Submitted:
06 April 2023
Posted:
07 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Population
2.2. Lipid indexes and genetic analyses
3. Results
3.1. Population and lipid characteristics
3.2. Genetic analyses
3.3. Genotype-phenotype associations
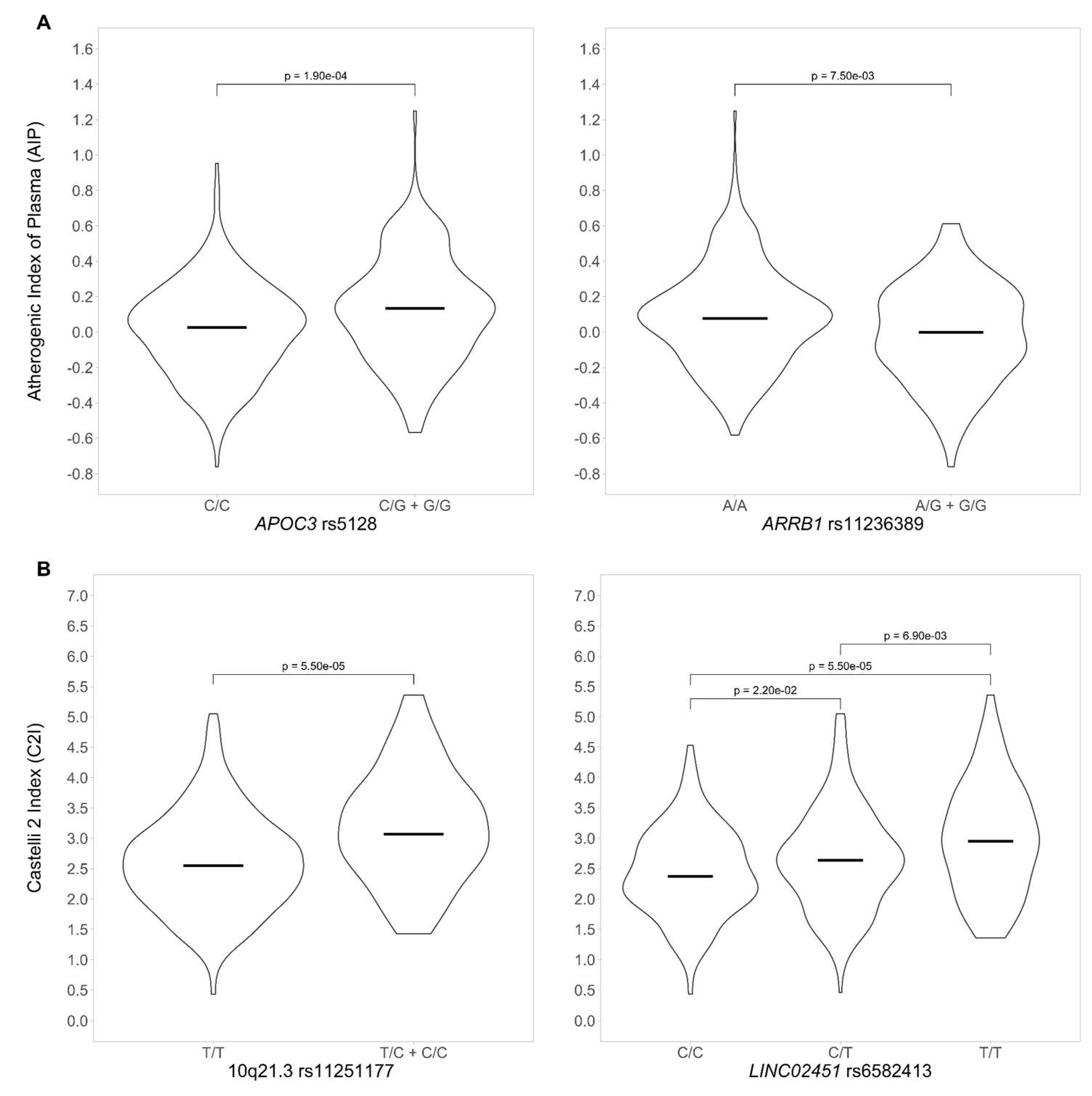
3.3.1. Genotype-phenotype association for AIP
3.3.2. Genotype-phenotype association for CI2
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lange LA, Lange EM, Bielak LF, Langefeld CD, Kardia SL, Royston P, Turner ST, Sheedy PF, Boerwinkle E, Peyser PA. Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler Thromb Vasc Biol (2002) 22:418– 423. [CrossRef]
- Herrington W, Lacey B, Sherliker P, Armitage J, Lewington S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ Res (2016) 118:535–546. [CrossRef]
- Araújo JA, Lusis AJ. “Atherosclerosis.,” Encyclopedia of Endocrine Diseases. Elsevier (2004). p. 282–288. [CrossRef]
- Scipione CA, Cybulsky MI. Early atherogenesis: new insights from new approaches. Curr Opin Lipidol (2022) 2022:2–12. [CrossRef]
- Huang Y, Gao L, Xie X, Tan SC. Epidemiology of dyslipidemia in Chinese adults: meta- analysis of prevalence, awareness, treatment, and control. Popul Health Metr (2014) 12:28. [CrossRef]
- Bayram F, Kocer D, Gundogan K, Kaya A, Demir O, Coskun R, Sabuncu T, Karaman A, Cesur M, Rizzo M, et al. Prevalence of dyslipidemia and associated risk factors in Turkish adults. J Clin Lipidol (2014) 8:206–216. [CrossRef]
- Urina-Jassir M, Pacheco-Paez T, Paez-Canro C, Urina-Triana M. Statin associated adverse reactions in Latin America: A scoping review. BMJ Open (2021) 11:. [CrossRef]
- Pu J, Romanelli R, Zhao B, Azar KMJ, Hastings KG, Nimbal V, Fortmann SP, Palaniappan LP. Dyslipidemia in Special Ethnic Populations. Endocrinol Metab Clin North Am (3AD) 45:205–216. [CrossRef]
- Gao P, Wen X, Ou Q, Zhang J. Which one of LDL-C /HDL-C ratio and non-HDL-C can better predict the severity of coronary artery disease in STEMI patients. BMC Cardiovasc Disord (2022) 22:318. [CrossRef]
- Teslovich TM, Musunuru K, Smith A V, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature (2010) 466:707–713. [CrossRef]
- Hindy G, Dornbos P, Chaffin MD, Liu DJ, Wang M, Selvaraj MS, Zhang D, Park J, Aguilar- Salinas CA, Antonacci-Fulton L, et al. Rare coding variants in 35 genes associate with circulating lipid levels—A multi-ancestry analysis of 170,000 exomes. Am J Hum Genet (2022) 109:81–96. [CrossRef]
