rufipogon (IR105491) | 1±0 | 32.5±1.8 | 1294±131 | 1.4±0.13 | 1±0 | 33.1±1.3 |
| O. rufipogon (CR100334A) | 1±0 | 31.6±0.8 | 1119±79 | 1.5±0.21 | 1±0 | 30.9±0.9 |
| O. rufipogon (CR100334B) | 1±0 | 31.2±1.1 | 1261±113 | 2.1±0.19 | 1±0 | 33.9±1.3 |
| O. rufipogon (CR100436) | 1±0 | 32.7±1.9 | 1664±143 | 1.4±0.11 | 1.3±0.47 | 30.2±1.6 |
| O. rufipogon (CR100462) | 1±0 | 32.8±2.1 | 1042±92 | 1.4±0.16 | 1±0 | 32.4±1.7 |
| O. rufipogon (IR80762A) | 1±0 | 32.9±0.4 | 1229±75 | 1.8±0.09 | 1±0 | 34.3±0.9 |
| O. rufipogon (CR100030) | 1±0 | 33.3±1.4 | 2127±193 | 1.2±0.13 | 1±0 | 32.8±1.6 |
| O. rufipogon (CR100001) | 1±0 | 35.3±1.3 | 1056±69 | 2.2±0.10 | 1.3±0.47 | 34.6±1.6 |
| O. rufipogon (CR100484) | 1±0 | 32.5±1 | 1085±82 | 1.4±0.09 | 1±0 | 33.1±1.7 |
| O. nivara (CR100454) | 1±0 | 31±0.6 | 1005±63 | 1.1±0.14 | 1±0 | 31.8±1.6 |
| O. nivara (CR100363) | 1±0 | 32.3±2.2 | 2101±173 | 1.4±0.12 | 1±0 | 31.2±1.7 |
| O. nivara (CR100371) | 1±0 | 33.8±1.3 | 1055±103 | 1.1±0.12 | 1±0 | 33.2±1.1 |
| O. nivara (CR100003) | 1±0 | 36±1.6 | 1802±144 | 1.5±0.16 | 1±0 | 34.7±1.9 |
| O. nivara (IR82018) | 1±0 | 29.7±2.5 | 1287±103 | 1.6±0.17 | 1.66±0.47 | 31.9±1.9 |
| O. nivara (CR100124A) | 1±0 | 29.8±0.8 | 1081±89 | 1.5±0.10 | 1±0 | 31.5±0.9 |
| O. nivara (CR100127) | 1±0 | 30.3±1.4 | 1662±123 | 1.6±0.12 | 1±0 | 29.8±1.1 |
| O. nivara (CR100337) | 1±0 | 33.3±1.1 | 1299±91 | 1.3±0.12 | 1±0 | 33.9±0.8 |
| O. nivara (CR100426) | 1±0 | 31.6±1.9 | 1079±59 | 1.2±0.08 | 1±0 | 32.3±1.7 |
| O. nivara (CR100360) | 1±0 | 33.5±0.9 | 1508±179 | 1±0.10 | 1±0 | 31.5±1.1 |
| O. nivara (CR100429) | 1±0 | 29.4±1.6 | 1318±132 | 1.3±0.07 | 1±0 | 30.1±1.5 |
| PAU 201 | 2.7±0.27 | 27.9±1.2 | 565±51 | 1.7±0.11 | 3±0 | 31.7±0.9 |
| Rasi | 4±0 | 29.2±1.7 | 560±60 | 1.2±0.09 | 3.3±0.27 | 29.7±1.3 |
| Basmati 370 | 1.67±0.47 | 23.9±0.9 | 580±44 | 1.2±0.07 | 2±0.0 | 27.5±2.1 |
| Cheng-Hui 448 | 4±0 | 26.9±2.1 | 682±77 | 1.2±0.10 | 4±0 | 26.9±1.1 |
| Feng-ai-zai | 5±0 | 28.2±2.8 | 847±92 | 1.1±0.11 | 5±0 | 29.1±1.9 |
| IR 64 | 3.3±0.27 | 28.3±1.7 | 734±55 | 1.2±0.10 | 4±0 | 26.8±1.9 |
| Lemont | 1.67±0.47 | 31.1±0.8 | 851±73 | 1.6±0.08 | 2±0.0 | 32.1±1.8 |
| Nagina 22 | 3±0 | 23.6±1.9 | 765±61 | 0.9±0.08 | 3.3±0.27 | 26.1±1.4 |
| SPAD value | LAI | |||||
|---|---|---|---|---|---|---|
| Species | Mean | Maximum | Minimum | Mean | Maximum | Minimum |
| O. nivara | 30.6 | 36.0 (CR100003) | 22.2 (CR100123) | 1.3 | 2.6 (CR100007) |
0.4 (CR100399) |
| O. rufipogon | 30.8 | 35.3 (CR100001) | 25.5 (CR100461) | 1.5 | 2.2 (CR100001) |
0.8 (CR100465B) |
| O. glaberrima | 26.4 | 30.3 (IR102980) | 22.8 (IR102489) | 1.5 | 1.7 (IR102489) |
1.2 (IR 102980) |
| O. barthii | 24.9 | 24.9 (IR103580) | 24.9 (IR103580) | 1.4 | 1.4 (IR103580) | 1.4 (IR103580) |
| O. sativa | 28.3 | 31.1 (Lemont) | 23.6 (Nagina 22) |
1.2 | 1.7 (PAU 201) |
0.9 (Nagina 22) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





