1. Introduction
With the outbreak of COVID-19 in 2019, many major food-producing countries have taken measures to restrict grain exports. As tomatoes are the most widely planted and consumed vegetable crop globally, and China is one of the world's largest producers and consumers of tomatoes, tomato production is an important means for farmers to increase income and generate export revenue. However, various diseases severely affect tomato yields, especially considering the increasingly serious issue of food safety. Therefore, accurate identification of tomato diseases and timely treatment has become an urgent problem that needs to be addressed [
1,
2,
3,
4].
Many deep learning-based methods have been proposed for crop disease identification [
5,
6,
7,
8,
9,
10,
11,
12,
13,
14,
15,
16,
17,
18]. For example, li et al. [
5] proposed the OplusVNet, a 13-layer convolutional neural network that achieved 99% prediction accuracy on a dataset collected from the field using VGG16 network modules for transfer learning. Nguyen et al. [
6] proposed a neural network model that combines image segmentation with transfer learning, segments the image and uses HSV to extract the original leaf area and black background, and feeds it into a VGG-19 model for transfer learning, achieving an accuracy of 99.72%, with a training time of 275000s. These networks have effectively improved the recognition of crop diseases and pests. However, due to their complex structure and large model size, they are difficult to deploy on current mainstream devices for real-time disease and pest identification.
Many researchers have realized the inconvenience brought by complex models and began designing models with simple structures but powerful functions. For example, Ding et al. [
19] proposed a model similar to the VGG inference time backbone, consisting of a series of 3x3 convolutions and ReLU, and proposed a multi-branch topology efficient model to reduce training time. Zeng et al. [
20] proposed a self-attention convolutional neural network (SACNN) to address confusion caused by small disease areas, low contrast between disease areas and backgrounds, and background complexity in crop disease images. The recognition accuracy on AES-CD9214 and MK-D2 were 95.33% and 98%, respectively. These studies focus on the structure and performance of the model to ensure that the model has efficient performance under a simple structure, but they do not fully consider the impact of data on the model. Deng et al. [
21] explored this issue from the perspective of data and found that the difficulty in obtaining data samples is the main challenge to improving disease recognition performance. Therefore, they proposed a new data augmentation method based on generative adversarial networks (GAN), called RAHC_GAN, for tomato leaf data augmentation and disease recognition. The results showed that RAHC_GAN can generate leaves with clear disease features, and the generated extended dataset can significantly improve the classifier's recognition performance. Data augmentation is a commonly used method in deep learning to prevent model overfitting. To address the problem of noise samples that may be introduced by data augmentation, which may damage the performance of unorganized data during the inference process, Gong et al. [
22] proposed KeepAugment, which uses saliency maps to detect important areas in the original image, and then preserves these information areas during the augmentation process to generate more realistic training images. The results showed that this method can improve the training effect on different datasets. Many studies have shown that the combination of attention mechanisms and deep learning models can effectively improve model performance [
23,
24,
25,
26,
27]. However, most studies did not investigate the impact of different attention mechanisms on the model. Different attention mechanisms have different characteristics, so their impact on the model must be different. One of the objectives of this study is to investigate the impact of different attention mechanisms on LBFNet to improve the model's generalization and practicality [
28].
Therefore, in response to the current problems of neural network models used to identify tomato leaf diseases, such as large model size, complex structure, slow inference speed, and insufficient accuracy, and the fact that most publicly available datasets do not include tomato leaves in the real world, and the data in these datasets has been processed, resulting in weak generalization ability of models trained on them. We propose a convolutional neural network model, LBFNet, based on the VGG series, which has a simple structure but powerful functions. We use different attention mechanisms to extract deep features from images and reduce the influence of factors such as background information on model accuracy, while using a cascade structure to preserve the original information of the image and improve model performance. We use various data augmentation techniques to enhance the ten types of tomato leaf data in PlantVillage and maintain sample balance, adding tomato leaf image data from different sources to construct the LBFtomato dataset. The training set and testing set were divided in a 7:3 ratio. The model trained on LBFtomato exhibited improved generalization and accuracy. Compared with previous studies on tomato disease and pest identification, the model proposed in this paper considers more diverse influencing factors and can be fully applicable to the identification of tomato leaf diseases in practical environments [
29,
30,
31,
32,
33,
34,
35,
36].
2. Materials and Methods
2.1. LBFtomato Leaf Image Datasets
The original experimental data used the publicly available Plant Village dataset, which consists of 10 types of tomato leaves, including nine types of tomato diseases and one type of healthy tomato leaves, namely early blight, late blight, powdery mildew, leaf mold, septoria leaf spot, bacterial spot, spider mites, yellow leaf curl virus, brown spot, and healthy leaves, totaling 18,835 images. The dataset was divided into a training set and a test set in a 7:3 ratio. Data augmentation techniques such as flipping, translation, and brightness adjustment were first applied to the training data set in Plant Village to increase the number of samples in each class to about 1000. To maintain data balance, the data set was then cleaned by removing some interfering samples, resulting in a Plant Village training data set of 13,062 images. Finally, a new dataset named LBFtomato was created by adding real tomato leaf images taken in the Changsha tomato planting base and real-world tomato leaf images obtained from Kaggle and github. LBFtomato consists of 10 types of tomato leaves, including nine types of tomato diseases and one type of healthy tomato leaves, and has real-world tomato leaf data, which can better verify the effectiveness of the model.
Table 1 and
Table 2 respectively show the number of images in the PlantVillage dataset and the LBFtomato dataset. the initial tomato leaf images were resized to the size of 256 × 256 as shown in
Figure 1 [
37].
The data augmentation was performed using the ImageDataGenerator function in Keras, with specific parameter settings: 40 degrees of rotation; 0.2 of horizontal and vertical translation; 0.2 of perspective transformation; 0.2 scaling; horizontal flip; and padding and random brightness in the nearest mode.
2.2. Test environment
The experimental environment of this paper is Windows 64-bit system, solid-state drive 500G, mechanical hard disk 2T, processor uses core i5-11400H, with RAM 16.0GB, graphics card is RTX 3050Ti, software environment uses Anaconda 4.8.4, CUDA11.0, python 3.7, and programming language tensorflow 2.2 is used for model construction and training.
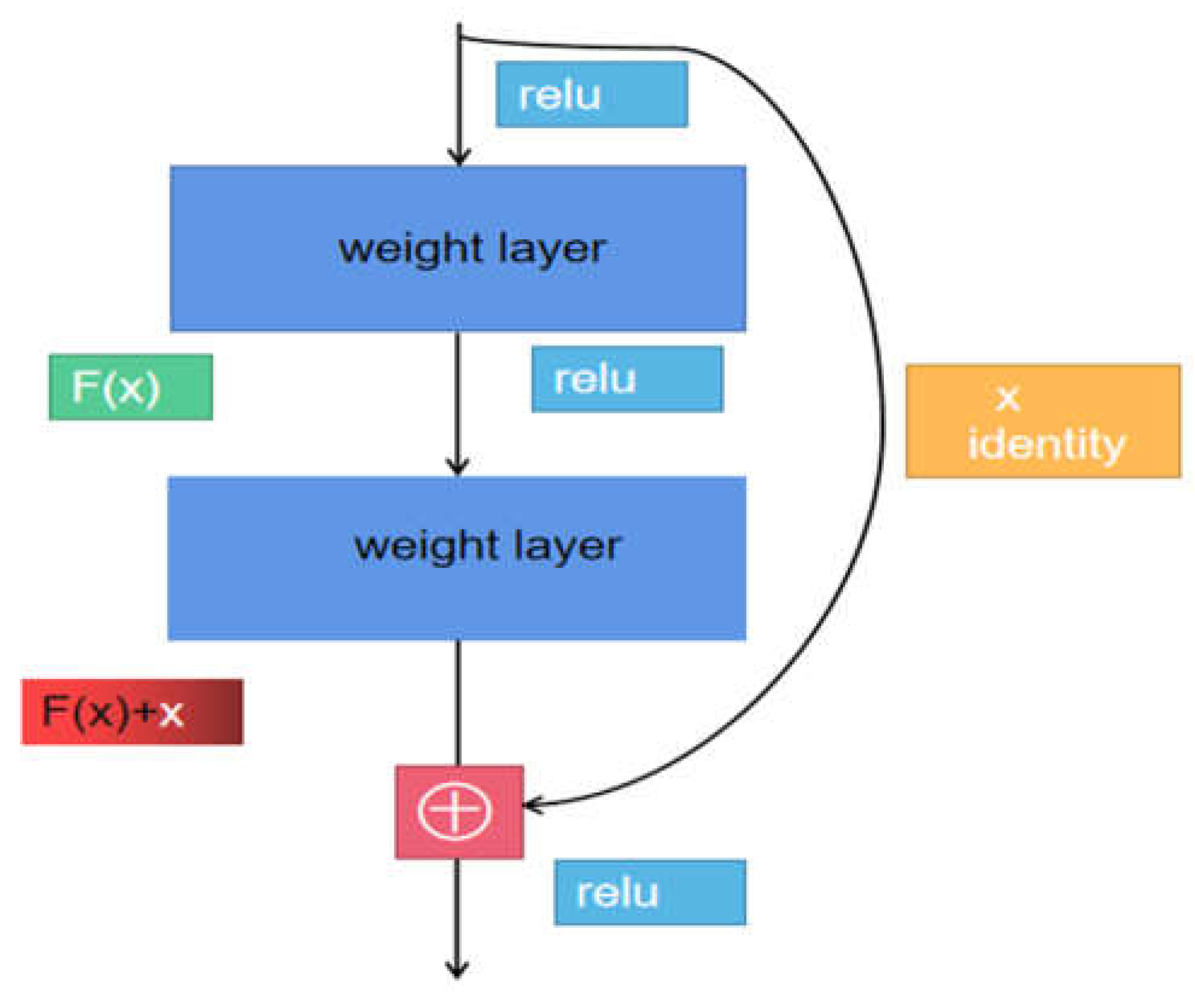
2.3. Use cascading structures to reduce model loss
Convolutional neural network models often encounter problems of gradient vanishing and explosion during the training process. The traditional solution is to initialize and regularize the data, but this can easily lead to network performance degradation and an increase in error rate as the depth increases. Therefore, a cascading structure is introduced to solve the problems of gradient vanishing and explosion. At the same time, the cascading structure can also prevent the loss of disease information caused by deepening and small tomato leaf images during the training of the tomato leaf disease classifier. It inputs the original information into a specific layer for information supplementation, so that the model can effectively supplement disease information during training, improve model accuracy and performance, and avoid gradient vanishing and explosion caused by deepening. The cascading structure is shown in
Figure 2.