- Weissglas-Volkov D, Aguilar-Salinas CA, Nikkola E, Deere KA, Cruz-Bautista I, Arellano- Campos O, Muñoz-Hernandez LL, Gomez-Munguia L, Ordoñez-Sánchez ML, Linga Reddy PMV, et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J Med Genet (2013) 50:298. [CrossRef]
- Weissglas-Volkov D, Aguilar-Salinas CA, Sinsheimer JS, Riba L, Huertas-Vazquez A, Ordoñez-Sánchez ML, Rodriguez-Guillen R, Cantor RM, Tusie-Luna T, Pajukanta P. Investigation of Variants Identified in Caucasian Genome-Wide Association Studies for Plasma High-Density Lipoprotein Cholesterol and Triglycerides Levels in Mexican Dyslipidemic Study Samples. Circ Cardiovasc Genet (2010) 3:31–38. [CrossRef]
- Olamoyegun M, Oluyombo R, Asaolu S. Evaluation of dyslipidemia, lipid ratios, and atherogenic index as cardiovascular risk factors among semi-urban dwellers in Nigeria. Ann Afr Med (2016) 15:194–199. [CrossRef]
- Liu H, Liu K, Pei L, Li S, Zhao J, Zhang K, Zong C, Zhao L, Fang H, Wu J, et al. Atherogenic Index of Plasma Predicts Outcomes in Acute Ischemic Stroke. Front Neurol (2021) 12:. [CrossRef]
- Dobiášová M, Frohlich J. The plasma parameter log (TG/HDL-C) as an atherogenic index: correlation with lipoprotein particle size and esterification rate in apoB-lipoprotein-depleted plasma (FER(HDL)). Clin Biochem (2001) 34:583–588. [CrossRef]
- Rivera-Mancía S, Colín-Ramírez E, Cartas-Rosado R, Infante O, Vargas-Barrón J, Vallejo M. Indicators of accumulated fat are stronger associated with prehypertension compared with indicators of circulating fat: A cross-sectional study. Medicine (Baltimore) (2018) 97:. [CrossRef]
- Colín-Ramírez E, Rivera-Manciá S, Infante-Vázquez O, Cartas-Rosado R, Vargas-Barrón J, Madero M, Vallejo M. Protocol for a prospective longitudinal study of risk factors for hypertension incidence in a Mexico City population: The Tlalpan 2020 cohort. BMJ Open (2017) 7:. [CrossRef]
- Rivera-Mancía S, Colín-Ramírez E, Cartas-Rosado R, Infante O, Vargas-Barrón J, Vallejo M. Indicators of accumulated fat are stronger associated with prehypertension compared with indicators of circulating fat: A cross-sectional study. Medicine (Baltimore) (2018) 97:. [CrossRef]
- Liu T, Liu J, Wu Z, Lv Y, Li W. Predictive value of the atherogenic index of plasma for chronic total occlusion before coronary angiography. Clin Cardiol (2021) 44:518–525. [CrossRef]
- Fernández-Macías JC, Ochoa-Martínez AC, Varela-Silva JA, Pérez-Maldonado IN. Atherogenic Index of Plasma: Novel Predictive Biomarker for Cardiovascular Illnesses. Arch Med Res (2019) 50:285–294. [CrossRef]
