Submitted:
10 April 2023
Posted:
11 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
2.1. Identification of UFO Genes
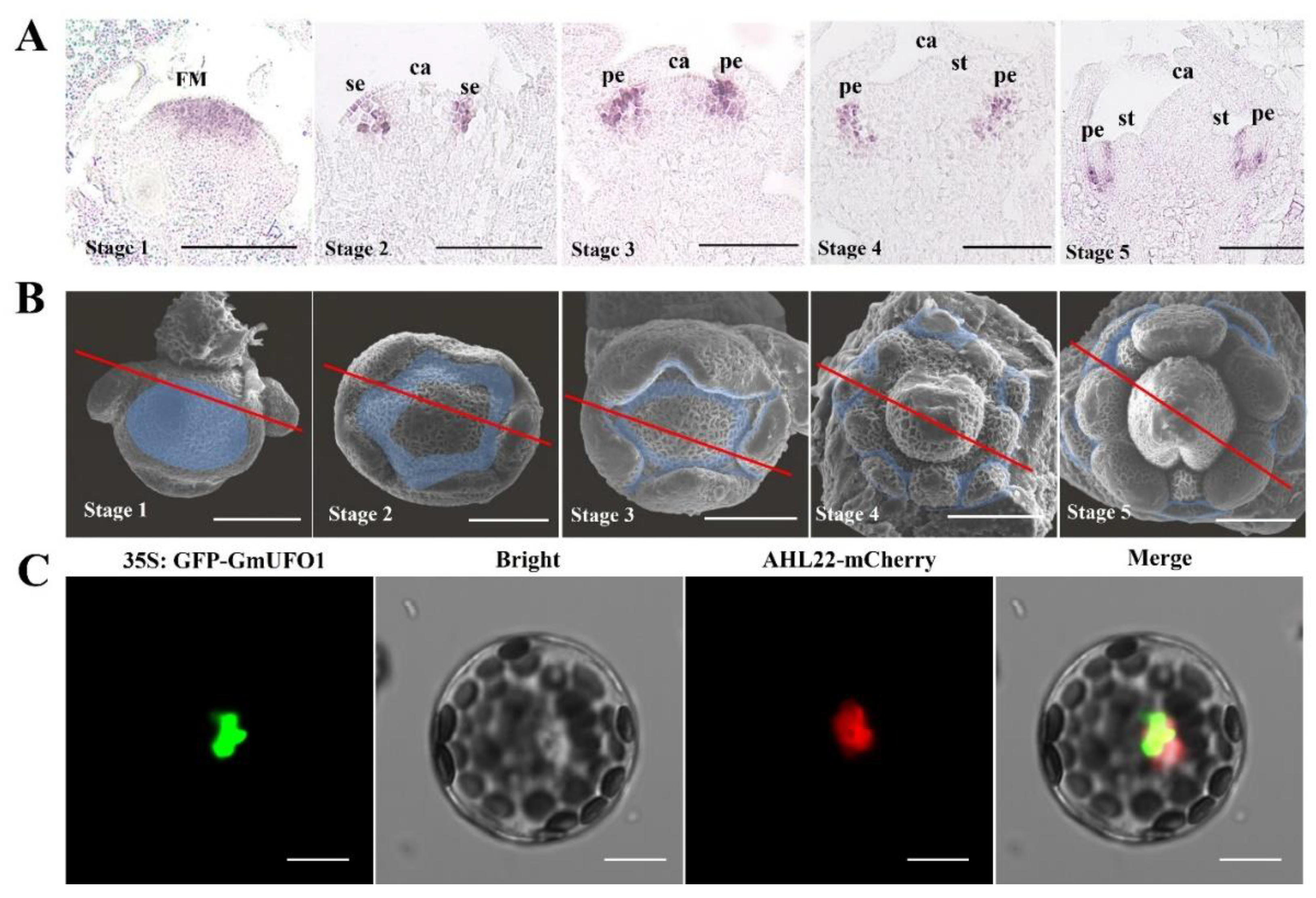
2.2. Spatial and Temporal Expression Profiles of the GmUFO1 Gene in Soybean Flower Development
2.3. CRISPR/Cas9 System Generated Target Mutations for GmUFO Genes
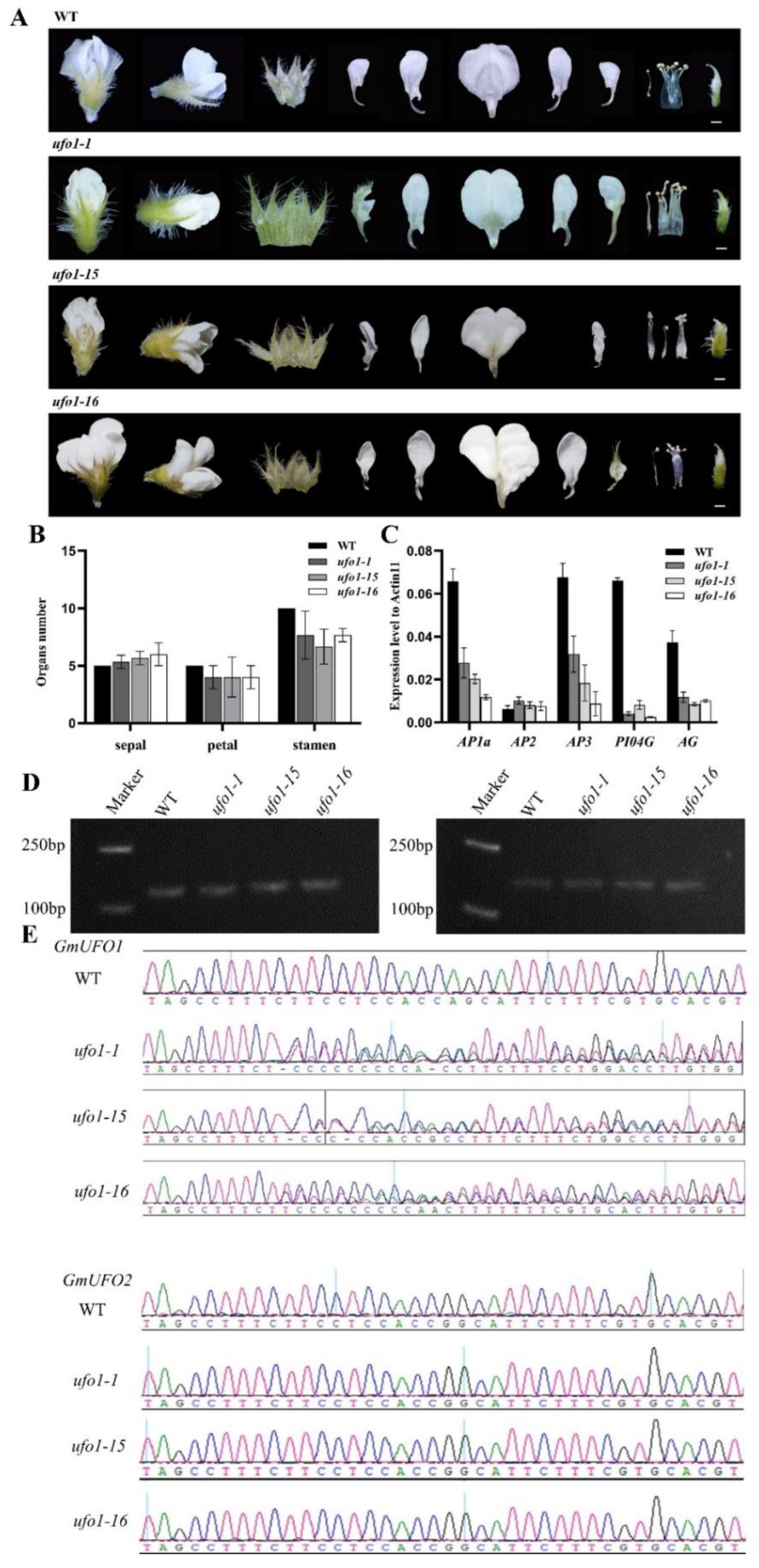
2.4. Knockout of the GmUFO1 Gene Caused Abnormal Flower Organs
2.5. Knockout of the GmUFO2 Gene Had no Effect on Flower Organs
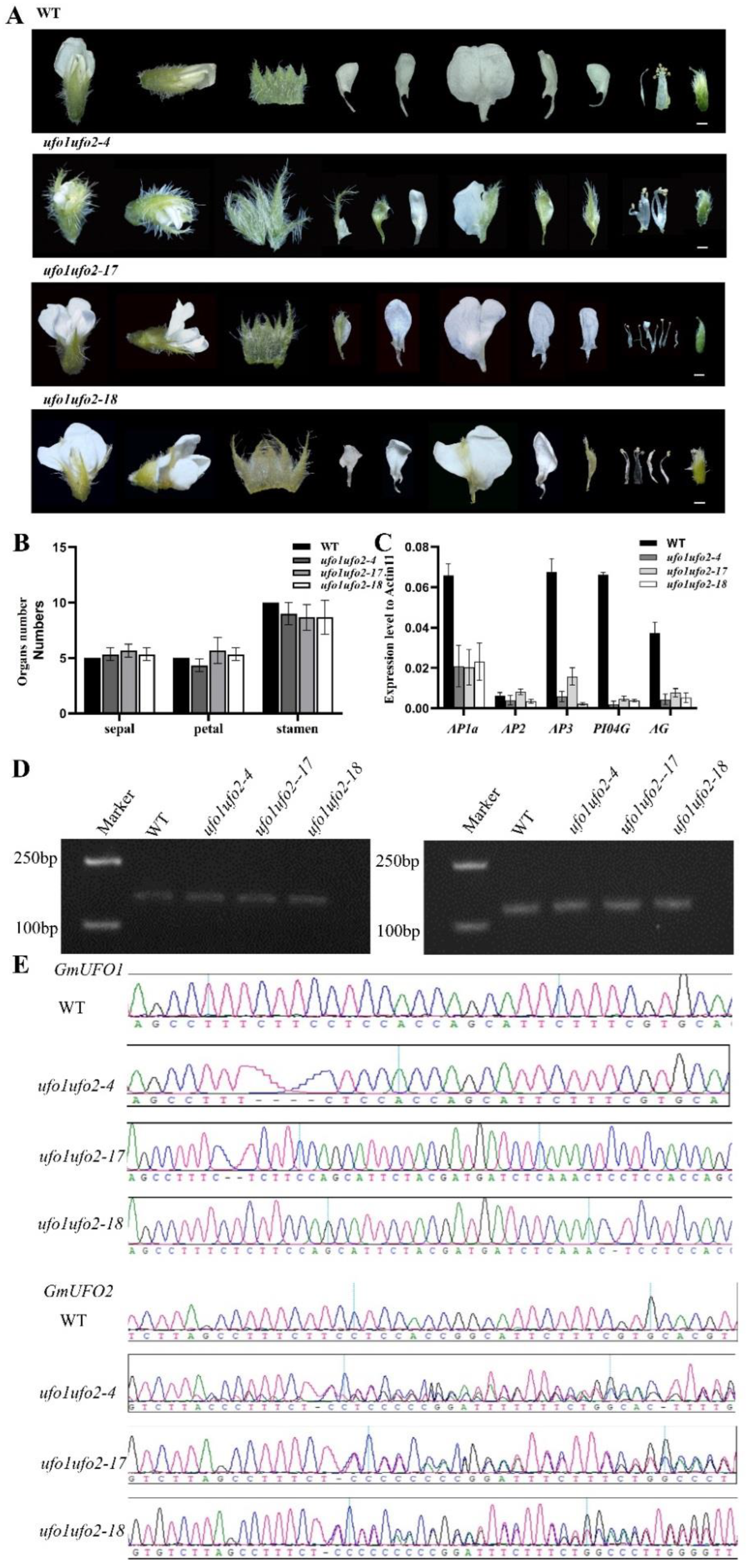
2.6. Double Knockout of GmUFO1 and GmUFO2 Had More Serious Defects in Organ Number and Shape
3. Discussion
4. Materials and Methods
4.1. GmUFO Identification and Cloning
4.2. Phylogenetic Analysis and Physical Properties of UFO Genes
4.3. In Situ Hybridization
4.4. Subcellular Localization
4.5. Cytological Analysis
4.6. Gene Knockout and Material Identification
4.7. Total RNA Extraction and qPCR
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Levin, J.Z.; Meyerowitz, E.M. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995, 7, 529–548. [Google Scholar] [PubMed]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls - genetic interactions controlling flower development. Nature. 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar] [PubMed]
- Sun, L.Y.; Nie, T.J.; Chen, Y.; Li, J.; Yang, A.X.; Yin, Z.F. Gene identification and tissue expression analysis inform the floral organization and color in the basal angiosperm Magnolia polytepala (Magnoliaceae). Planta. 2023, 257, 4. [Google Scholar] [CrossRef] [PubMed]
