Submitted:
31 March 2023
Posted:
07 April 2023
You are already at the latest version
Abstract
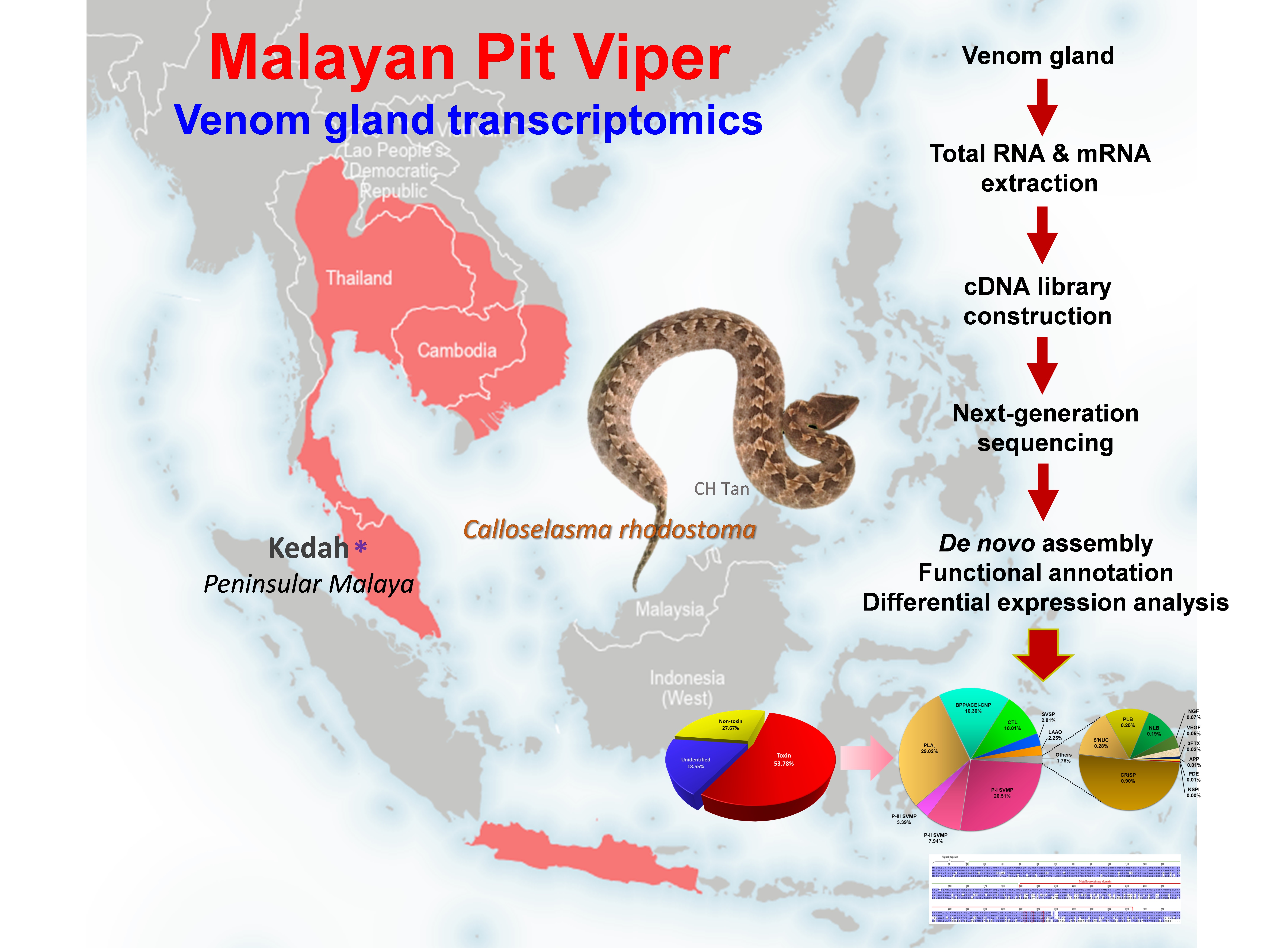
Keywords:
1. Introduction
2. Results and Discussion
2.1. De novo transcriptome assembly, transcripts categorization and expression
| Parameter | Value |
|---|---|
| Total raw reads | 47,365,132 |
| Total clean reads | 46,636,384 |
| Total clean nucleotides (nt) | 4,663,638,400 |
| Q20 percentage | 97.17% |
| N percentage | 0.00% |
| GC percentage | 44.52% |
| Contigs created | 146,916 |
| Total length (nt) | 47,039,053 |
| Mean length (nt) | 320 |
| N50 | 632 |
| Unigenes/transcripts assembled | 74,445 |
| Total length (nt) | 52,612,410 |
| Mean length (nt) | 707 |
| N50 | 1636 |
| Unigene/transcripts assembled (FPKM>1) | 59,348 |
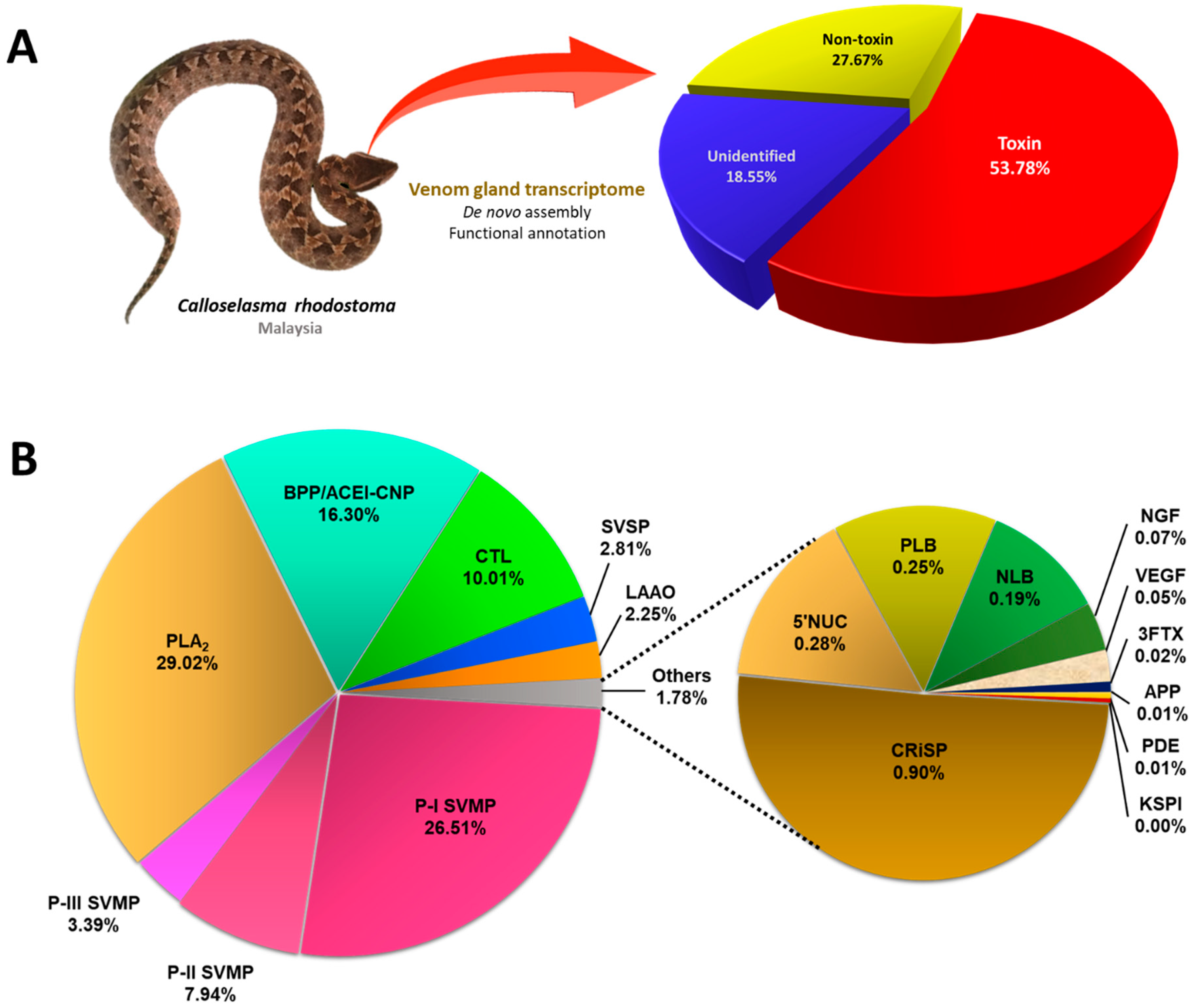
| Unidentified | 38,289 |
| Non-toxin | 20,962 |
| Toxin | 97 |
2.2. Overview of toxin gene expression in C. rhodostoma venom gland transcriptome
| Protein family/Protein subtype | Accession no. (Species) | Relative abundance % (subtype) |
|---|---|---|
| Snake venom metalloproteinase (SVMP) | 37.84 (22) | |
| P-I SVMP | 26.51 (4) | |
| Snake venom metalloproteinase kistomin | P0CB14 (Calloselasma rhodostoma) | 26.29 |
| Snake venom metalloproteinase BpirMP | P0DL29 (Bothrops pirajai) | 0.17 |
| Zinc metalloproteinase/disintegrin ussurin | Q7SZD9 (Gloydius ussuriensis) | 0.03 |
| Group I snake venom metalloproteinase | Q2UXQ3 (Echis ocellatus) | 0.02 |
| P-II SVMP | 7.94 (4) | |
| Zinc metalloproteinase/disintegrin | P30403 (Calloselasma rhodostoma) | 7.89 |
| Zinc metalloproteinase/disintegrin ussurin | Q7SZD9 (Gloydius ussuriensis) | 0.04 |
| Metalloprotease PIIa | V5IWE4 (Trimeresurus gracilis) | 0.01 |
| Zinc metalloproteinase-disintegrin VMP-II | J9Z332 (Crotalus adamanteus) | < 0.01 |
| P-III SVMP | 3.39 (14) | |
| Zinc metalloproteinase-disintegrin-like halysase | Q8AWI5 (Gloydius halys) | 3.32 |
| Metalloprotease P-III | A0A077L6L9 (Protobothrops elegans) | 0.03 |
| Zinc metalloproteinase-disintegrin-like NaMP | A8QL59 (Naja atra) | 0.01 |
| Metalloprotease P-III | A0A077L6L9 (Protobothrops elegans) | 0.01 |
| Flavorase | G1UJB2 (Protobothrops flavoviridis) | 0.01 |
| Zinc metalloproteinase-disintegrin-like NaMP | A8QL59 (Naja atra) | 0.01 |
| Metalloprotease P-III 5 | A0A077L7M5 (Protobothrops flavoviridis) | < 0.01 |
| Metalloproteinase | A0A2Z4N9U9 (Boiga irregularis) | < 0.01 |
| Zinc metalloproteinase-disintegrin-like NaMP | A8QL59 (Naja atra) | < 0.01 |
| Metalloprotease P-III 5 | A0A077L7M5 (Protobothrops flavoviridis) | < 0.01 |
| Metalloproteinase | A0A2Z4N9U9 (Boiga irregularis) | < 0.01 |
| Zinc metalloproteinase-disintegrin-like NaMP | A8QL59 (Naja atra) | < 0.01 |
| Metalloprotease P-III 5 | A0A077L7M5 (Protobothrops flavoviridis) | < 0.01 |
| Zinc metalloproteinase-disintegrin-like NaMP | A8QL59 (Naja atra) | < 0.01 |
| Phospholipase A2 (PLA2) | 29.02 (15) | |
| Phospholipase A2 | A0A0H3U266 (Calloselasma rhodostoma) | 16.47 |
| Acidic phospholipase A2 S1E6-c | Q9PVE9 (Calloselasma rhodostoma) | 5.19 |
| K49 phospholipase A2 | A8Y7N3 (Deinagkistrodon acutus) | 3.62 |
| Acidic phospholipase A2 Ts-A4 | Q6H3C7 (Trimeresurus stejnegeri) | 2.74 |
| Phospholipase A2 homolog | P0DMT1 (Echis pyramidum leakeyi) | 0.91 |
| Phospholipase A2 | A0A0H3U279 (Ovophis makazayazaya) | 0.06 |
| Phospholipase A2 group IIE | A0A2H4N3A5 (Bothrops moojeni) | 0.02 |
| Phospholipase A2, group IIE | A0A1J0R065 (Crotalus atrox) | 0.01 |
| Group 3 secretory phospholipase A2 | A0A223PK36 (Daboia russelii) | < 0.01 |
| Basic phospholipase A2 beta-bungarotoxin A4 chain | Q75S51 (Bungarus candidus) | < 0.01 |
| Phospholipase A2 isoform 2 | H8PG83 (Parasuta nigriceps) | < 0.01 |
| Group 15 secretory phospholipase A2 | A0A223PK35 (Daboia russelii) | < 0.01 |
| Acidic phospholipase A2 homolog | P29601 (Bungarus fasciatus) | < 0.01 |
| Acidic phospholipase A2 | P00606 (Bungarus multicinctus) | < 0.01 |
| Group 3 secretory phospholipase A2 | A0A223PK36 (Daboia russelii) | < 0.01 |
| Bradykinin-potentiating/Angiotensin-converting enzyme inhibitor/C-type natriuretic peptide (BPP/ACEI-CNP) | 16.30 (3) | |
| Angiotensin converting enzyme inhibitor and C-type natriuretic peptide | M5A7D0 (Calloselasma rhodostoma) | 5.77 |
| Angiotensin converting enzyme inhibitor and C-type natriuretic peptide | M5A7D0 (Calloselasma rhodostoma) | 5.51 |
| Angiotensin converting enzyme inhibitor and C-type natriuretic peptide | M5A7D0 (Calloselasma rhodostoma) | 5.02 |
| Snake C-type lectin (CTL) | 10.01 (7) | |
| Snaclec rhodocytin subunit beta | Q9I840 (Calloselasma rhodostoma) | 4.33 |
| C-type lectin | G8FML6 (Calloselasma rhodostoma) | 3.26 |
| Snaclec rhodocytin subunit alpha | Q9I841 (Calloselasma rhodostoma) | 1.76 |
| Snaclec rhodocetin subunit delta | D2YW40 (Calloselasma rhodostoma) | 0.37 |
| C-type lectin beta subunit | T2HPS7 (Protobothrops flavoviridis) | 0.27 |
| Lectoxin-Enh9 | A7XQ58 (Pseudoferania polylepis) | 0.01 |
| C-type lectin 3 | A0A346CLX6 (Ahaetulla prasina) | 0.01 |
| Snake venom serine proteinase (SVSP) | 2.81 (14) | |