2.3. Using Three-Channel Attention Mechanism to Enhance Model Robustness.
The tomato leaf diseases have problems such as small disease targets, small differences between different types of tomato leaf diseases, and unclear disease features. Therefore, a three-channel attention mechanism module is constructed to obtain subtle features of tomato leaf diseases, allowing the model to focus on the specific location of the tomato leaf disease and learn its specific characteristics. It suppresses the negative impact of interfering information such as leaves and background on the model, and enhances the robustness of the model. The three-channel attention mechanism module, as shown in
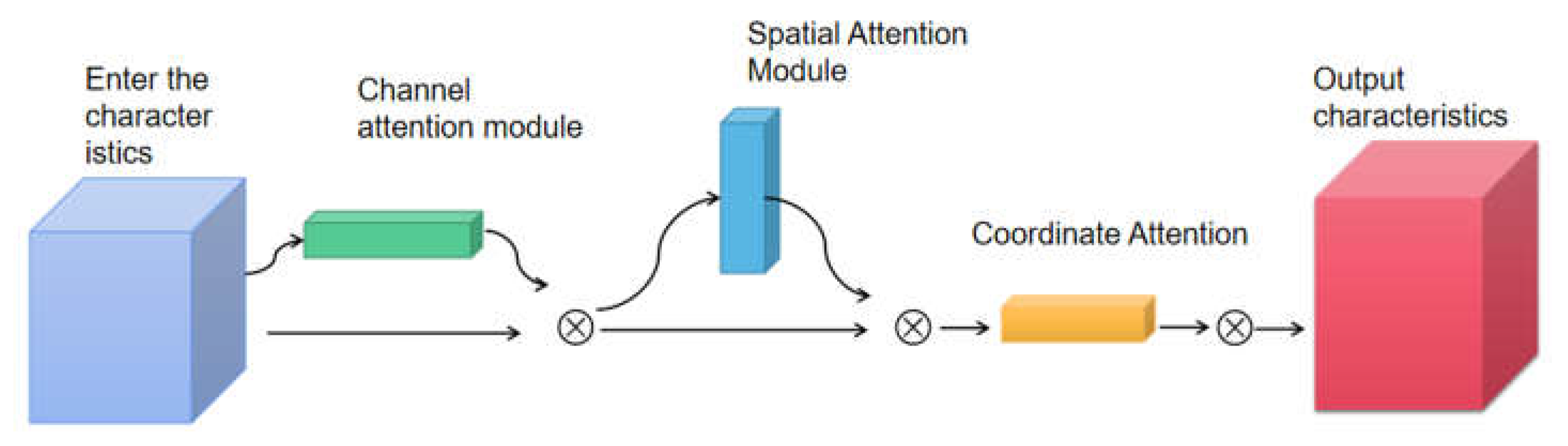
Figure 3,includes a spatial attention module, a channel attention module, and a coordinate attention module, which is the product of the connection of the three.
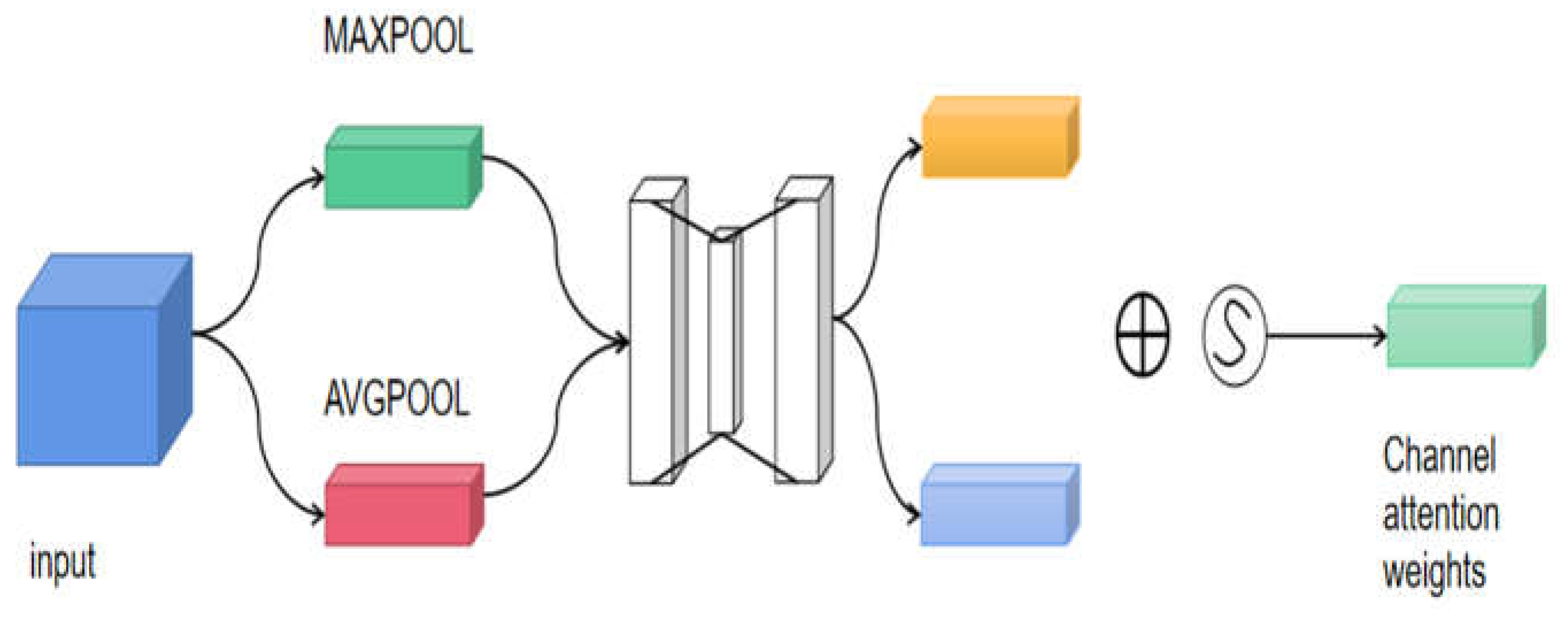
The spatial attention mechanism is an adaptive spatial region selection mechanism that selects the positions the model needs to focus on in order to obtain the specific location of the disease in the image. The channel attention mechanism weights the convolutional features of the channels, thereby enhancing the expression ability of the disease parts. The channel attention mechanism can be expressed as formula (1), as shown in detail in
Figure 4.
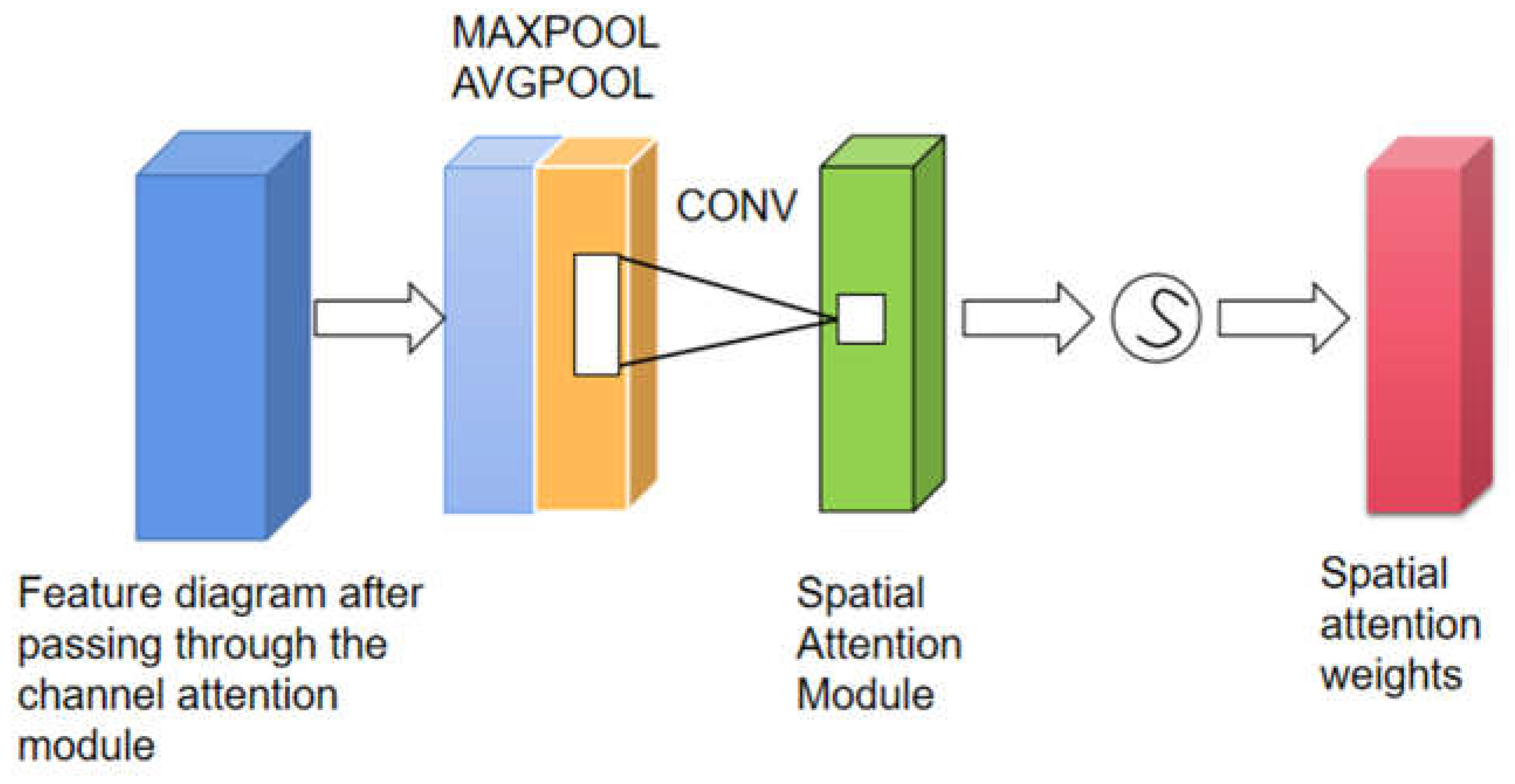
The Spatial Attention Module is a compression of channels, where average pooling and max pooling are performed separately in the channel dimension. The MaxPool operation extracts the maximum value in the channel, with the number of extractions being the product of height and width. The AvgPool operation extracts the average value in the channel, with the number of extractions being the product of height and width. Then, the feature maps extracted earlier (each with a single channel) are combined to obtain a 2-channel feature map, which is used to locate the specific position of the disease. The module structure is shown in
Figure 4. The Spatial Attention Module can be expressed as Formula (2).