- Laufs, P.; Coen, E.; Kronenberger, J.; Traas, J.; Doonan, J. Separable roles of UFO during floral development revealed by conditional restoration of gene function. Development. 2003, 130, 785–796. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.X.; Li, K.P.; Zhang, Z.R.; Li, J.G.; Wang, B.; Zhang, Z.M.; Zhu, Y.Y.; Pan, C.C.; Sun, K.; He, C.Y. Transcriptomic comparison sheds new light on regulatory networks for dimorphic flower development in response to photoperiod in Viola prionantha. BMC Plant Biol. 2022, 22, 336. [Google Scholar] [CrossRef]
- Wang, X.P.; Feng, S.H.; Nakayama, N.; Crosby, W.L.; Irish, V.; Deng, X.W.; Wei, N. The COP9 signalosome interacts with SCFUFO and participates in Arabidopsis flower development. Plant Cell. 2003, 15, 1071–1082. [Google Scholar] [CrossRef]
- Rieu, P.; Turchi, L.; Thévenon, E.; Zarkadas, E.; Nanao, M.; Chahtane, H.; Tichtinsky, G.; Lucas, J.; Blanc-Mathieu, R.; Zubieta, C. The F-box protein UFO controls flower development by redirecting the master transcription factor LEAFY to new cis-elements. Nat. Plants. 2023, 9, 315–329. [Google Scholar] [CrossRef]
- Pelosi, J.A.; Sessa, E.B. From genomes to populations: a meta-analysis and review of fern population genetics. Int. J. Mol. Sci. 2021, 182, 325–343. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, M.; Solava, J.; Ma, H. The ASK1 gene regulates development and interacts with the UFO gene to control floral organ identity in Arabidopsis. Dev. Genet. 1999, 25, 209–223. [Google Scholar] [CrossRef]
- Levin, J.Z.; Meyerowitz, E.M. UFO: an Arabidopsis gene involved in both floral meristem and floral organ development. Plant Cell. 1995, 7, 529–548. [Google Scholar] [PubMed]
- Ingram, G.C.; Goodrich, J.; Wilkinson, M.D.; Simon, R.; Haughn, G.W.; Coen, E.S. Parallels between UNUSUAL FLORAL ORGANS and FIMBRIATA, genes controlling flower development in Arabidopsis and Antirrhinum. Plant Cell. 1995, 7, 1501–1510. [Google Scholar] [PubMed]