| Thrombin-like enzyme ancrod | P26324 (Calloselasma rhodostoma) | 1.92 |
| Snake venom serine protease 3 | O13058 (Protobothrops flavoviridis) | 0.20 |
| Snake venom serine protease ussurin | Q8UUJ2 (Gloydius ussuriensis) | 0.19 |
| Snake venom serine protease gussurobin | Q8UVX1 (Gloydius ussuriensis) | 0.14 |
| Venom thrombin-like enzyme | Q90Z47 (Deinagkistrodon acutus) | 0.12 |
| Thrombin-like enzyme | Q98TT5 (Deinagkistrodon acutus) | 0.08 |
| Thrombin-like enzyme stejnobin | Q8AY81 (Trimeresurus stejnegeri) | 0.08 |
| Snake venom serine protease 3 | O13063 (Trimeresurus gramineus) | 0.02 |
| Venom plasminogen activator GPV-PA | P0DJF5 (Trimeresurus albolabris) | 0.02 |
| Thrombin-like enzyme ancrod-2 | P47797 (Calloselasma rhodostoma) | 0.02 |
| Serine protease 3 | A0A286S0D3 (Gloydius intermedius) | 0.01 |
| Thrombin-like enzyme kangshuanmei | P85109 (Gloydius brevicaudus) | 0.01 |
| Serine proteinase isoform 7 | B0VXT9 (Sistrurus catenatus edwardsii) | < 0.01 |
| Thrombin-like protein DAV-WY | B3V4Z6 (Deinagkistrodon acutus) | < 0.01 |
| L-amino acid oxidase (LAAO) | 2.25 (1) | |
| L-amino-acid oxidase | P81382 (Calloselasma rhodostoma) | 2.25 |
| Cysteine-rich secretory protein (CRiSP) | 0.90 (2) | |
| Cysteine-rich secretory protein LCCL domain-containing 2 | V8NV17 (Ophiophagus hannah) | 0.90 |
| Cysteine-rich seceretory protein Bc-CRPa | F2Q6G3 (Bungarus candidus) | < 0.01 |
| 5’Nucleotidase (5’NUC) | 0.28 (5) | |
| Snake venom 5’-nucleotidase | F8S0Z7 (Crotalus adamanteus) | 0.27 |
| 5’ nucleotidase | A6MFL8 (Demansia vestigiata) | < 0.01 |
| 5’ nucleotidase | A6MFL8 (Demansia vestigiata) | < 0.01 |
| 5’ nucleotidase | A6MFL8 (Demansia vestigiata) | < 0.01 |
| 5’ nucleotidase 1 | A0A346CLX4 (Borikenophis portoricensis) | < 0.01 |
| Phospholipase B (PLB) | 0.25 (4) | |
| Phospholipase B-like | A0A2H4N395 (Bothrops moojeni) | 0.25 |
| Phospholipase B1, membrane-associated | V8NN21 (Ophiophagus hannah) | < 0.01 |
| Phospholipase B-like | V8NLQ9 (Ophiophagus hannah) | < 0.01 |
| Phospholipase B-like | V8NLQ9 (Ophiophagus hannah) | < 0.01 |
| Nucleobindin (NLB) | 0.19 (1) | |
| Nucleobindin-1 | V8P8E3 (Ophiophagus hannah) | 0.19 |
| Nerve growth factor | 0.07 (1) | |
| Nerve growth factor | B1Q3K2 (Protobothrops flavoviridis) | 0.07 |
| Snake venom vascular endothelial growth factor (VEGF) | 0.05 (1) | |
| Snake venom vascular endothelial growth factor toxin | P67862 (Protobothrops flavoviridis) | 0.05 |
| Three-finger toxin (3FTX) | 0.02 (9) | |
| Alpha-bungarotoxin isoform A31 | P60615 (Bungarus multicinctus) | 0.01 |
| Neurotoxin-like protein pMD18-NTL1/2/4/5 | Q7ZT13 (Bungarus multicinctus) | < 0.01 |
| Muscarinic toxin BM14 | Q8JFX7 (Bungarus multicinctus) | < 0.01 |
| Kappa-3-bungarotoxin | P15817 (Bungarus multicinctus) | < 0.01 |
| Gamma-bungarotoxin | Q9YGJ0 (Bungarus multicinctus) | < 0.01 |
| Three finger toxin 1 | A5X2W6 (Sistrurus catenatus edwardsii) | < 0.01 |
| Short neurotoxin homolog NTL4 | Q9YGI8 (Bungarus multicinctus) | < 0.01 |
| Three finger toxin 2 | A5X2W7 (Sistrurus catenatus edwardsii) | < 0.01 |
| Putative three finger toxin | F5CPD4 (Micrurus altirostris) | < 0.01 |
| Aminopeptidase A | 0.01 (1) | |
| Aminopeptidase | T2HQN1 (Ovophis okinavensis) | 0.01 |
| Phosphodiesterase (PDE) | 0.01 (5) | |
| Venom phosphodiesterase 1 | J3SEZ3 (Crotalus adamanteus) | < 0.01 |
| Venom phosphodiesterase 1 | J3SEZ3 (Crotalus adamanteus) | < 0.01 |
| Venom phosphodiesterase 2 | J3SBP3 (Crotalus adamanteus) | < 0.01 |
| Venom phosphodiesterase 1 | J3SEZ3 (Crotalus adamanteus) | < 0.01 |
| Venom phosphodiesterase 2 | J3SBP3 (Crotalus adamanteus) | < 0.01 |
| Kunitz-type serine proteinase inhibitor (KSPI) | < 0.01 (1) | |
| Kunitz-type serine protease inhibitor homolog beta-bungarotoxin B2a chain | Q8AY45 (Bungarus candidus) | < 0.01 |
| Protein family/Protein ID | Annotated accession | Species | Amino acid chain | Mature chain of accession ID | Coverage (mature chain) | Coverage percentage (%) | |
|---|---|---|---|---|---|---|---|
| Snake venom metalloproteinase (SVMP) | |||||||
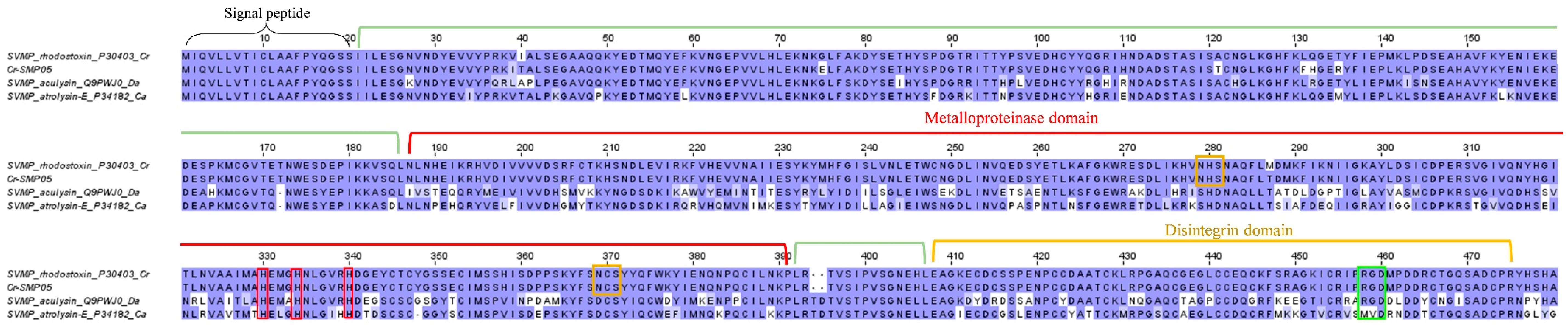
| Cr-SVMP01 | Snake venom metalloproteinase kistomin | P0CB14 | Calloselasma rhodostoma | 417 | 417 | 1-417 |
100 |
| Cr-SVMP05 | Zinc metalloproteinase/disintegrin | P30403 | Calloselasma rhodostoma | 478 | 478 | 1-478 | 100 |
| Phospholipase A2 (PLA2) | |||||||
| Cr-PLA04 |
Acidic phospholipase A2 Ts-A4 | Q6H3C7 | Trimeresurus stejnegeri | 139 | 139 | 1-139 | 100 |
| Cr-PLA10 |
Basic phospholipase A2 beta-bungarotoxin A4 chain | Q75S51 |
Bungarus candidus |
147 | 147 | 1-147 | 100 |
| Cr-PLA11 |
Phospholipase A2 isoform 2 | H8PG83 | Parasuta nigriceps | 136 | 136 | 1-136 | 100 |
| Cr-PLA12 |
Group 15 secretory phospholipase A2 | A0A223PK35 | Daboia russelii | 362 | 393 | 1-341 | 92 |
| Snake C-type lectin (Snaclec) | |||||||
| Cr-CTL01 | Snaclec rhodocytin subunit beta | Q9I840 | Calloselasma rhodostoma | 146 | 146 | 1-146 | 100 |
| Cr-CTL02 | C-type lectin | G8FML6 |
Agkistrodon piscivorus leucostoma |
157 | 158 | 1-158 | 99 |