The coordinate attention module uses two 1D global pooling operations to aggregate input features along the vertical and horizontal directions into two separate direction-aware feature maps. these two feature maps with embedded direction-specific information are then encoded into two attention maps, each Each attention map captures the long-range dependencies of the input feature maps along a spatial direction. The location information can thus be stored in the generated attention maps. Both attention maps are then applied to the input feature maps by multiplication to emphasize the representation of the attention region.
For applications in mobile environments, the new transformation should be as simple as possible and be able to utilize the captured position information in order to accurately capture the region of interest and efficiently capture the relationship between channels, therefore a three-channel attention mechanism is constructed for fusing the information between different channels. The output of the coordinate attention module is shown in Equation (3).
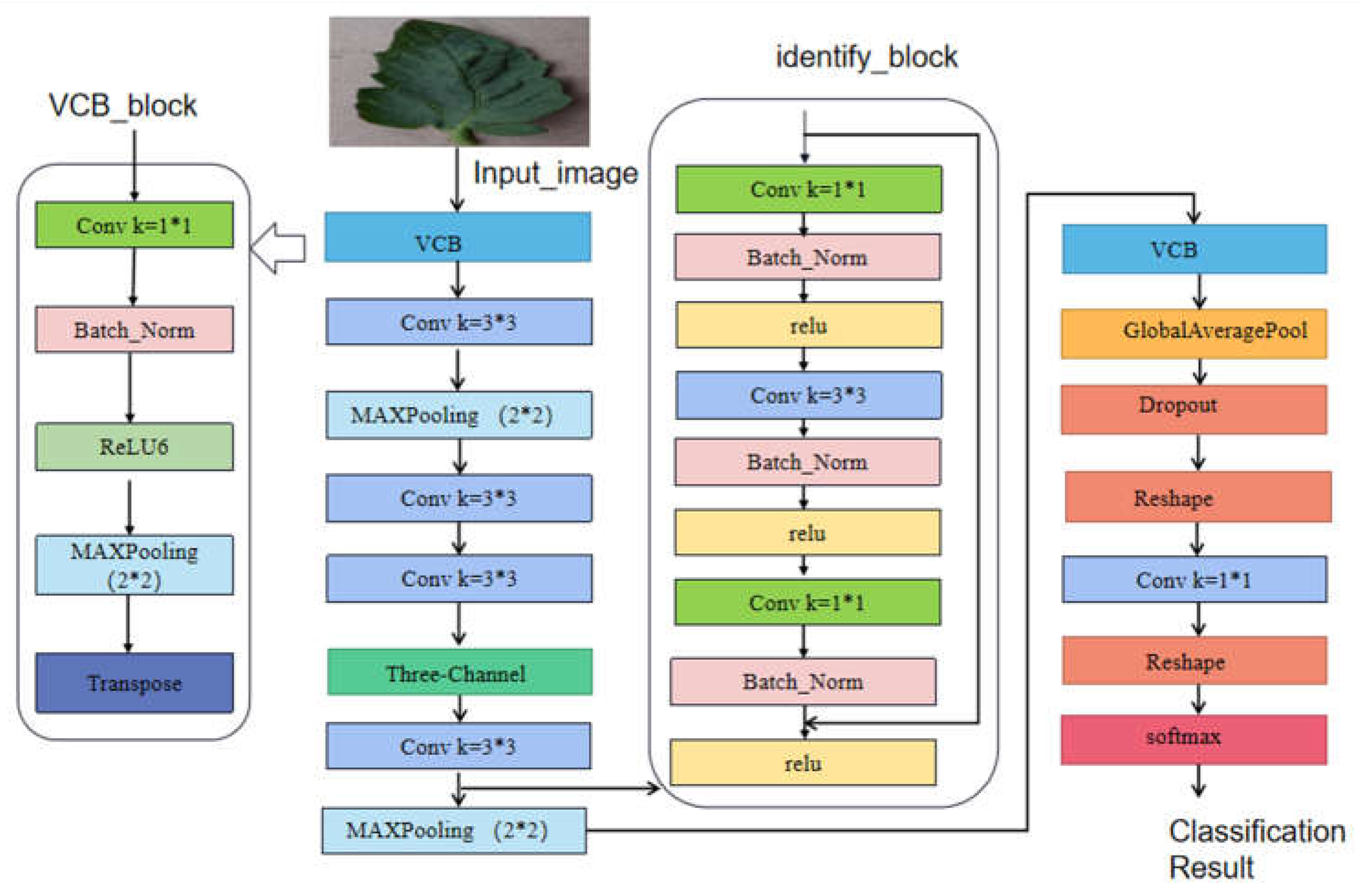
2.4. Reducing model parameters using Vgg-style convolutional neural network
The current convolutional neural network models are well-designed but have made the models extremely complex. These complex models occupy a large amount of memory and decrease the inference speed while achieving high accuracy. The classical VGG convolutional neural network uses a simple system architecture consisting of convolutional layers, Relu activation functions, and pooling layers, which has extremely fast inference speed and good detection ability. However, VGG loses some original information as the structure becomes deeper and cannot fully obtain the original image information. Therefore, we propose a simple but powerful convolutional neural network structure called LBFNet, which has a linear structure similar to VGG and has the advantages of cascading structure and attention mechanism, allowing deep models to obtain complementary original information and learn important parts of disease images,reducing the influence of background noise and improving model performance. To further improve the model's generalization ability, a Dropout layer is added to the model to prevent overfitting and enhance model robustness. The basic module of LBFNet, LBFB,as shown in
Figure 6, uses a 1×1 convolutional kernel to obtain image information. The 1×1 convolutional kernel can observe finer features of diseases, which is beneficial for recognizing small targets such as tomato leaves. At the same time, a BN layer is added to prevent model overfitting, and relu6 is used as the activation function. ReLU6 is the same as ordinary ReLU but limits the maximum output value to 6 (clipping the output value),which enables the model to have good numerical resolution even with low-precision float16 on mobile devices, allowing the model to perform better when deployed on mobile devices. Meanwhile, in order to supplement more information and improve the representation of disease information, we add a deconvolution layer at the end to map the low-dimensional features into high-dimensional inputs to further improve the performance of the model and perfect the low-cost and high-efficiency tomato leaf disease recognition under natural conditions.The structure of LBFNet is shown in
Figure 6.
3. Results
3.1. Research on Tomato Leaf Disease Classification Based on LBFNet Model.
3.1.1. The impact of different optimizers on the model.
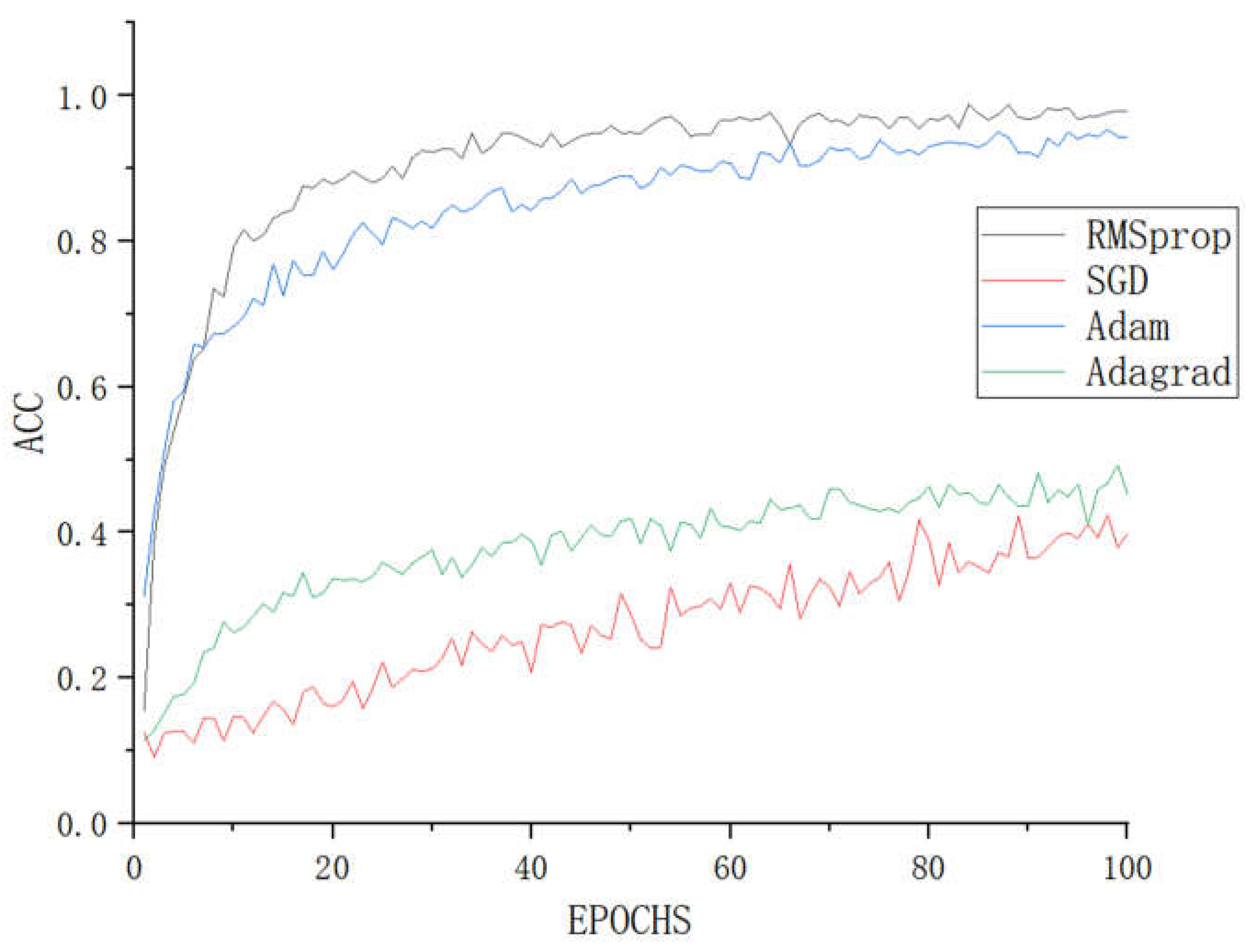
The translation optimizer guides the various parameters of the loss function to update in the correct direction with an appropriate size during the backpropagation process of deep learning. This enables the updated parameters to continuously approach the global minimum of the loss function. In order to achieve the minimum loss value and ensure optimal performance of the model in identifying tomato leaf diseases, different optimizers were used to train the LBFNet model, with the training results shown in
Figure 7.