- Koca TT, Tugan CB, Seyithanoglu M, Kocyigit BF. The Clinical Importance of the Plasma Atherogenic Index, Other Lipid Indexes, and Urinary Sodium and Potassium Excretion in Patients with Stroke. Eurasian J Med (2019) 51:171–175. [CrossRef]
- Team RC. A Language and Environment for Statistical Computing. https://www.R-project.org. (2014).
- Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics (2011) 12:246. [CrossRef]
- Turner S, Armstrong LL, Bradford Y, Carlson CS, Crawford DC, Crenshaw AT, de Andrade M, Doheny KF, Haines JL, Hayes G, et al. Quality Control Procedures for Genome-Wide Association Studies. Curr Protoc Hum Genet (2011) 68:1.19.1-1.19.18. [CrossRef]
- Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics (2011) 12:1–6. [CrossRef]
- Gonzalez-Covarrubias V. Lipidomics in longevity and healthy aging. Biogerontology (2013) 14:663–672. [CrossRef]
- Boyle AP, Hong EL, Hariharan M, Cheng Y, Schaub MA, Kasowski M, Karczewski KJ, Park J, Hitz BC, Weng S, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res (2012) 22:1790–1797. [CrossRef]
- Afsin A, Kaya H, Suner A, Uzel KE, Bursa N, Hosoglu Y, Yavuz F, Asoglu R. Plasma atherogenic indices are independent predictors of slow coronary flow. BMC Cardiovasc Disord (2021) 21:608. [CrossRef]
- Millán J, Pintó X, Muñoz A, Zúñiga M, Rubiés-Prat J, Pallardo LF, Masana L, Mangas A, Hernández-Mijares A, González-Santos P, et al. Lipoprotein ratios: Physiological significance and clinical usefulness in cardiovascular prevention. Vasc Health Risk Manag (2009) 5:757– 765. [CrossRef]
- Kammar-García A, López-Moreno P, Hernández-Hernández ME, Ortíz-Bueno AM, Martínez- Montaño M de LC. Atherogenic index of plasma as a marker of cardiovascular risk factors in Mexicans aged 18 to 22 years. Proc (Bayl Univ Med Cent) (2020) 34:22–27. [CrossRef]
- Ahn SS, Lee LE, Pyo JY, Song JJ, Park YB, Lee SW. Atherogenic index of plasma predicts cerebrovascular accident occurrence in antineutrophil cytoplasmic antibody-associated vasculitis. Lipids Health Dis (2020) 19:1–11. [CrossRef]
- Du R, Li M, Wang X, Wang S, Li S, Tian H, Wu Y, Zhang C. LDL-C/HDL-C ratio associated with carotid intima-media thickness and carotid plaques in male but not female patients with type 2 diabetes. Clin Chim Acta (2020) 511:215–220. [CrossRef]
- Talmud PJ, Hawe E, Miller GJ, Humphries SE. Nonfasting Apolipoprotein B and Triglyceride Levels as a Useful Predictor of Coronary Heart Disease Risk in Middle-Aged UK Men. Arter Thromb Vasc Biol (2002) 22:1918–1923. [CrossRef]