| Cr-CTL03 | Snaclec rhodocytin subunit alpha | Q9I841 | Calloselasma rhodostoma | 136 | 136 | 1-136 | 100 |
| Cr-CTL04 | Snaclec rhodocetin subunit beta | P81398 | Calloselasma rhodostoma | 129 | 129 | 1-129 | 100 |
| Cr-CTL05 | Snaclec rhodocetin subunit delta | D2YW40 | Calloselasma rhodostoma | 150 | 150 | 1-150 | 100 |
| Snake venom serine proteinase (SVSP) | |||||||
| Cr-SSP01 |
Thrombin-like enzyme ancrod | P26324 | Calloselasma rhodostoma | 234 | 234 | 1-234 | 100 |
| Cr-SSP03 |
Snake venom serine protease ussurin | Q8UUJ2 |
Gloydius ussuriensis |
224 | 236 | 13-236 | 95 |
| L-amino acid oxidase (LAAO) | |||||||
| Cr-LAO01 |
L-amino-acid oxidase | P81382 | Calloselasma rhodostoma | 516 | 516 | 1-516 | 100 |
| Cysteine-richvenom protein (CRiSP) | |||||||
| Cr-CRP01 | Cysteine-rich secretory protein LCCL domain-containing 2 | V8NV17 | Ophiphagus Hannah | 495 | 472 | 1-472 | 100 |
| 5’Nucleotidase (5’NUC) | |||||||
| Cr-NUC01 |
Snake venom 5’-nucleotidase | F8S0Z7 |
Crotalus adamanteus |
588 | 588 | 1-588 | 100 |
| Phospholipase B (PLB) | |||||||
| Cr-PLB01 | Phospholipase B-like | A0A2H4N395 |
Bothrops moojeni |
553 | 558 | 1-553 | 99 |
| Cr-PLB03 | Phospholipase B-like | V8NLQ9 | Ophiophagus Hannah | 321 | 300 | 87-299 | 100 |
| Nucleobindin (NLB) | |||||||
| Cr-NLB01 |
Nucleobindin-1 | V8P8E3 | Ophiophagus Hannah | 452 | 397 | 22-372 | 100 |
| Nerve growth factor (NGF) | |||||||
| Cr-NGF01 |
Nerve growth factor | B1Q3K2 | Protobothrops flavoviridis | 237 | 241 | 1-237 | 98 |
| Snake venom vascular endothelial growth factor (VEGF) | |||||||
| Cr-VGF01 |
Snake venom vascular endothelial growth factor toxin | P67862 | Protobothrops flavoviridis | 145 | 146 | 1-145 | 99 |
| Three finger toxin (3FTX) | |||||||
| Cr-FTX01 |
Alpha-bungarotoxin isoform A31 | P60615 | Bungarus multicinctus | 95 | 95 | 1-95 | 100 |
| Cr-FTX02 |
Neurotoxin-like protein pMD18-NTL1/2/4/5 | Q7ZT13 | Bungarus multicinctus | 86 | 86 | 1-86 | 100 |
| Cr-FTX03 |
Muscarinic toxin BM14 | Q8JFX7 | Bungarus multicinctus | 97 | 103 | 7-103 | 94 |
| Aminopeptidase A | |||||||
| Cr-APP01 | Aminopeptidase | T2HQN1 |
Ovophis okinavensis |
953 | 953 | 1-953 | 100 |
| Phosphodiesterase (PDE) | |||||||
| Cr-PDE02 |
Venom phosphodiesterase 1 | J3SEZ3 |
Crotalus adamanteus |
844 | 851 | 6-849 | 99 |
| Cr-PDE03 |
Venom phosphodiesterase 2 | J3SBP3 |
Crotalus adamanteus |
808 | 810 | 1-808 | 99 |
| Cr-PDE04 |
Venom phosphodiesterase 1 | J3SEZ3 |
Crotalus adamanteus |
849 | 851 | 1-849 | 99 |
| Cr-PDE05 |
Venom phosphodiesterase 2 | J3SBP3 |
Crotalus adamanteus |
803 | 810 | 6-808 | 99 |
| Kunitz-type serine proteinase inhibitor (KSPI) | |||||||
| Cr-KUN01 |
Kunitz-type serine protease inhibitor homolog beta-bungarotoxin B2a chain | Q8AY45 |
Bungarus candidus |
84 | 85 | 2-85 | 99 |
2.3. Toxin gene diversity and implication on bioactivity of snake venom
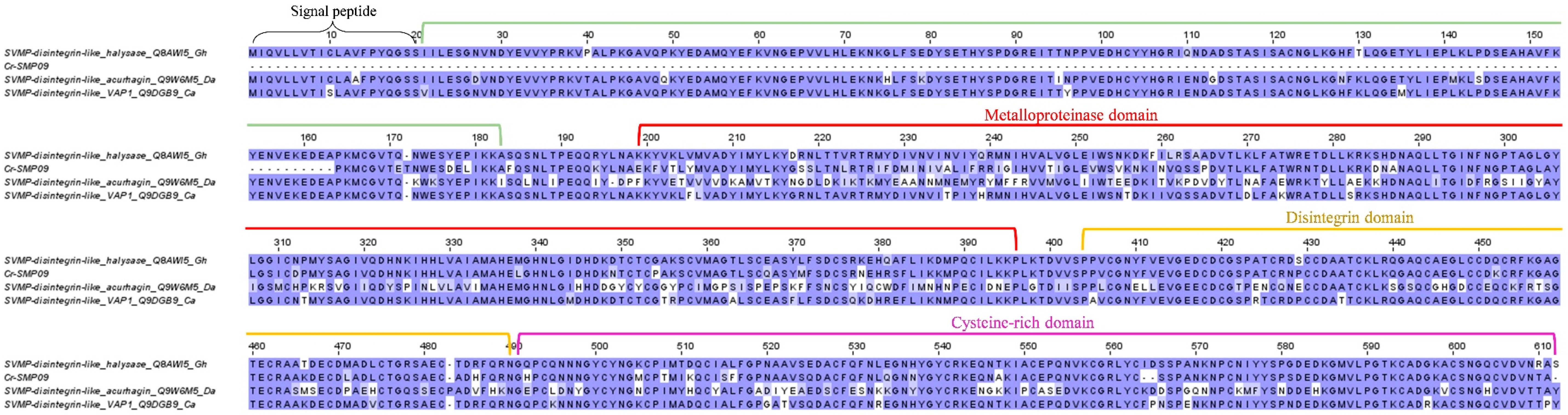
2.3.1. Snake venom metalloproteinase (SVMP)

2.3.2. Phospholipase A2 (PLA2)
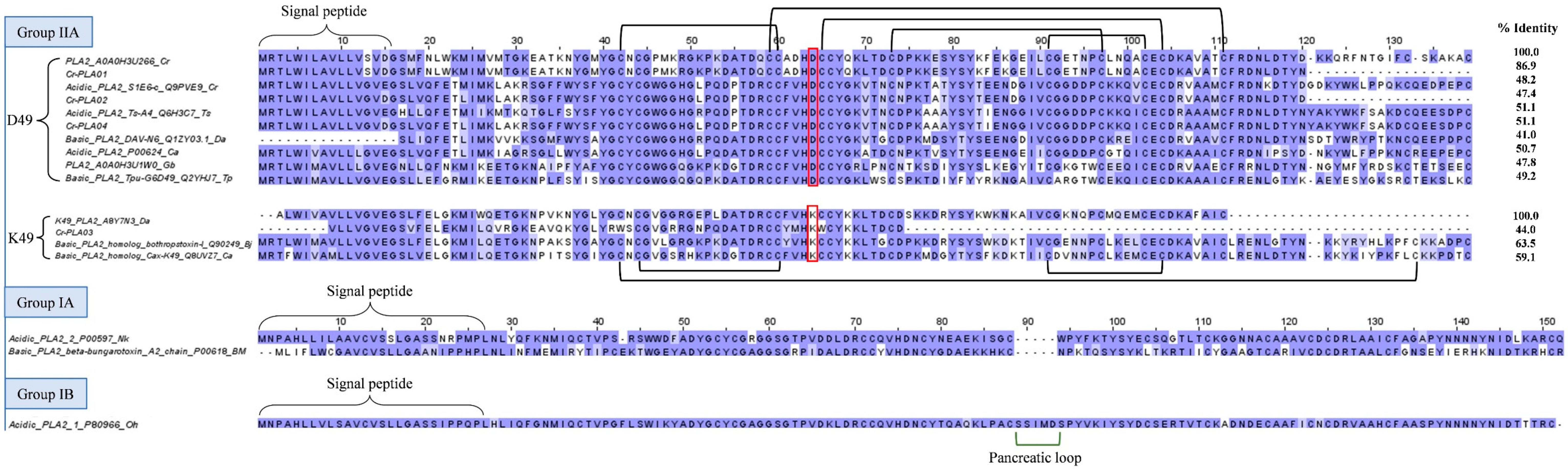
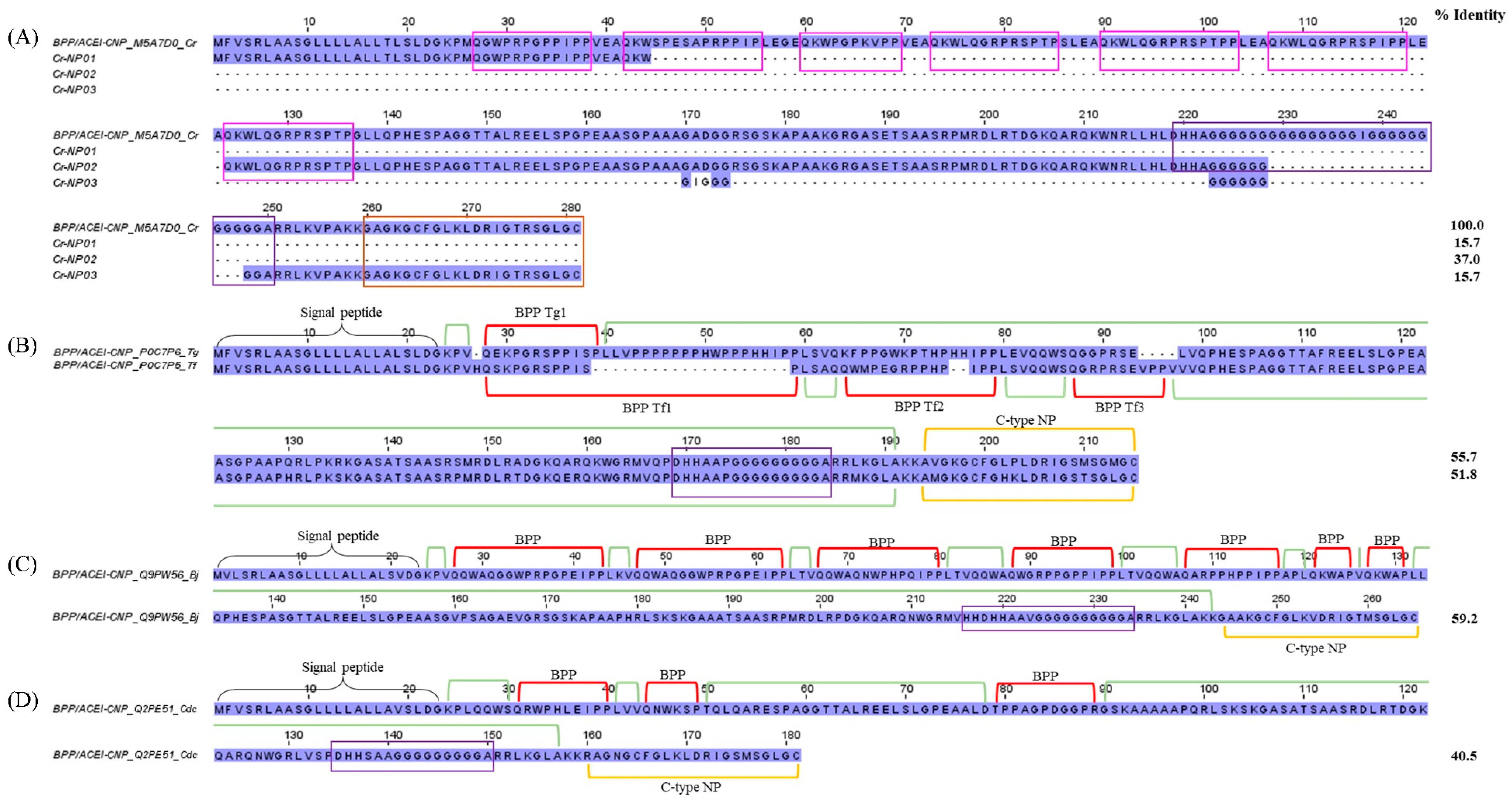
2.3.3. Bradykinin-potentiating peptide (BPP)/Angiotensinogen-converting enzyme inhibitor (ACEI) and Natriuretic peptide (NP)
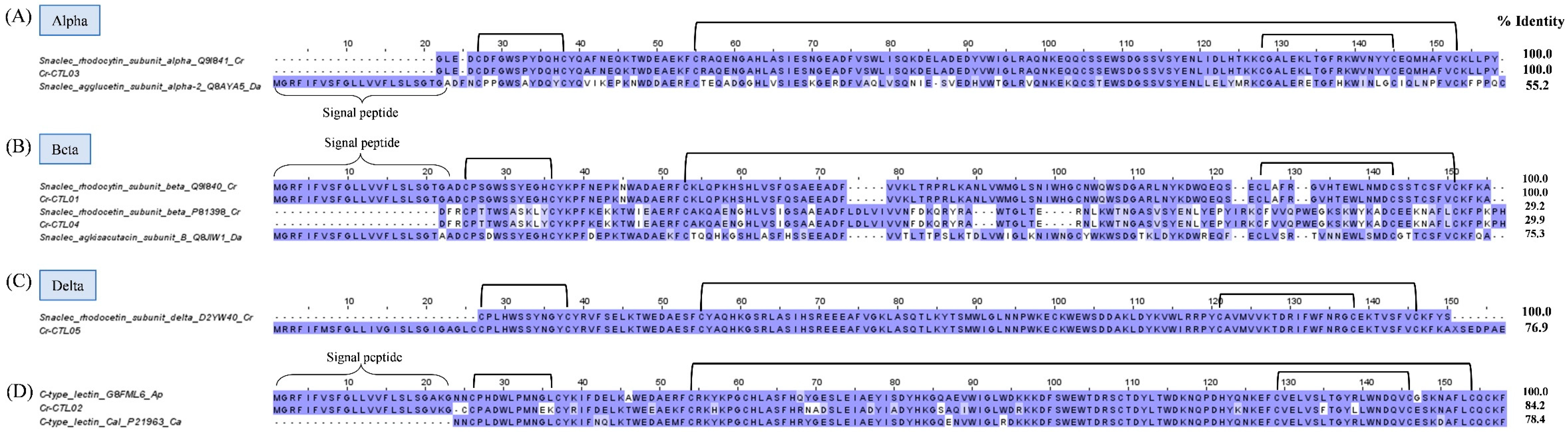
2.3.4. Snake C-type lectins
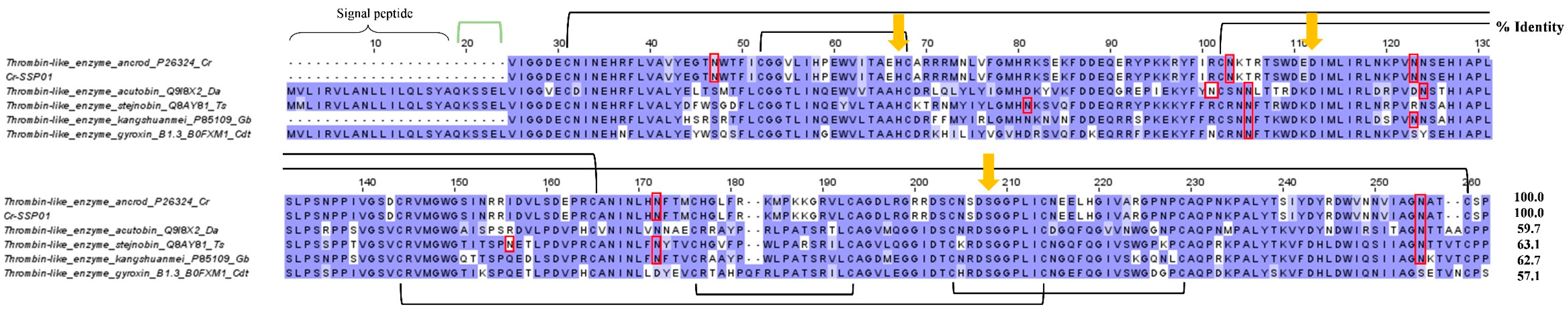
2.3.5. Snake venom serine proteinase (SVSP)
2.3.6. L-amino acid oxidase (LAAO)
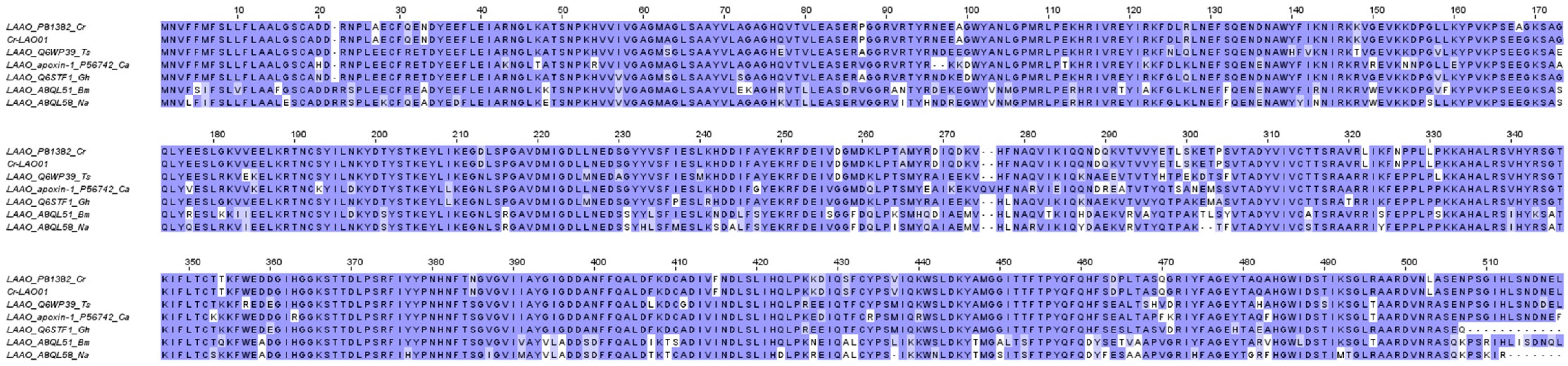
2.4. Low-abundance toxin transcripts
2.4.1. Toxins detected in both venom gland transcriptome and venom proteome
2.4.2. Toxins detected exclusively in venom gland transcriptome
3. Conclusion
5. Materials and Methods
5.1. Preparation of C. rhodostoma venom gland tissue
5.2. RNA extraction and mRNA purification
5.3. Filtration of raw sequenced reads
5.4. De novo transcriptome assembly
5.5. Clustering and functional annotation of transcripts
5.6. Quantifying transcript abundance
5.7. Categorization of transcripts
5.8. Sequence alignment and analysis
5.9. Supporting data
5.10. Statistical analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Gutiérrez, J.M.; Calvete, J.J.; Habib, A.G.; Harrison, R.A.; Williams, D.J.; Warrell, D.A. Snakebite envenoming. Nature Reviews Disease Primers 2017, 3, 17063. [Google Scholar] [CrossRef]
- Sanders, K.L.; Lee, M.S. Molecular evidence for a rapid late-Miocene radiation of Australasian venomous snakes (Elapidae, Colubroidea). Molecular Phylogenetics and Evolution 2008, 46, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Alencar, L.R.V.; Martins, M.; Greene, H.W. Evolutionary History of Vipers. eLS 2018. [Google Scholar] [CrossRef]
- WHO, W.H.O. Guidelines for the management of snakebites, 2nd ed. WHO Regional Office for South-East Asia. 2016.
- Wüster, W.; Peppin, L.; Pook, C.E.; Walker, D.E. A nesting of vipers: phylogeny and historical biogeography of the Viperidae (Squamata: Serpentes). Molecular Phylogenetics and Evolution 2008, 49, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Das, I. Field guide to the reptiles of South-East Asia; Bloomsbury Publishing: 2015.