Figure 7.
Accuracy comparison of different optimizers.
Figure 7.
Accuracy comparison of different optimizers.
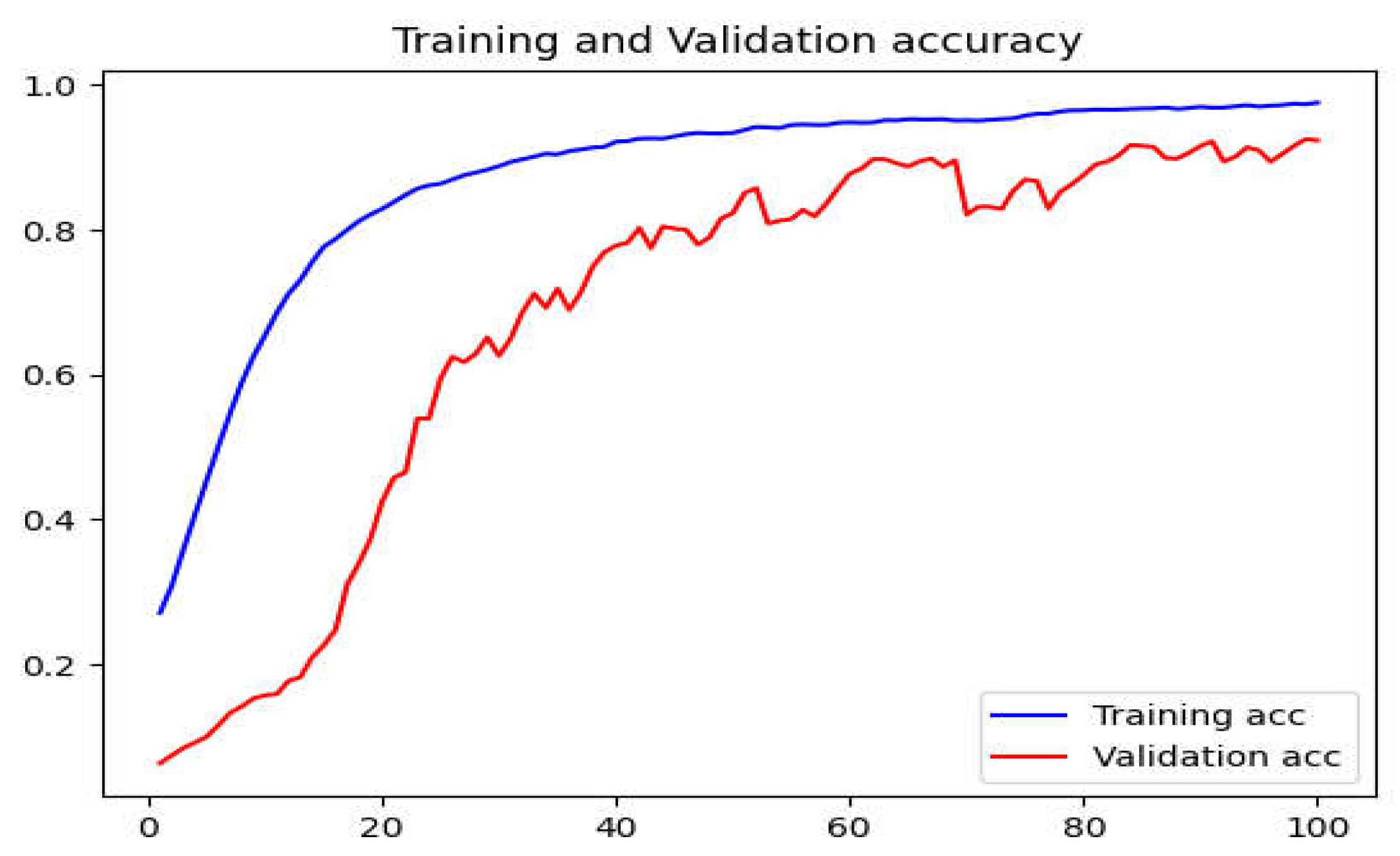
Figure 8.
The accuracy graph using the SGD optimizer with a learning rate of 0.01.
Figure 8.
The accuracy graph using the SGD optimizer with a learning rate of 0.01.
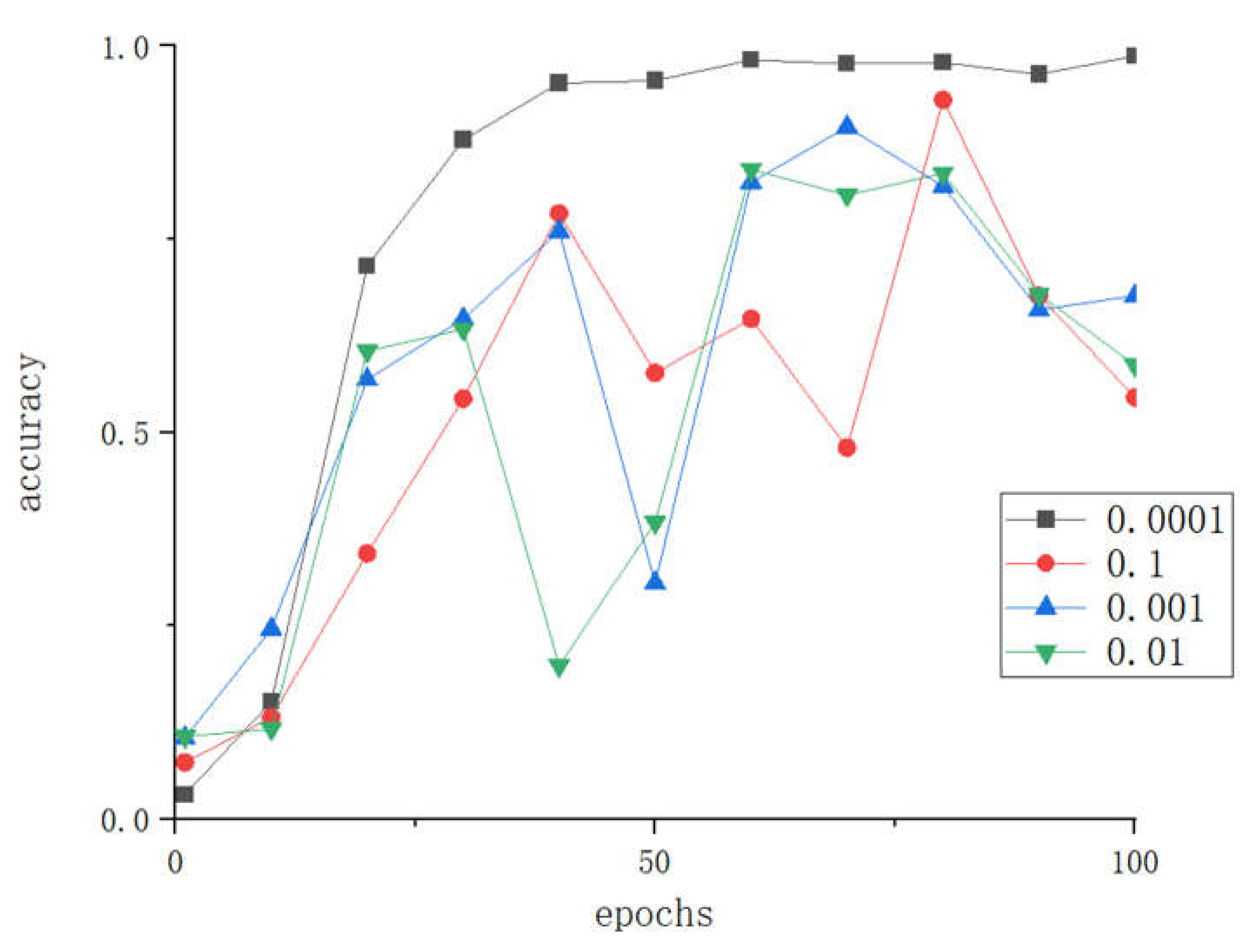
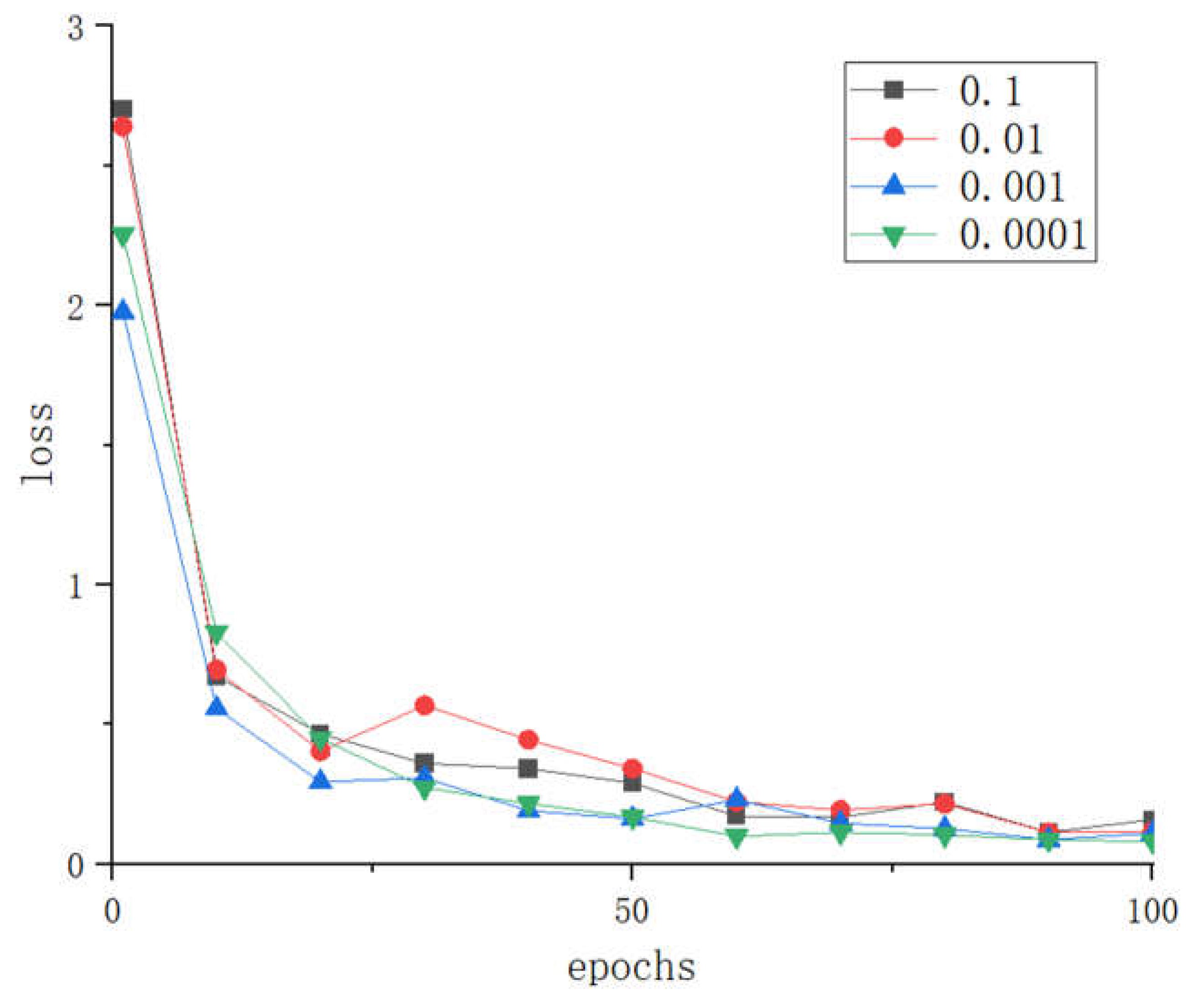
3.1.2. The impact of different learning rate parameters on the model
The learning rate is an important hyperparameter of convolutional neural networks. A learning rate that is too large can cause the loss function to miss the global optimal point, while a learning rate that is too small can increase the convergence complexity of the network. Therefore, to explore the optimal learning rate for LBFNet in tomato disease recognition, we conducted experiments with learning rates set to 0.0001, 0.001, 0.01, and 0.1.