- Peréz-Mesa, P.; Suárez-Baron, H.; Ambrose, B.A.; González, F.; Pabón-Mora, N.J.E. Floral MADS-box protein interactions in the early diverging angiosperm Aristolochia fimbriata Cham.(Aristolochiaceae: Piperales). Evol. Dev. 2019, 21, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.J.; Ye, L.X.; Ai, X.Y.; Hu, C.G.; Cheng, Z.P.; Zhang, J.Z. Functional analysis of a PISTILLATA-like gene CcMADS20 involved in floral organs specification in citrus. Plant Sci. 2022, 6, 319. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.Y.; Meng, Q.; Tan, X.M.; Yang, L.; Zhang, K.L.; Xu, Z.Q. Functional identification of the different regions in B-class floral homeotic MADS-box proteins IiAP3 and IiPI from Isatis indigotica. Physiol. Plantarum. 2022, 174, e13713. [Google Scholar] [CrossRef]
- Zhang, A.J.; He, H.B.; Li, Y.; Wang, L.X.; Liu, Y.X.; Luan, X.C.; Wang, J.X.; Liu, H.J.; Liu, S.Y.; Zhang, J.; et al. MADS-Box subfamily gene GmAP3 from Glycine max regulates early flowering and flower development. Int. J. Mol. Sci. 2023, 24, 2751. [Google Scholar] [CrossRef]
- Krishnan, H.B.; Kim, W.S.; Oehrle, N.W.; Smith, J.R.; Gillman, J.D. Effect of heat stress on seed protein composition and ultrastructure of protein storage vacuoles in the Cotyledonary Parenchyma cells of soybean genotypes that are either tolerant or sensitive to elevated temperatures. Int. J. Mol. Sci. 2020, 21, 4775. [Google Scholar] [CrossRef]