- Uetz, P.; Freed, P.; Aguilar, R.; Reyes, F.; Hošek, J. The Reptile Database, http://www.reptile-database.org, accessed [20 March 2022]. Available online: http://www.reptile-database.org (accessed on. 20 March.
- Warrell, D.A. Tropical snakebite-clinical-studies in South East-Asia. Toxicon 1985, 23. [Google Scholar]
- Reid, H.A.; Chan, K.; Thean, P. Prolonged coagulation defect (defibrination syndrome) in Malayan viper bite. Lancet 1963, 621–626. [Google Scholar] [CrossRef]
- Reid, H.; Thean, P.; Chan, K.; Baharom, A. Clinical Effects of Bites by Malayan Viper (Ancistrodon rhodostomd). Lancet 1963, 617–621. [Google Scholar] [CrossRef]
- Wongtongkam, N.; Wilde, H.; Sitthi-Amorn, C.; Ratanabanangkoon, K. A study of 225 Malayan pit viper bites in Thailand. Military medicine 2005, 170, 342–348. [Google Scholar] [CrossRef]
- Kunalan, S.; Othman, I.; Syed Hassan, S.; Hodgson, W. Proteomic Characterization of Two Medically Important Malaysian Snake Venoms, Calloselasma rhodostoma (Malayan Pit Viper) and Ophiophagus hannah (King Cobra). Toxins 2018, 10, 434. [Google Scholar] [CrossRef]
- Tang, E.L.H.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics of Calloselasma rhodostoma, the Malayan pit viper: A complex toxin arsenal unraveled. Journal of Proteomics 2016, 148, 44–56. [Google Scholar] [CrossRef]
- Vejayan, J.; Khoon, T.L.; Ibrahim, H. Comparative analysis of the venom proteome of four important Malaysian snake species. Journal of venomous animals and toxins including tropical diseases 2014, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.A.; Baumann, K.; Jackson, T.N.W.; Wood, K.; Mason, S.; Undheim, E.A.B.; Nouwens, A.; Koludarov, I.; Hendrikx, I.; Jones, A.; et al. Proteomic comparison of Hypnale hypnale (Hump-Nosed Pit-Viper) and Calloselasma rhodostoma (Malayan Pit-Viper) venoms. Journal of Proteomics 2013, 91, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.L.H.; Tan, N.H.; Fung, S.Y.; Tan, C.H. Comparative proteomes, immunoreactivities and neutralization of procoagulant activities of Calloselasma rhodostoma (Malayan pit viper) venoms from four regions in Southeast Asia. Toxicon 2019, 169, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.L.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics of Calloselasma rhodostoma, the Malayan pit viper: A complex toxin arsenal unraveled. J Proteomics 2016. [Google Scholar] [CrossRef]
- Ismail, A.K. Snakebite and envenomation management in Malaysia. Toxinology: Clinical Toxinology in Asia Pacific and Africa 2015, 2, 71–102. [Google Scholar]
- Tan, C.H. Snake Venomics: Fundamentals, Recent Updates, and a Look to the Next Decade. Toxins 2022, 14, 247. [Google Scholar] [CrossRef]
- Neiva, M.; Arraes, F.B.; de Souza, J.V.; Radis-Baptista, G.; da Silva, A.R.P.; Walter, M.E.M.; de Macedo Brigido, M.; Yamane, T.; Lopez-Lozano, J.L.; Astolfi-Filho, S. Transcriptome analysis of the Amazonian viper Bothrops atrox venom gland using expressed sequence tags (ESTs). Toxicon 2009, 53, 427–436. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Liu, Q.; Yin, W.; Zhang, X.; Huang, Y.; Luo, Y.; Qiu, P.; Su, X.; Yu, J.; Hu, S.; et al. Transcriptome analysis of Deinagkistrodon acutus venomous gland focusing on cellular structure and functional aspects using expressed sequence tags. BMC genomics 2006, 7, 152. [Google Scholar] [CrossRef]
- Yang, Z.-M.; Yang, Y.-E.; Chen, Y.; Cao, J.; Zhang, C.; Liu, L.-L.; Wang, Z.-Z.; Wang, X.-M.; Wang, Y.-M.; Tsai, I.-H. Transcriptome and proteome of the highly neurotoxic venom of Gloydius intermedius. Toxicon 2015, 107, 175–186. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Chanhome, L.; Tan, N.H. Comparative venom gland transcriptomics of Naja kaouthia (monocled cobra) from Malaysia and Thailand: elucidating geographical venom variation and insights into sequence novelty. PeerJ 2017, 5, e3142. [Google Scholar] [CrossRef]
- Tan, C.H.; Tan, K.Y.; Fung, S.Y.; Tan, N.H. Venom-gland transcriptome and venom proteome of the Malaysian king cobra (Ophiophagus hannah). BMC genomics 2015, 16, 687. [Google Scholar] [CrossRef] [PubMed]
- Chong, H.P.; Tan, K.Y.; Tan, N.H.; Tan, C.H. Exploring the diversity and novelty of toxin genes in Naja sumatrana, the Equatorial spitting cobra from Malaysia through de novo venom-gland transcriptomics. Toxins 2019, 11, 104. [Google Scholar] [CrossRef] [PubMed]
- Palasuberniam, P.; Tan, K.Y.; Tan, C.H. De novo venom gland transcriptomics of Calliophis bivirgata flaviceps: uncovering the complexity of toxins from the Malayan blue coral snake. Journal of Venomous Animals and Toxins including Tropical Diseases 2021, 27. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y. De Novo venom-gland transcriptomics of spine-bellied sea snake (Hydrophis curtus) from Penang, Malaysia—next-generation sequencing, functional annotation and toxinological correlation. Toxins 2021, 13, 127. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wagstaff, S.C.; Wüster, W.; Cook, D.A.N.; Bolton, F.M.S.; King, S.I.; Pla, D.; Sanz, L.; Calvete, J.J.; Harrison, R.A. Medically important differences in snake venom composition are dictated by distinct postgenomic mechanisms. 2014, 111, 9205-9210. https://doi.org/10.1073/pnas.1405484111 %J Proceedings of the National Academy of Sciences. [CrossRef]