Figure 9.
Accuracy comparison of different learning rates.
Figure 9.
Accuracy comparison of different learning rates.
Figure 10.
Loss comparison of different learning rates.
Figure 10.
Loss comparison of different learning rates.
3.1.3. The impact of different Attention mechanism on the model.
The attention mechanism has evolved in recent years, further improving the performance of deep learning models. In order to explore the influence of different attention mechanisms and modules on the LBFNet model, SEnet attention mechanism module, three-channel attention mechanism module, DUAL attention mechanism module, CA attention mechanism module, ECA attention mechanism module, CBAM attention mechanism module,cascade module and hybrid structure were added for ablation experiments.
Table 3.
Add renderings of different modules.
Table 3.
Add renderings of different modules.
| Module |
Accuracy |
Loss |
Parameters |
Train time/s |
| LBFB |
0.6267 |
1.0625 |
689,034 |
4633 |
| LBFB+cascade |
0.9567 |
0.1513 |
955,722 |
2158 |
| LBFB+three-channel attention mechanism |
0.9688 |
0.1034 |
532,900 |
1347 |
| LBFB+cascade+three-channel attention mechanism |
0.9906 |
0.0408 |
897,188 |
966 |
| LBFB+SE |
0.5578 |
1.2754 |
691,098 |
5194 |
| LBFB+cascade+SE |
0.9465 |
0.1703 |
957,786 |
2879 |
| LBFB+CA |
0.8922 |
0.3146 |
776,914 |
1552 |
| LBFB+CA+cascade |
0.9683 |
0.1220 |
962,386 |
2312 |
| LBFB+ECA |
0.8745 |
0.3650 |
773,584 |
1432 |
| LBFB+ECA+cascade |
0.9615 |
0.1405 |
955,728 |
1786 |
| LBFB+DUAL |
0.8853 |
0.3411 |
794,060 |
2434 |
| LBFB+DUAL+cascade |
0.9588 |
0.1261 |
976,204 |
2755 |
| LBFB+CBAM |
0.9089 |
0.3053 |
794,940 |
1537 |
| LBFB+cascade+CBAM |
0.9790 |
0.0815 |
777,468 |
1172 |
3.2. Model performance comparison
3.2.1. Parameter settings
Set the parameters for the comparative experiment as follows: the original size of the image is 256×256 pixels, so the model input is also adjusted to 256×256×3. The training set and test set are set at a ratio of 7:3. The batch size is set to 32,the number of epochs is set to 100, and the initial learning rate is adjusted by comparison and finally set to 0.0001. The optimizer used is RMSprop, the loss function used is categorical_crossentropy, and softmax activation function is used. Categorical_crossentropy is shown in formula (4):
In the formula, "output size" represents the number of classification categories, and "y_i" represents the true label for the i-th category.
3.2.2. Evaluation Metrics
In this study, we used F1 score, precision, recall and accuracy as metrics to evaluate the effectiveness of different network models in tomato leaf disease image classification.The F1 score is defined as:
where Precision is the ratio between the number of correctly identified disease images and the number of correctly predicted disease images; Recall is the ratio between the number of correctly identified disease images and the number of all correct disease images in that category.TP is true positive, FP is false positive,FN is false negative.Accuracy is defined as:
3.2.3. comparative analysis result
To explore the effect of data increment on model accuracy, different models were trained on PlantVillage and LBFtomato datasets, and the results are presented in
Table 4 and
Table 5. It can be seen that the performance of all models improved on the LBFtomato dataset, proving that balancing positive and negative samples optimizes the dataset to improve model performance. Meanwhile, LBFNet achieves excellent results on different datasets, and the convergence speed and model accuracy, although lower than the two large models, ConvNeXth and vit-transformer, the training time is tens of times lower than these two models, and maintains the minimum number of parameters, and the accuracy rate is only 0.01 compared to the large models, while the training time is the shortest. And for the problems of large parameters, long training time and low accuracy of traditional models such as VGG and ResNet, LBFNet is a good solution that can be flexibly applied to tomato leaf disease identification in modern agriculture.
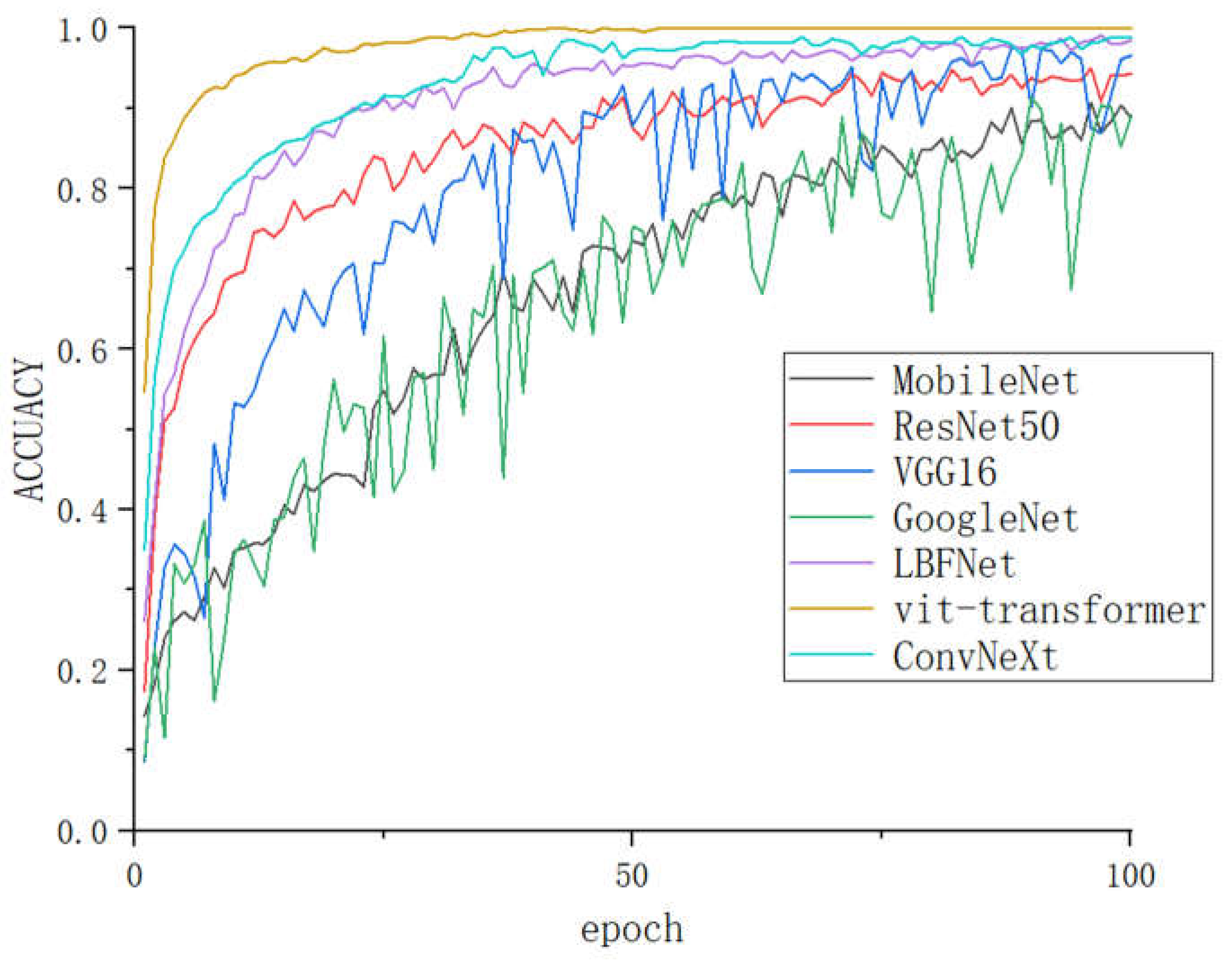
As shown in
Figure 11, GoogleNert and MobileNet, as early lightweight models, have fewer parameters and simple structures, but the same training time increases accordingly and cannot achieve a complete fit after 100 rounds, while VGG16, although simple but with a huge number of parameters and large fluctuations, also does not achieve a fit, and ResNet has the loss ResNet has the disadvantage of excessive loss, while vit-transformer, ConvNeXt and LBFNet can reach saturation within a very short number of rounds, but vit-transformer and ConvNeXt have the disadvantages of large number of parameters, complex structure and long training time of the model, while LBFNet has an accuracy of 0.99, while the fitting speed is fast, and the parameters The minimum number of parameters and very simple structure can effectively solve the current problems of low accuracy, large model, long time and difficult to deploy on mobile devices for tomato leaf disease recognition models.
Figure 11.
The comparison of the accuracy of different models on LBFtomato.
Figure 11.
The comparison of the accuracy of different models on LBFtomato.
Figure 12.
Confusion matrix of LBFNet:0:'Bacterial_spot',1: 'Early_blight', 2:'healthy',3:'Late_blight',4:'Leaf_Mold',5:'Septoria_leaf_spot',6:'Spider_mites',7:'Target_Spot',8:'mosaic_virus',9:'yellow_Leaf_Curl_Virus'.
Figure 12.