- Ko A, Cantor RM, Weissglas-Volkov D, Nikkola E, Reddy PMVL, Sinsheimer JS, Pasaniuc B, Brown R, Alvarez M, Rodriguez A, et al. Amerindian-specific regions under positive selection harbour new lipid variants inLatinos. Nat Commun (2014) 5:. [CrossRef]
- Luo H, Zhang X, Shuai P, Miao Y, Ye Z, Lin Y. Genetic variants influencing lipid levels and risk of dyslipidemia in Chinese population. J Genet (2017) 96:985–992. [CrossRef]
- Song Y, Zhu L, Richa M, Li P, Yang Y, Li S. Associations of the APOC3 rs5128 polymorphism with plasma APOC3 and lipid levels: a meta-analysis. (2015). [CrossRef]
- Wu J, Zhou Q, Wei Z, Wei J, Cui M. Atherogenic Index of Plasma and Coronary Artery Disease in the Adult Population: A Meta-Analysis. Front Cardiovasc Med (2021) 0:1927. [CrossRef]
- Rocco CA, Mecikovsky D, Aulicino P, Bologna R, Sen L. Hypercholesterolemia Is Associated with the Apolipoprotein C-III (APOC3) Genotype in Children Receiving HAART: An Eight- Year Retrospective Study. PLoS One (2012) 7:39678. [CrossRef]
- Hsueh W-C, Nair AK, Kobes S, Chen P, Göring HHH, Pollin TI, Malhotra A, Knowler WC, Baier LJ, Hanson RL. Identity-by-Descent Mapping Identifies Major Locus for Serum Triglycerides in Amerindians Largely Explained by an APOC3 Founder Mutation. Circ Cardiovasc Genet (2017) 10:. [CrossRef]
- Jurado-Camacho PA, Cid-Soto MA, Barajas-Olmos F, García-Ortíz H, Baca-Peynado P, Martínez-Hernández A, Centeno-Cruz F, Contreras-Cubas C, González-Villalpando ME, Saldaña-Álvarez Y, et al. Exome Sequencing Data Analysis and a Case-Control Study in Mexican Population Reveals Lipid Trait Associations of New and Known Genetic Variants in Dyslipidemia-Associated Loci. Front Genet (2022) 13:807381. [CrossRef]
- Davis JP, Vadlamudi S, Roman TS, Zeynalzadeh M, Iyengar AK, Mohlke KL. Enhancer deletion and allelic effects define a regulatory molecular mechanism at the VLDLR cholesterol GWAS locus. Hum Mol Genet (2019) 28:888. [CrossRef]
- Van Den Berg ME, Warren HR, Cabrera CP, Verweij N, Mifsud B, Haessler J, Bihlmeyer NA, Fu YP, Weiss S, Lin HJ, et al. Discovery of novel heart rate-associated loci using the Exome Chip. Hum Mol Genet (2017) 26:2346. [CrossRef]
- Savarese M, Maggi L, Vihola A, Jonson PH, Tasca G, Ruggiero L, Bello L, Magri F, Giugliano T, Torella A, et al. Interpreting Genetic Variants in Titin in Patients With Muscle Disorders. JAMA Neurol (2018) 75:557–565. [CrossRef]