- Margres, M.J.; McGivern, J.J.; Wray, K.P.; Seavy, M.; Calvin, K.; Rokyta, D.R. Linking the transcriptome and proteome to characterize the venom of the eastern diamondback rattlesnake (Crotalus adamanteus). Journal of Proteomics 2014, 96, 145–158. [Google Scholar] [CrossRef]
- Bjarnason, J.B.; Fox, J.W. Hemorrhagic metalloproteinases from snake venoms. Pharmacology & Therapeutics 1994, 62, 325–372. [Google Scholar] [CrossRef]
- Takeda, S.; Takeya, H.; Iwanaga, S. Snake venom metalloproteinases: Structure, function and relevance to the mammalian ADAM/ADAMTS family proteins. Biochim Biophys Acta Proteins Proteom 2012, 1824, 164–176. [Google Scholar] [CrossRef]
- Huang, T.-F.; Chang, M.-C.; Teng, C.-M. Antiplatelet protease, kistomin, selectively cleaves human platelet glycoprotein Ib. Biochimica et Biophysica Acta (BBA) - General Subjects 1993, 1158, 293–299. [Google Scholar] [CrossRef]
- Hsu, C.; Wu, W.; Huang, T. A snake venom metalloproteinase, kistomin, cleaves platelet glycoprotein VI and impairs platelet functions. Journal of ThrombosisHaemostasis 2008, 6, 1578–1585. [Google Scholar] [CrossRef]
- Casewell, N.R.; Wagstaff, S.C.; Harrison, R.A.; Renjifo, C.; Wüster, W. Domain loss facilitates accelerated evolution and neofunctionalization of duplicate snake venom metalloproteinase toxin genes. Molecular biology and evolution 2011, 28, 2637–2649. [Google Scholar] [CrossRef]
- Au, L.-C.; Huang, Y.-B.; Huang, T.-F.; Teh, G.-W.; Lin, H.-H.; Choo, K.-B. A common precursor for a putative hemorrhagic protein and rhodostomin, a platelet aggregation inhibitor of the venom of Calloselasma rhodostoma: molecular cloning and sequence analysis. Biochemical and biophysical research communications 1991, 181, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Chung, M.C.M.; Ponnudurai, G.; Kataoka, M.; Shimizu, S.; Tan, N.-H. Structural Studies of a Major Hemorrhagin (Rhodostoxin) from the Venom ofCalloselasma rhodostoma (Malayan Pit Viper). Archives of biochemistry and biophysics 1996, 325, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Tan, N.-H.; Ponnudurai, G.; Chung, M.C.M. Proteolytic specificity of rhodostoxin, the major hemorrhagin of Calloselasma rhodostoma (Malayan pit viper) venom. Toxicon 1997, 35, 979–984. [Google Scholar] [CrossRef]
- Yeh, C.-H.; Peng, H.-C.; Yang, R.-S.; Huang, T.-F. Rhodostomin, a snake venom disintegrin, inhibits angiogenesis elicited by basic fibroblast growth factor and suppresses tumor growth by a selective αvβ3 blockade of endothelial cells. Molecular pharmacology 2001, 59, 1333–1342. [Google Scholar] [CrossRef] [PubMed]
- Au, L.-C.; Chou, J.-S.; Chang, K.-J.; Teh, G.-W.; Lin, S.-B. Nucleotide sequence of a full-length cDNA encoding a common precursor of platelet aggregation inhibitor and hemorrhagic protein from Calloselasma rhodostoma venom. Biochimica et Biophysica Acta (BBA)-Gene Structure and Expression 1993, 1173, 243–245. [Google Scholar] [CrossRef]
- Fujii, Y.; Okuda, D.; Fujimoto, Z.; Horii, K.; Morita, T.; Mizuno, H. Crystal structure of trimestatin, a disintegrin containing a cell adhesion recognition motif RGD. Journal of molecular biology 2003, 332, 1115–1122. [Google Scholar] [CrossRef]
- Ponnudurai, G.; Chung, M.C.M.; Tan, N.-H. Isolation and characterization of a hemorrhagin from the venom of Calloselasma rhodostoma (Malayan pit viper). Toxicon 1993, 31, 997–1005. [Google Scholar] [CrossRef]
- Tan, N.-H.; Ponnudurai, G. The toxinology of Calloselasma rhodostoma (Malayan pit viper) venom. Journal of Toxicology: Toxin Reviews 1996, 15, 1–17. [Google Scholar]
- Gutiérrez, J.M.; Escalante, T.; Rucavado, A.; Herrera, C.; Fox, J.W. A comprehensive view of the structural and functional alterations of extracellular matrix by snake venom metalloproteinases (SVMPs): novel perspectives on the pathophysiology of envenoming. Toxins 2016, 8, 304. [Google Scholar] [CrossRef]
- Chang, H.-H.; Chang, C.-P.; Chang, J.-C.; Dung, S.-Z.; Lo, S.J. Application of recombinant rhodostomin in studying cell adhesion. Journal of Biomedical Science 1997, 4, 235–243. [Google Scholar] [CrossRef]
- Tan, C.H.; Liew, J.L.; Navanesan, S.; Sim, K.S.; Tan, N.H.; Tan, K.Y. Cytotoxic and anticancer properties of the Malaysian mangrove pit viper (Trimeresurus purpureomaculatus) venom and its disintegrin (purpureomaculin). Journal of Venomous Animals and Toxins including Tropical Diseases 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Liew, J.L.; Tan, N.H.; Tan, C.H. Proteomics and preclinical antivenom neutralization of the mangrove pit viper (Trimeresurus purpureomaculatus, Malaysia) and white-lipped pit viper (Trimeresurus albolabris, Thailand) venoms. Acta tropica 2020, 209, 105528. [Google Scholar] [CrossRef] [PubMed]
- Moura-da-Silva, A.; Almeida, M.; Portes-Junior, J.; Nicolau, C.; Gomes-Neto, F.; Valente, R. Processing of snake venom metalloproteinases: generation of toxin diversity and enzyme inactivation. Toxins 2016, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Tasoulis, T.; Isbister, G.K. A review and database of snake venom proteomes. Toxins 2017, 9, 290. [Google Scholar] [CrossRef]