Confusion matrix of LBFNet:0:'Bacterial_spot',1: 'Early_blight', 2:'healthy',3:'Late_blight',4:'Leaf_Mold',5:'Septoria_leaf_spot',6:'Spider_mites',7:'Target_Spot',8:'mosaic_virus',9:'yellow_Leaf_Curl_Virus'.
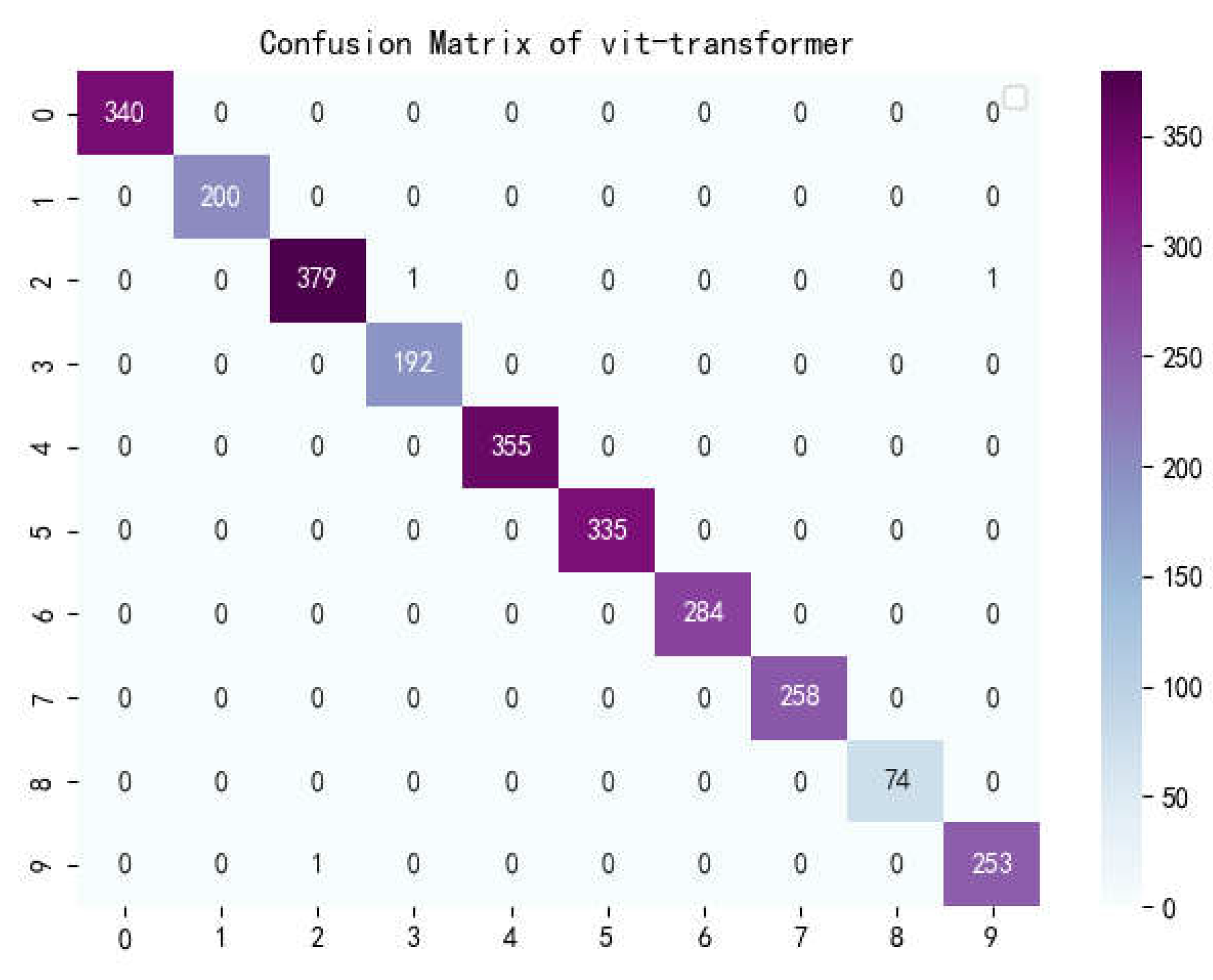
Figure 13.
Confusion matrix of vit-transformer.
Figure 13.
Confusion matrix of vit-transformer.
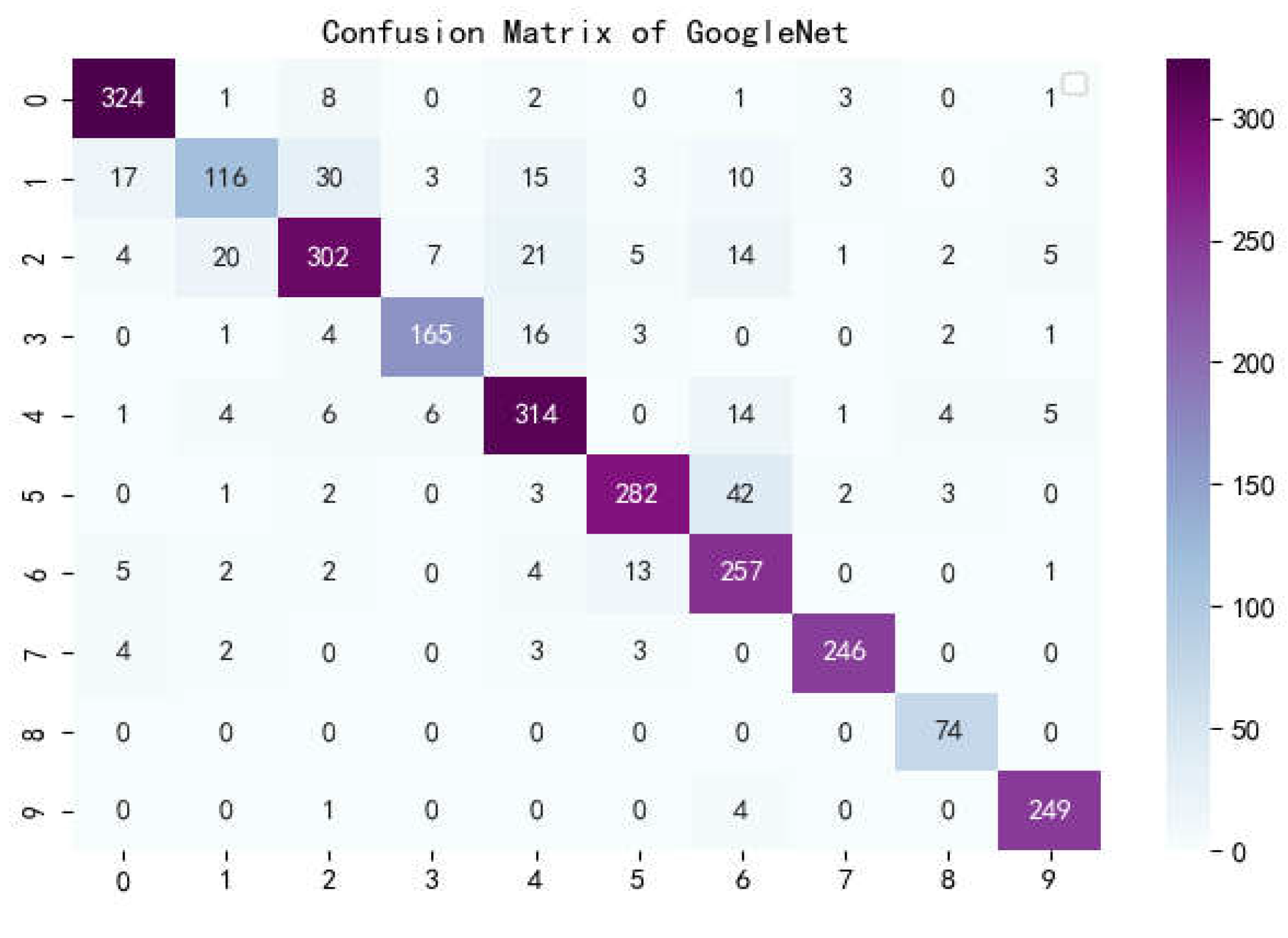
Figure 14.
Confusion matrix of GoogleNet.
Figure 14.
Confusion matrix of GoogleNet.
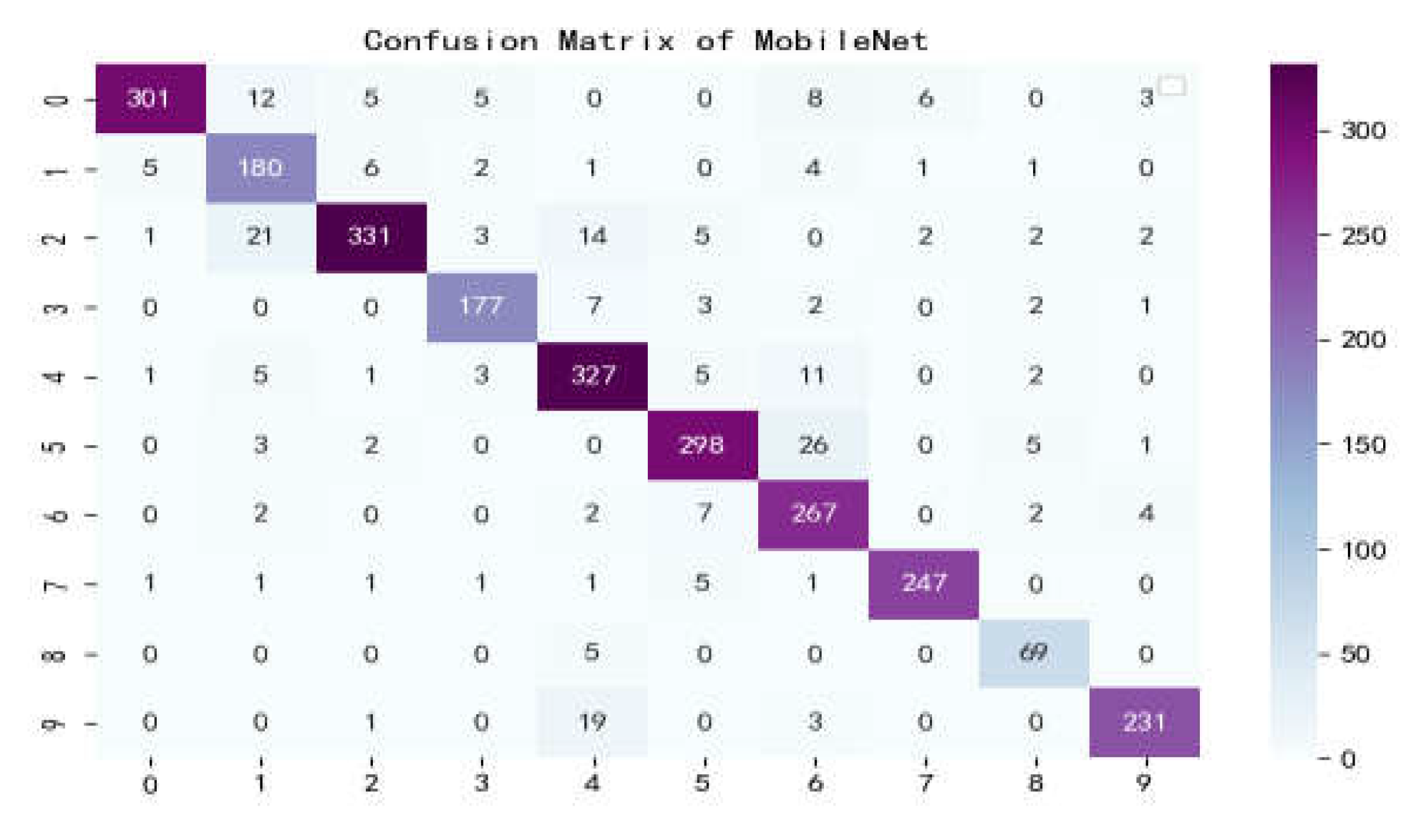
Figure 15.