- Lyu, J.; Cai, Z.D.; Li, Y.H.; Suo, H.C.; Yi, R.; Zhang, S.; Nian, H. The floral repressor GmFLC-like is involved in regulating flowering time mediated by low temperature in soybean. Int. J. Mol. Sci. 2020, 21, 1322. [Google Scholar] [CrossRef]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Haughn, G.W. UNUSUAL FLORAL ORGANS controls meristem identity and organ primordia fate in Arabidopsis. Plant Cell. 1995, 7, 1485–1499. [Google Scholar] [CrossRef]
- Zhang, S.; Sandal, N.; Polowick, P.L.; Stiller, J.; Stougaard, J.; Fobert, P.R. Proliferating Floral Organs (Pfo), a Lotus japonicus gene required for specifying floral meristem determinacy and organ identity, encodes an F-box protein. Plant J. 2003, 33, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Jofuku, K.D.; den Boer. B.G.; Van Montagu.M.; Okamuro J.K. Control of Arabidopsis flower and seed development by the homeotic gene APETALA2. Plant Cell. 1994, 6, 1211–1225. [Google Scholar]
- Bradley, D.; Ratcliffe, O.; Vincent, C.; Carpenter, R.; Coen, E. Inflorescence commitment and architecture in Arabidopsis. Science. 1997, 275, 80–83. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Yanofsky, M.F. Activation of the Arabidopsis B class homeotic genes by APETALA1. Plant Cell. 2001, 13, 739–753. [Google Scholar] [CrossRef] [PubMed]
- Busch, M.A.; Bomblies, K.; Weigel, D. Activation of a floral homeotic gene in Arabidopsis. Science. 1999, 285, 585–587. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Mena, C.; de Folter, S.; Costa, M.M.R.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development. 2005, 132, 429–438. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Kim, Y.J.; Muller, R.; Yumul, R.E.; Liu, C.Y. Pan Y.Y.; Cao X.F.; Goodrich J.; Chena X.M. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell. 2011, 23, 3654–3670. [Google Scholar] [CrossRef]