- Esnouf, M.P.; Tunnah, G.W. The isolation and properties of the thrombin-like activity from Ancistrodon rhodostoma Venom. British Journal of Haematology 1967, 13, 581–590. [Google Scholar] [CrossRef]
- Shin, Y.; Morita, T. Rhodocytin, a functional novel platelet agonist belonging to the heterodimeric C-type lectin family, induces platelet aggregation independently of glycoprotein Ib. Biochemical and biophysical research communications 1998, 245, 741–745. [Google Scholar] [CrossRef]
- Fox, J.W.; Serrano, S.M.T. Insights into and speculations about snake venom metalloproteinase (SVMP) synthesis, folding and disulfide bond formation and their contribution to venom complexity. The FEBS journal 2008, 275, 3016–3030. [Google Scholar] [CrossRef]
- Kini, R.M. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon 2003, 42, 827–840. [Google Scholar] [CrossRef]
- Ferraz, C.R.; Arrahman, A.; Xie, C.; Casewell, N.R.; Lewis, R.J.; Kool, J.; Cardoso, F.C. Multifunctional toxins in snake venoms and therapeutic implications: From pain to hemorrhage and necrosis. Frontiers in ecology and evolution 2019, 7, 218. [Google Scholar] [CrossRef]
- Faisal, T.; Tan, K.Y.; Tan, N.H.; Sim, S.M.; Gnanathasan, C.A.; Tan, C.H. Proteomics, toxicity and antivenom neutralization of Sri Lankan and Indian Russell’s viper (Daboia russelii) venoms. Journal of Venomous Animals and Toxins including Tropical Diseases 2021, 27. [Google Scholar] [CrossRef]
- Pla, D.; Sanz, L.; Quesada-Bernat, S.; Villalta, M.; Baal, J.; Chowdhury, M.A.W.; León, G.; Gutiérrez, J.M.; Kuch, U.; Calvete, J.J. Phylovenomics of Daboia russelii across the Indian subcontinent. Bioactivities and comparative in vivo neutralization and in vitro third-generation antivenomics of antivenoms against venoms from India, Bangladesh and Sri Lanka. Journal of proteomics 2019, 207, 103443. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Wong, K.Y.; Huang, L.-K.; Tan, K.Y.; Tan, N.H.; Wu, W.-G. Snake venomics and antivenomics of Cape Cobra (Naja nivea) from South Africa: insights into venom toxicity and cross-neutralization activity. Toxins 2022, 14, 860. [Google Scholar] [CrossRef]
- Tan, K.Y.; Wong, K.Y.; Tan, N.H.; Tan, C.H. Quantitative proteomics of Naja annulifera (sub-Saharan snouted cobra) venom and neutralization activities of two antivenoms in Africa. International journal of biological macromolecules 2020, 158, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y.; Tan, K.Y.; Tan, N.H.; Tan, C.H. A neurotoxic snake venom without phospholipase A2: proteomics and cross-neutralization of the venom from Senegalese Cobra, Naja senegalensis (Subgenus: Uraeus). Toxins 2021, 13, 60. [Google Scholar] [CrossRef]
- Tan, N.-H.; Kanthimathi, M.; Tan, C.-S. Enzymatic activities of Calloselasma rhodostoma (Malayan pit viper) venom. Toxicon 1986, 24, 626–630. [Google Scholar] [CrossRef]
- Tan, K.Y.; Tan, C.H.; Fung, S.Y.; Tan, N.H. Venomics, lethality and neutralization of Naja kaouthia (monocled cobra) venoms from three different geographical regions of Southeast Asia. J Proteomics 2015, 120, 105–125. [Google Scholar] [CrossRef] [PubMed]
- Yap, M.K.; Fung, S.Y.; Tan, K.Y.; Tan, N.H. Proteomic characterization of venom of the medically important Southeast Asian Naja sumatrana (Equatorial spitting cobra). Acta Trop 2014, 133, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.H.; Tan, K.Y.; Ng, T.S.; Sim, S.M.; Tan, N.H. Venom proteome of spine-bellied sea snake (Hydrophis curtus) from Penang, Malaysia: Toxicity correlation, immunoprofiling and cross-neutralization by sea snake antivenom. Toxins (Basel) 2019, 11, 3. [Google Scholar] [CrossRef]
- Tsai, I.H.; Wang, Y.M.; Au, L.C.; Ko, T.P.; Chen, Y.H.; Chu, Y.F. Phospholipases A2 from Callosellasma rhodostoma venom gland: Cloning and sequencing of 10 of the cDNAs, three-dimensional modelling and chemical modification of the major isozyme. European journal of biochemistry 2000, 267, 6684–6691. [Google Scholar] [CrossRef]
- Tsai, I.-H.; Wang, Y.-M.; Chen, Y.-H.; Tsai, T.-S.; Tu, M.-C. Venom phospholipases A2 of bamboo viper (Trimeresurus stejnegeri): molecular characterization, geographic variations and evidence of multiple ancestries. Biochemical Journal 2004, 377, 215–223. [Google Scholar] [CrossRef]
- Van Den Bergh, C.J.; Slotboom, A.J.; Verheij, H.M.; De Haas, G.H. The role of aspartic acid-49 in the active site of phospholipase A2: A site-specific mutagenesis study of porcine pancreatic phospholipase A2 and the rationale of the enzymatic activity of [Iysine49] phospholipase A2 from Agkistrodon piscivorus piscivorus venom. European journal of biochemistry 1988, 176, 353–357. [Google Scholar] [PubMed]
- Lomonte, B. Lys49 myotoxins, secreted phospholipase A2-like proteins of viperid venoms: A comprehensive review. Toxicon 2023, 107024. [Google Scholar] [CrossRef] [PubMed]
- De Bold, A.J.; Borenstein, H.B.; Veress, A.T.; Sonnenberg, H. A rapid and potent natriuretic response to intravenous injection of atrial myocardial extract in rats. Life sciences 1981, 28, 89–94. [Google Scholar] [CrossRef] [PubMed]