Confusion matrix of MobileNet.
Figure 15.
Confusion matrix of MobileNet.
Figure 16.
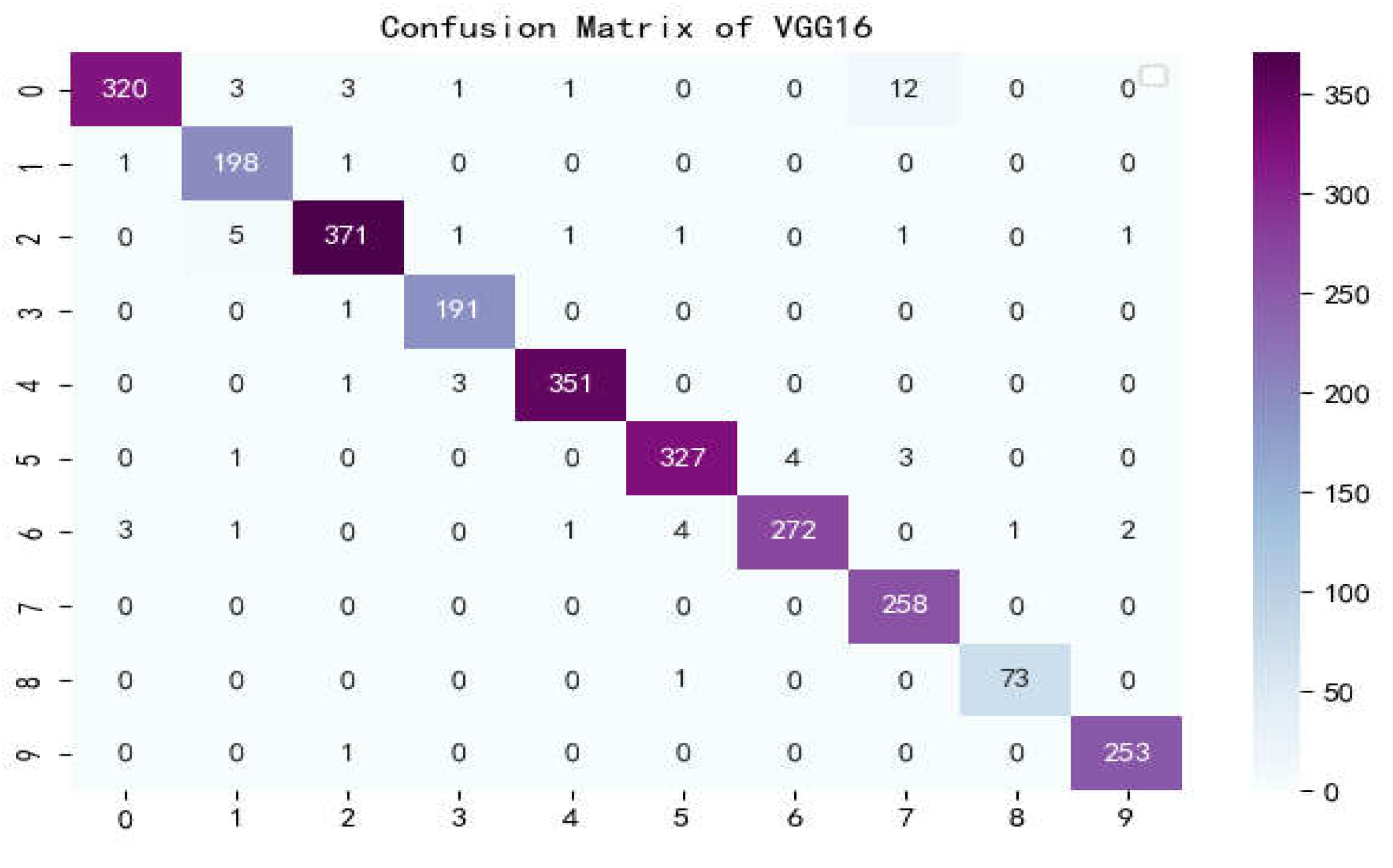
Confusion matrix of VGG16.
Figure 16.
Confusion matrix of VGG16.
Figure 17.
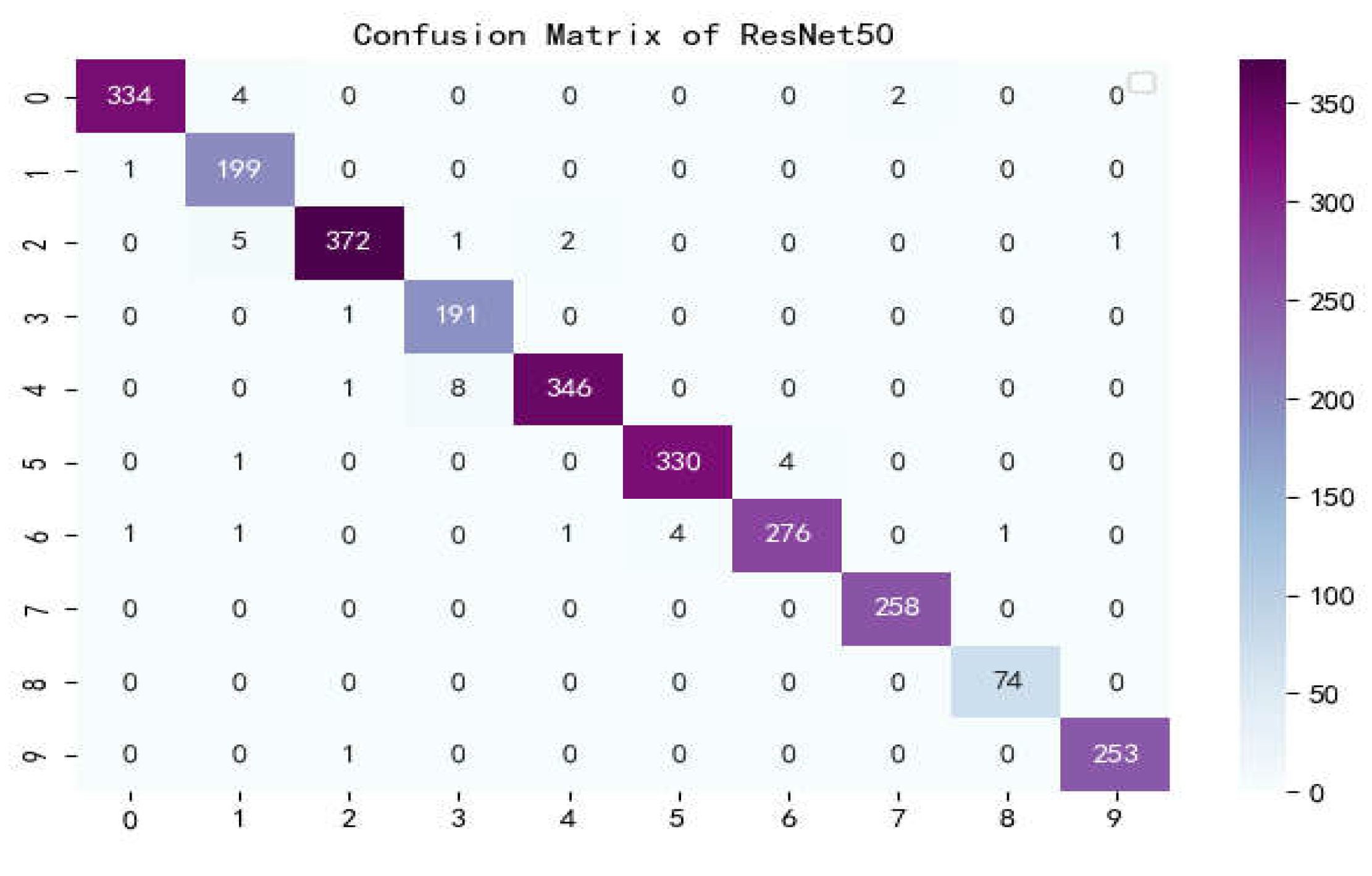
Confusion matrix of ResNet50.
Figure 17.
Confusion matrix of ResNet50.
Figure 18.
Confusion matrix of ConvNeXt.
Figure 18.
Confusion matrix of ConvNeXt.
3.2.4. Reduce model size using quantitative pruning
To enable the model to be deployed on any device, the model is further processed by first pruning the model, performing normal model training using LBFNET until basic convergence, then pruning the lower weight layers starting at a sparsity of 0.5 and ending at a sparsity of 0.9, finally qu antizing and compressing the model, and retraining the pruned network again to recover accuracy until convergence. Quantization pruning can effectively reduce the complexity of the model, reduce memory, and reduce overfitting to a certain extent.The results after quantized pruning are shown in
Table 6. It can be seen that the original model is 6.85 MB, and after quantized pruning, the model parameters are only 3.46 MB.Also, by using the quantified model weight file for training, the model effect reached 97.66%. The model accuracy only decreases by 1.4%, but the model size is reduced by half, which is an acceptable cost.