- Tharp CA, Haywood ME, Sbaizero O, Taylor MRG, Mestroni L. The Giant Protein Titin’s Role in Cardiomyopathy: Genetic, Transcriptional, and Post-translational Modifications of TTN and Their Contribution to Cardiac Disease. Front Physiol (2019) 10:1436. [CrossRef]
- Ye D, Zhou W, Hamrick SK, Tester DJ, Kim CSJ, Barajas-Martinez H, Hu D, Giudicessi JR, Antzelevitch C, Ackerman MJ. Acacetin, a Potent Transient Outward Current Blocker, May Be a Novel Therapeutic for KCND3 -Encoded Kv4.3 Gain-of-Function-Associated J-Wave Syndromes. Circ Genomic Precis Med (2022) 15:419–428. [CrossRef]
- Zhu Z, Guo Y, Shi H, Liu CL, Panganiban RA, Chung W, O’Connor LJ, Himes BE, Gazal S, Hasegawa K, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol (2020) 145:537–549. [CrossRef]
- El-Ansary A, Chirumbolo S, Bhat RS, Dadar M, Ibrahim EM, Bjørklund G. The Role of Lipidomics in Autism Spectrum Disorder. Mol Diagn Ther (2020) 24:31–48. [CrossRef]
- Tierney E, Remaley AT, Thurm A, Jager LR, Wassif CA, Kratz LE, Bailey-Wilson JE, Bukelis I, Sarphare G, Jung ES, et al. Sterol and lipid analyses identifies hypolipidemia and apolipoprotein disorders in autism associated with adaptive functioning deficits. Transl Psychiatry (2021) 11:. [CrossRef]
- Yano Y, Butler KR, Hall ME, Schwartz GL, Knopman DS, Lirette ST, Jones DW, Wilson JG, Hall JE, Correa A, et al. Associations of Nocturnal Blood Pressure With Cognition by Self- Identified Race in Middle-Aged and Older Adults: The GENOA (Genetic Epidemiology Network of Arteriopathy) Study. J Am Heart Assoc (2017) 6:. [CrossRef]
- Zanni G, Hsiao C-T, Fu S-J, Tang C-Y, Capuano A, Bosco L, Graziola F, Bellacchio E, Servidei S, Primiano G, et al. Novel KCND3 Variant Underlying Nonprogressive Congenital Ataxia or SCA19/22 Disrupt K V 4.3 Protein Expression and K+ Currents with Variable Effects on Channel Properties. Int J Mol Sci Artic Int J Mol Sci (2021) 22:4986. [CrossRef]
- Pollini L, Galosi S, Tolve M, Caputi C, Carducci C, Angeloni A, Leuzzi V. Molecular Sciences KCND3-Related Neurological Disorders: From Old to Emerging Clinical Phenotypes. [CrossRef]
- Feitosa MF, Myers RH, Pankow JS, Province MA, Borecki IB. LIPC variants in the promoter and intron 1 modify HDL-C levels in a sex-specific fashion. Atherosclerosis (2009) 204:171– 177. [CrossRef]
- Talmud PJ, Drenos F, Shah S, Shah T, Palmen J, Verzilli C, Gaunt TR, Pallas J, Lovering R, Li K, et al. Gene-centric association signals for lipids and apolipoproteins identified via the HumanCVD BeadChip. Am J Hum Genet (2009) 85:628–642. [CrossRef]
| Males N = 170 |