- Drews, N.; Bowman, J.L.; Meyerowitz, E.M. Negative regulation of the Arabidopsis homeotic gene AGAMOUS by the APETALA2 product. Cell. 1991, 65, 991–1002. [Google Scholar] [CrossRef]
- Chae, E.; Tan, Q.K.; Hill, T.A.; Irish, V.F. An Arabidopsis F-box protein acts as a transcriptional co-factor to regulate floral development. Development. 2008, 135, 1235–1245. [Google Scholar] [CrossRef]
- Ng, M.; Yanofsky, M.F. Function and evolution of the plant MADS-box gene family. Nat. Rev. Genet. 2001, 2, 186–195. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.Z.; Zhao, Z.; Tian, Z.X.; Xu, S.L.; Luo, Y.H.; Cai, Z.G.; Wang, Y.M.; Yang, J.; Wang, Z.; Weng, L.; et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA. 2006, 103, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. Plant Cell. 2005, 17, 1979–1993. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.S.; Jiang, L.W. Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2007, 2, 2348–2353. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Shen, W.J.; Chen, H.F.; Chu, L.T.; Xu, Y.C.; Zhou, X.C.; Liu, C.L.; Chen, C.M.; Zeng, J.H.; Liu, J.; et al. Characterization and subcellular localization of histone deacetylases and their roles in response to abiotic stresses in soybean. BMC Plant Biol. 2018, 18, 226. [Google Scholar] [CrossRef]
- Tetsuya, Y.; Satoshi, W.; Maiko, A.; Kyuya, H.; Keisuke, K. Cotyledonary node pre-wounding with a micro-brush increased frequency of Agrobacterium-mediated transformation in soybean. Plant Biotechnol. 2010, 27, 217–220. [Google Scholar]
- Rio, D.C.; Ares, M.J.; Hannon, G.J.; Nilsen, T.W. Preparation of cytoplasmic and nuclear RNA from tissue culture cells. Cold. Spring. Harb. Protoc. 2010, 6, 5441. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