- Vink, S.; Jin, A.H.; Poth, K.J.; Head, G.A.; Alewood, P.F. Natriuretic peptide drug leads from snake venom. Toxicon 2012, 59, 434–445. [Google Scholar] [CrossRef]
- Pimenta, D.C.; Spencer, P.J. Bradykinin-potentiating and related peptides from reptile venoms. In Handbook of Venoms and Toxins of Reptiles; CRC Press: 2021; pp. 241-250.
- Lameu, C.; Neiva, M.; Hayashi, M. Venom Bradykinin-related peptides (BRPs) and its multiple biological roles. An Integrated View of the Molecular Recognition and Toxinology—From Analytical Procedures to Biomedical Applications 2013, 119-151.
- Higuchi, S.; Murayama, N.; Saguchi, K.; Ohi, H.; Fujita, Y.; Camargo, A.C.; Ogawa, T.; Deshimaru, M.; Ohno, M. Bradykinin-potentiating peptides and C-type natriuretic peptides from snake venom. Immunopharmacology 1999, 44, 129–135. [Google Scholar] [CrossRef]
- Hayashi, M.A.; Camargo, A.C. The Bradykinin-potentiating peptides from venom gland and brain of Bothrops jararaca contain highly site specific inhibitors of the somatic angiotensin-converting enzyme. Toxicon 2005, 45, 1163–1170. [Google Scholar] [CrossRef]
- Michel, G.H.; Murayama, N.; Sada, T.; Nozaki, M.; Saguchi, K.; Ohi, H.; Fujita, Y.; Koike, H.; Higuchi, S. Two N-terminally truncated forms of C-type natriuretic peptide from habu snake venom. Peptides 2000, 21, 609–615. [Google Scholar] [CrossRef]
- Fucase, T.M.; Sciani, J.M.; Cavalcante, I.; Viala, V.L.; Chagas, B.B.; Pimenta, D.C.; Spencer, P.J. Isolation and biochemical characterization of bradykinin-potentiating peptides from Bitis gabonica rhinoceros. Journal of Venomous Animals and Toxins including Tropical Diseases 2018, 23. [Google Scholar] [CrossRef]
- Ianzer, D.; Konno, K.; Marques-Porto, R.; Portaro, F.C.V.; Stöcklin, R.; de Camargo, A.C.M.; Pimenta, D.C. Identification of five new bradykinin potentiating peptides (BPPs) from Bothrops jararaca crude venom by using electrospray ionization tandem mass spectrometry after a two-step liquid chromatography. Peptides 2004, 25, 1085–1092. [Google Scholar] [CrossRef]
- Cheung, H.; Cushman, D. Inhibition of homogeneous angiotensin-converting enzyme of rabbit lung by synthetic venom peptides of Bothrops jararaca. Biochimica et Biophysica Acta (BBA)-Enzymology 1973, 293, 451–463. [Google Scholar] [CrossRef]
- Morita, T. Structures and functions of snake venom CLPs (C-type lectin-like proteins) with anticoagulant-, procoagulant-, and platelet-modulating activities. Toxicon 2005, 45, 1099–1114. [Google Scholar] [CrossRef] [PubMed]
- Arlinghaus, F.T.; Eble, J.A. C-type lectin-like proteins from snake venoms. Toxicon 2012, 60, 512–519. [Google Scholar] [CrossRef] [PubMed]
- Clemetson, K.; Morita, T.; Kini, R.M. Classification and nomenclature of snake venom C-type lectins and related proteins. Toxicon: official journal of the International Society on Toxinology 2009, 54, 83. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.A.; O’callaghan, C.A. Molecular analysis of the interaction of the snake venom rhodocytin with the platelet receptor CLEC-2. Toxins 2011, 3, 991–1003. [Google Scholar] [CrossRef]
- Suzuki-Inoue, K.; Fuller, G.L.; García, Á.; Eble, J.A.; Pöhlmann, S.; Inoue, O.; Gartner, T.K.; Hughan, S.C.; Pearce, A.C.; Laing, G.D. A novel Syk-dependent mechanism of platelet activation by the C-type lectin receptor CLEC-2. Blood 2006, 107, 542–549. [Google Scholar] [CrossRef]
- Wang, Y.M.; Liew, Y.F.; Chang, K.Y.; Tsai, I.H. Purification and characterization of the venom phospholipases A2 from Asian monotypic crotalinae snakes. Journal of Natural Toxins 1999, 8, 331–340. [Google Scholar]
- Eble, J.A.; Niland, S.; Bracht, T.; Mormann, M.; Peter-Katalinic, J.; Pohlentz, G.; Stetefeld, J. The α2β1 integrin-specific antagonist rhodocetin is a cruciform, heterotetrameric molecule. The FASEB Journal 2009, 23, 2917–2927. [Google Scholar] [CrossRef]
- Eble, J.A.; Beermann, B.; Hinz, H.-J.r.; Schmidt-Hederich, A. α2β1 integrin is not recognized by rhodocytin but is the specific, high affinity target of rhodocetin, an RGD-independent disintegrin and potent inhibitor of cell adhesion to collagen. Journal of Biological Chemistry 2001, 276, 12274–12284. [Google Scholar] [CrossRef]
- Sartim, M.A.; Sampaio, S.V. Snake venom galactoside-binding lectins: a structural and functional overview. Journal of Venomous Animals and Toxins including Tropical Diseases 2015, 21, 1–11. [Google Scholar] [CrossRef]
- Swenson, S.D.; Stack, S.; Markl, F.S. Thrombin-Like Serine Proteinases in Reptile Venoms. In Handbook of Venoms and Toxins of Reptiles; CRC Press: 2021; pp. 351-362.
- Burkhart, W.; Smith, G.F.; Su, J.-L.; Parikh, I.; LeVine, H. Amino acid sequence determination of Ancrod, the thrombin-like α-fibrinogenase from the venom of Akistrodon rhodostoma. FEBS letters 1992, 297, 297–301. [Google Scholar] [CrossRef]
- Rawlings, N.D.; Barrett, A.J. Serine peptidases and their clans. In Handbook of Proteolytic Enzymes, 2nd edition, edited by N.D. Rawlings, A.J. Barrett, J. F. Wossner. San Diego, CA: Academic Press Ltd., pp. 1417–39.; 2004.