Females N = 256 |
All N = 426 |
|
|---|---|---|---|
| Age, y | 38 (18-53) | 40 (17-52) | 39 (17-53) |
| Weight, kg | 78.1 (51.2 -125) | 63.1 (41.7 - 119) | 70 (41.7 - 125) |
| Height, m | 1.70 (1.50 - 1.99) | 1.57 (1.36 - 1.72) | 1.61 (1.36 - 1.99) |
| BMI, kg/m2 | 26.8 (16.9 - 40.3) | 26.2 (16.9 - 47.1) | 26.4 (16.8 - 47.1) |
| Waist circumference, cm | 94.0 (63.0 – 130) | 85.0 (54.0 – 126) | 89.0 (54.0 – 130) |
| Glucose, mg/dL | 94.0 (72.0 – 166) | 90.0 (74.0 – 241) | 92.0 (72.0 – 241) |
| Uric acid, mg/dL | 6.34 (1.82 – 10.0) | 4.62 (2.30 – 7.58) | 5.31 (1.82 – 10.0) |
| Creatinine, mg/dL | 0.95 (0.62 – 1.40) | 0.69 (0.44 – 1.19) | 0.77 (0.44 – 1.40) |
| Cholesterol, mmol/dL | 4.62 (2.96 – 8.30) | 4.39 (2.16 – 7.06) | 4.49 (2.16 – 8.30) |
| HDL-C, mmol/dL | 1.10 (0.60 – 2.12) | 1.22 (0.73 – 2.27) | 1.16 (0.60 – 2.27) |
| LDL-C, mmol/dL | 3.09 (0.98 – 6.87) | 2.93 (0.54 – 5.38) | 3.00 (0.54 – 6.87) |
| Triglycerides (TG), mmol/dL | 1.48 (0.47 – 15.4) | 1.22 (0.22 – 5.86) | 1.32 (0.22 – 15.34) |
| Dyslipidemia, n (%) | 119 (70%) | 85 (72%) | 304 (71%) |
| Castelli risk index 2 (CI2)1 | 2.91 (1.25 – 5.36) | 2.45 (0.43 – 4.92) | 2.60 (0.43 – 5.36) |
| Atherogenic index of plasma (AIP)2 | 0.48 (-0.18 – 1.61) | 0.38 (-0.40 – 1.17) | 0.42 (-0.40 – 1.61) |
| High TG > 1.9 mmol/L | High cholesterol > 5 mmol/L | High LDL-C > 3.9 mmol/L | Low HDL-C < 1.04 mmol/L | |
|---|---|---|---|---|
| Males %, n1 | 30%, 51 | 35.3%, 60 | 16.5%, 28 | 38%, 65 |
| Females %, n1 | 16%, 41 | 22.3%, 59 | 10.9%, 28 | 26%, 67 |
| p-value2 | 8.74E-06 | 2.32E-03 | 1.80E-03 | 2.59E-05 |
| Gene | Chr | rs identifier | Coefficient | p-value |
|---|---|---|---|---|
| Variants associated to AIP | ||||
| APOA1/APOC3 | 11 | rs5128 | 0.094 | 2.61E-06 |
| CYBA | 16 | rs12709102 | 0.078 | 3.91E-06 |
| ARRIB1 | 11 | rs11236389 | -0.102 | 6.63E-06 |
| TTN/CCDC141 | 2 | rs10497528 | 0.089 | 8.29E-06 |
| APOA1/APOC3 | 11 | rs5072 | 0.091 | 8.94E-06 |
| Variants associated to CI2 | ||||
| Intergenic | 10q21.3 | rs11251177 | 0.606 | 1.07E-07 |
| LINC02451 | 12 | rs6582413 | 0.259 | 5.19E-07 |
| LINC02451 | 12 | rs12817366 | 0.254 | 1.88E-06 |
| Intergenic | 12 | rs34115639 | 0.244 | 7.06E-06 |
| Intergenic | 12 | rs10880344 | -0.233 | 7.10E-06 |
| Intergenic | 6 | rs7762658 | -0.247 | 2.03E-06 |
| LIPC/ALDH1A2 | 15 | rs261342 | 0.227 | 1.10E-06 |
| DIPK2B | 23 | rs4294309 | 0.306 | 1.18E-05 |
| KCND3 | 1 | rs6703437 | -0.234 | 1.76E-05 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