4. Discussion
The differences in optimizer performance are quite significant. The reason why SGD's performance is unsatisfactory is that SGD itself performs stochastic gradient descent on the learning rate, and the original learning rate is set to 0.0001, which makes the learning rate too small during model training, resulting in a very slow convergence speed. To verify this, we used an initial learning rate of 0.01 to train with SGD, and the results are shown in
Figure 8. Adagrad also makes it difficult for the model to converge because it causes the learning rate to dynamically decrease. Using a higher initial learning rate can solve this problem, but ultimately, RMSprop has better performance under the same conditions. RMSprop was developed to address the rapid learning rate decrease issue in Adagrad, and as a result, it achieves the fastest fitting speed and highest accuracy. The idea behind Adam is to set the initial learning rate to a larger value and dynamically decrease it as the number of iterations increases, to achieve a balance between efficiency and effectiveness. Therefore, it also achieves good results. Taking all factors into consideration, we decided to use RMSprop as the optimizer for LBFNet.
From
Figure 9 and
Figure 10, it can be seen that when the learning rate is set to 0.0001, LBFNet has the best fitting speed, and the highest accuracy, and the loss is also the smallest, and the expected effect is achieved, so 0.0001 is chosen as a value of the learning rate of LBFNet.For RMSprop optimizer, the effect of excessive learning rate in tomato leaf disease identification cannot support the actual production needs.
Table 3 shows that in addition to the addition of SE attention mechanism alone, the addition of other modules has greatly improved the performance of the model, of which the cascade module has a huge effect on the improvement of the model, and for the SE attention mechanism, the loss of information in the process of Global Information Embedding will cause the model performance to decrease, and from the rest of the data can be seen that the attention mechanism and cascade module have a huge improvement for the model. The three-channel attention mechanism can achieve optimal results at a small cost.
The confusion matrix is a situation analysis table in machine learning that summarizes the prediction results of the classification model, and summarizes the records in the dataset in the form of a matrix according to the two criteria of real category and the category judgment predicted by the classification model. Among them, the rows of the matrix represent the true values, the list of the matrix shows the predicted values, the sum of the data of the matrix rows is the number of categories of the real value, and the sum of the column data is the number of categories after classification.
Figure 12,
Figure 13,
Figure 14,
Figure 15,
Figure 16 and
Figure 17 show the confusion matrix of the seven models, and it can be seen that LBFNet has good results not only for diseases with large differences between classes, but also for diseases with small differences between classes. From
Table 5, it can be seen that for LBFNet, the classification and recognition effects of all types of diseases meet the criteria of the actual situation, and all indicators are close to 1. Even for mosaic_virusy, which has the least samples, it can achieve 94% high accuracy, which is fully compatible with the needs of agricultural production, and there are no extreme cases.
5. Conclusions
In this paper, a novel VGG-like convolutional neural network model, LBFNet, is proposed for tomato leaf disease identification.The LBFNet model has a simple structure and efficient performance, which improves the problems of previous models with complex and poor accuracy.The LBFNet model combines the advantages of VGG networks, cascade networks and attention networks. After balancing the data, the model achieves 99.06% accuracy on the LBFtomato dataset, and after quantitative pruning and saving then further training, the model achieves 97.66% accuracy, and the parameter size is reduced by half, making it easy to deploy on mobile devices. The experimental results show that the model solves the problems of large model parameters, slow inference time, and low accuracy of current neural network models in tomato leaf disease identification. Compared with other models, LBFNet has high accuracy, fast inference time, and few parameters, which is outstanding in the field of tomato leaf disease recognition and can be applied to agricultural production activities to effectively improve agricultural production efficiency.
Author Contributions
Conceptualization, H.C. and R.Z.; methodology,H.C.; software, H.C.and J.P.;validation, H.C.,R.Z. and J.P.; resources, H.C.;data curation,Y.W.; writing—original draft preparation, H.C. and Y.W.; writing—review and editing, H.C., J.P., R.Z. and Y.W.; supervision, Y.W.;project administration, Y.W.; funding acquisition, Y.W.; investigation, H.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research is funded by the Ministry of Science and Technology of the People's Republic of China and belongs to the National Key Research and Development Program, number 2022YFD2002001.
Data Availability Statement
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu J, Wang X. Plant diseases and pests detection based on deep learning: a review[J]. Plant Methods, 2021, 17(1): 1-18.
- Abade A, Ferreira P A, de Barros Vidal F. Plant diseases recognition on images using convolutional neural networks: A systematic review[J]. Computers and Electronics in Agriculture, 2021, 185: 106125.
- Kamilaris A, Prenafeta-Boldú F X. A review of the use of convolutional neural networks in agriculture[J]. The Journal of Agricultural Science, 2018, 156(3): 312-322.
- Gui P, Dang W, Zhu F, et al. Towards automatic field plant disease recognition[J]. Computers and Electronics in Agriculture, 2021, 191: 106523.
- Yang C, Teng Z, Dong C, et al. In-Field Citrus Disease Classification via Convolutional Neural Network from Smartphone Images[J]. Agriculture, 2022, 12(9): 1487.
- Nguyen T H, Nguyen T N, Ngo B V. A VGG-19 Model with Transfer Learning and Image Segmentation for Classification of Tomato Leaf Disease[J]. AgriEngineering, 2022, 4(4): 871-887.
- Chen S, Xiong J, Jiao J, et al. Citrus fruits maturity detection in natural environments based on convolutional neural networks and visual saliency map[J]. Precision Agriculture, 2022, 23(5): 1515-1531.
- Mishra S, Sachan R, Rajpal D. Deep convolutional neural network based detection system for real-time corn plant disease recognition[J]. Procedia Computer Science, 2020, 167: 2003-2010.
- Dong C, Zhang Z, Yue J, et al. Automatic recognition of strawberry diseases and pests using convolutional neural network[J]. Smart Agricultural Technology, 2021, 1: 100009.
- Xu W, Zhao L, Li J, et al. Detection and classification of tea buds based on deep learning[J]. Computers and Electronics in Agriculture, 2022, 192: 106547.
- Liu C, Zhu H, Guo W, et al. EFDet: An efficient detection method for cucumber disease under natural complex environments[J]. Computers and Electronics in Agriculture, 2021, 189: 106378.
- Chen J, Liu Q, Gao L. Visual tea leaf disease recognition using a convolutional neural network model[J]. Symmetry, 2019, 11(3): 343.
- Singh A K, Sreenivasu S V N, Mahalaxmi U, et al. Hybrid feature-based disease detection in plant leaf using convolutional neural network, bayesian optimized SVM, and random forest classifier[J]. Journal of Food Quality, 2022, 2022: 1-16.
- Zhang S W, Shang Y J, Wang L. Plant disease recognition based on plant leaf image[J]. J. Anim. Plant Sci, 2015, 25(3): 42-45.
- Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, inception-resnet and the impact of residual connections on learning[C].AAAI. 2017, 4: 12.
- Abbas A, Jain S, Gour M, et al. Tomato plant disease detection using transfer learning with C-GAN synthetic images[J]. Computers and Electronics in Agriculture, 2021, 187: 106279.
- Huang G, Liu Z, Weinberger K Q, et al. Densely connected convolutional networks[C].Proceedings of the IEEE conference on computer vision and pattern recognition. 2017, 1(2): 3.
- Howard A G, Zhu M, Chen B, et al. Mobilenets: Efficient convolutional neural networks for mobile vision applications[J]. arXiv preprint arXiv:1704.04861, 2017.
- Ding X, Zhang X, Ma N, et al. Repvgg: Making vgg-style convnets great again[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 13733-13742.
- Zeng W, Li M. Crop leaf disease recognition based on Self-Attention convolutional neural network[J]. Computers and Electronics in Agriculture, 2020, 172: 105341.
- Deng H, Luo D, Chang Z, et al. RAHC_GAN: A Data Augmentation Method for Tomato Leaf Disease Recognition[J]. Symmetry, 2021, 13(9): 1597.
- Gong C, Wang D, Li M, et al. KeepAugment: A simple information-preserving data augmentation approach[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 1055-1064.
- Wang H, Song H, Wu H, et al. Multilayer feature fusion and attention-based network for crops and weeds segmentation[J]. Journal of Plant Diseases and Protection, 2022, 129(6): 1475-1489.
- Fang W, Guan F, Yu H, et al. Identification of wormholes in soybean leaves based on multi-feature structure and attention mechanism[J]. Journal of Plant Diseases and Protection, 2022: 1-12.
- Wang S H, Fernandes S L, Zhu Z, et al. AVNC: attention-based VGG-style network for COVID-19 diagnosis by three-channel attention mechanism[J]. IEEE Sensors Journal, 2021, 22(18): 17431-17438.
- Fukui H, Hirakawa T, Yamashita T, et al. Attention branch network: Learning of attention mechanism for visual explanation[C]//Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2019: 10705-10714.
- Chefer H, Gur S, Wolf L. Transformer interpretability beyond attention visualization[C]//Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition. 2021: 782-791.
- Guo M H, Xu T X, Liu J J, et al. Attention mechanisms in computer vision: A survey[J]. Computational Visual Media, 2022, 8(3): 331-368.
- Gadekallu T R, Rajput D S, Reddy M P K, et al. A novel PCA–whale optimization-based deep neural network model for classification of tomato plant diseases using GPU[J]. Journal of Real-Time Image Processing, 2021, 18: 1383-1396.
- Gonzalez-Huitron V, León-Borges J A, Rodriguez-Mata A E, et al. Disease detection in tomato leaves via CNN with lightweight architectures implemented in Raspberry Pi 4[J]. Computers and Electronics in Agriculture, 2021, 181: 105951.
- Rangarajan A K, Purushothaman R, Ramesh A. Tomato crop disease classification using pre-trained deep learning algorithm[J]. Procedia computer science, 2018, 133: 1040-1047.
- Wang Q, Qi F, Sun M, et al. Identification of tomato disease types and detection of infected areas based on deep convolutional neural networks and object detection techniques[J]. Computational intelligence and neuroscience, 2019, 2019.
- Zhang Y, Song C, Zhang D. Deep learning-based object detection improvement for tomato disease[J]. IEEE access, 2020, 8: 56607-56614.
- Chakravarthy A S, Raman S. Early blight identification in tomato leaves using deep learning[C]//2020 International conference on contemporary computing and applications (IC3A). IEEE, 2020: 154-158.
- Ahmed S, Hasan M B, Ahmed T, et al. Less is more: lighter and faster deep neural architecture for tomato leaf disease classification[J]. IEEE Access, 2022, 10: 68868-68884.
- Yu H, Liu J, Chen C, et al. Optimized deep residual network system for diagnosing tomato pests[J]. Computers and Electronics in Agriculture, 2022, 195: 106805.
- Singh D, Jain N, Jain P, et al. PlantDoc: A dataset for visual plant disease detection[M]//Proceedings of the 7th ACM IKDD CoDS and 25th COMAD. 2020: 249-253.
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).