- Pfeiffer, G.; Linder, D.; Strube, K.; Geyer, R. Glycosylation of the thrombin-like serine protease ancrod fromAgkistrodon rhodostoma venom. Oligosaccharide substitution pattern at eachN-glycosylation site. Glycoconjugate journal 1993, 10, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.; Zuber, C.; Park, S.; Jang, I.; Lee, Y.; Kysela, K.G.; Le Fourn, V.; Santimaria, R.; Guhl, B.; Cho, J.W. Protein N-glycosylation, protein folding, and protein quality control. Molecules and cells 2010, 30, 497–506. [Google Scholar] [CrossRef]
- Lin, C.-W.; Chen, J.-M.; Wang, Y.-M.; Wu, S.-W.; Tsai, I.-H.; Khoo, K.-H. Terminal disialylated multiantennary complex-type N-glycans carried on acutobin define the glycosylation characteristics of the Deinagkistrodon acutus venom. Glycobiology 2011, 21, 530–542. [Google Scholar] [CrossRef] [PubMed]
- Wormald, M.R.; Dwek, R.A. Glycoproteins: glycan presentation and protein-fold stability. Structure 1999, 7, R155–R160. [Google Scholar] [CrossRef]
- Lannoo, N.; Van Damme, E.J. Review/N-glycans: The making of a varied toolbox. Plant Science 2015, 239, 67–83. [Google Scholar] [CrossRef]
- Hennerici, M.G.; Kay, R.; Bogousslavsky, J.; Lenzi, G.L.; Verstraete, M.; Orgogozo, J.M. Intravenous ancrod for acute ischaemic stroke in the European Stroke Treatment with Ancrod Trial: a randomised controlled trial. The Lancet 2006, 368, 1871–1878. [Google Scholar] [CrossRef] [PubMed]
- Sherman, D.G.; Atkinson, R.P.; Chippendale, T.; Levin, K.A.; Ng, K.; Futrell, N.; Hsu, C.Y.; Levy, D.E. Intravenous ancrod for treatment of acute ischemic stroke: the STAT study: a randomized controlled trial. Jama 2000, 283, 2395–2403. [Google Scholar] [CrossRef]
- Maduwage, K.; Isbister, G.K. Current treatment for venom-induced consumption coagulopathy resulting from snakebite. PLoS neglected tropical diseases 2014, 8, e3220. [Google Scholar] [CrossRef]
- Li, Z.-Y.; Yu, T.-F.; Lian, E.C.Y. Purification and characterization of l-amino acid oxidase from king cobra (Ophiophagus hannah) venom and its effects on human platelet aggregation. Toxicon 1994, 32, 1349–1358. [Google Scholar] [CrossRef]
- Samel, M.; Tõnismägi, K.; Rönnholm, G.; Vija, H.; Siigur, J.; Kalkkinen, N.; Siigur, E. l-Amino acid oxidase from Naja naja oxiana venom. Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology 2008, 149, 572–580. [Google Scholar] [CrossRef]
- Moustafa, I.M.; Foster, S.; Lyubimov, A.Y.; Vrielink, A. Crystal structure of LAAO from Calloselasma rhodostoma with an L-phenylalanine substrate: insights into structure and mechanism. Journal of molecular biology 2006, 364, 991–1002. [Google Scholar] [CrossRef] [PubMed]
- Paloschi, M.V.; Boeno, C.N.; Lopes, J.A.; Rego, C.M.A.; Silva, M.D.S.; Santana, H.M.; Serrath, S.N.; Ikenohuchi, Y.J.; Farias, B.J.C.; Felipin, K.P. Reactive oxygen species-dependent-NLRP3 inflammasome activation in human neutrophils induced by l-amino acid oxidase derived from Calloselasma rhodostoma venom. Life Sciences 2022, 308, 120962. [Google Scholar] [CrossRef]
- Lee, M.L.; Tan, N.H.; Fung, S.Y.; Sekaran, S.D. Antibacterial action of a heat-stable form of L-amino acid oxidase isolated from king cobra (Ophiophagus hannah) venom. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 2011, 153, 237–242. [Google Scholar]
- Yamazaki, Y.; Morita, T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon 2004, 44, 227–231. [Google Scholar] [CrossRef]
- Dhananjaya, B.L.; D’souza, C.J.M. An overview on nucleases (DNase, RNase, and phosphodiesterase) in snake venoms. Biochemistry (Moscow) 2010, 75, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Coronado, M.A.; da Silva Olivier, D.; Eberle, R.J.; do Amaral, M.S.; Arni, R.K. Modeling and molecular dynamics indicate that snake venom phospholipase B-like enzymes are Ntn-hydrolases. Toxicon 2018. [Google Scholar] [CrossRef]
- Takasaki, C.; Tamiya, N. Isolation and properties of lysophospholipases from the venom of an Australian elapid snake, Pseudechis australis. Biochemical Journal 1982, 203, 269–276. [Google Scholar] [CrossRef]
- Bernheimer, A.; Weinstein, S.; Linder, R. Isoelectric analysis of some Australian elapid snake venoms with special reference to phospholipase B and hemolysis. Toxicon 1986, 24, 841–849. [Google Scholar] [CrossRef]
- Bernheimer, A.; Linder, R.; Weinstein, S.; Kim, K.-S. Isolation and characterization of a phospholipase B from venom of Collett’s snake, Pseudechis colletti. Toxicon 1987, 25, 547–554. [Google Scholar] [CrossRef]
- Lavin, M.; Earl, S.; Birrel, G.; St Pierre, L.; Guddat, L.; de Jersey, J.; Masci, P. Snake venom nerve growth factors. In Handbook of Venoms and Toxins of Reptiles, SP, M., Ed.; Taylor and Francis Group, CRC Press: Boca Raton, 2009. [Google Scholar]
- Sunagar, K.; Fry, B.G.; Jackson, T.N.; Casewell, N.R.; Undheim, E.A.; Vidal, N.; Ali, S.A.; King, G.F.; Vasudevan, K.; Vasconcelos, V. Molecular evolution of vertebrate neurotrophins: co-option of the highly conserved nerve growth factor gene into the advanced snake venom arsenalf. PloS one 2013, 8, e81827. [Google Scholar] [CrossRef]
- Vaiyapuri, S.; Wagstaff, S.C.; Watson, K.A.; Harrison, R.A.; Gibbins, J.M.; Hutchinson, E.G. Purification and functional characterisation of rhiminopeptidase A, a novel aminopeptidase from the venom of Bitis gabonica rhinoceros. PLoS neglected tropical diseases 2010, 4, e796. [Google Scholar] [CrossRef]
- Nicolau, C.A.; Carvalho, P.C.; Junqueira-de-Azevedo, I.L.M.; Teixeira-Ferreira, A.; Junqueira, M.; Perales, J.; Neves-Ferreira, A.G.C.; Valente, R.H. An in-depth snake venom proteopeptidome characterization: Benchmarking Bothrops jararaca. Journal of proteomics 2017, 151, 214–231. [Google Scholar] [CrossRef] [PubMed]
- Lingam, T.M.C.; Tan, K.Y.; Tan, C.H. Proteomics and antivenom immunoprofiling of Russell’s viper (Daboia siamensis) venoms from Thailand and Indonesia. Journal of Venomous Animals and Toxins including Tropical Diseases 2020, 26. [Google Scholar] [CrossRef] [PubMed]
- Lingam, T.M.C.; Tan, K.Y.; Tan, C.H. Capillary leak syndrome induced by the venoms of Russell’s Vipers (Daboia russelii and Daboia siamensis) from eight locales and neutralization of the differential toxicity by three snake antivenoms. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology 2021, 250, 109186. [Google Scholar]
- Rucavado, A.; Escalante, T.; Camacho, E.; Gutiérrez, J.M.; Fox, J.W. Systemic vascular leakage induced in mice by Russell’s viper venom from Pakistan. Scientific Reports 2018, 8, 16088. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, J.; Mackessy, S.P.; Fry, B.G.; Bhatia, M.; Mourier, G.; Fruchart-Gaillard, C.; Servent, D.; Ménez, R.; Stura, E.; Ménez, A. Denmotoxin, a three-finger toxin from the colubrid snake Boiga dendrophila (Mangrove Catsnake) with bird-specific activity. Journal of Biological Chemistry 2006, 281, 29030–29041. [Google Scholar] [CrossRef]
- Pahari, S.; Mackessy, S.P.; Kini, R.M. The venom gland transcriptome of the Desert Massasauga Rattlesnake (Sistrurus catenatus edwardsii): towards an understanding of venom composition among advanced snakes (Superfamily Colubroidea). BMC molecular biology 2007, 8, 115. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.K.; Mackessy, S.P.; Dutta, S. Characterization of a Kunitz-type protease inhibitor peptide (Rusvikunin) purified from Daboia russelii russelii venom. International journal of biological macromolecules 2014, 67, 154–162. [Google Scholar] [CrossRef]
- Rotenberg, D.; Bamberger, E.; Kochva, E. Studies on ribonucleic acid synthesis in the venom glands of Vipera palaestinae (Ophidia, Reptilia). Biochemical Journal 1971, 121, 609–612. [Google Scholar] [CrossRef]
- Wery, M.; Descrimes, M.; Thermes, C.; Gautheret, D.; Morillon, A. Zinc-mediated RNA fragmentation allows robust transcript reassembly upon whole transcriptome RNA-Seq. Methods 2013, 63, 25–31. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X. A survey of best practices for RNA-seq data analysis. Genome biology 2016, 17, 1–19. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nature protocols 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics (Oxford, England) 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nature methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC bioinformatics 2011, 12, 1–16. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics (Oxford, England) 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





