Submitted:
14 April 2023
Posted:
17 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Viruses, cells, and chemicals
2.2. Plasmids
2.3. RNA isolation and real-time PCR
2.4. Western blot (WB) analysis
2.5. Immunoprecipitation (IP)
2.6. Statistical analysis
3. Results
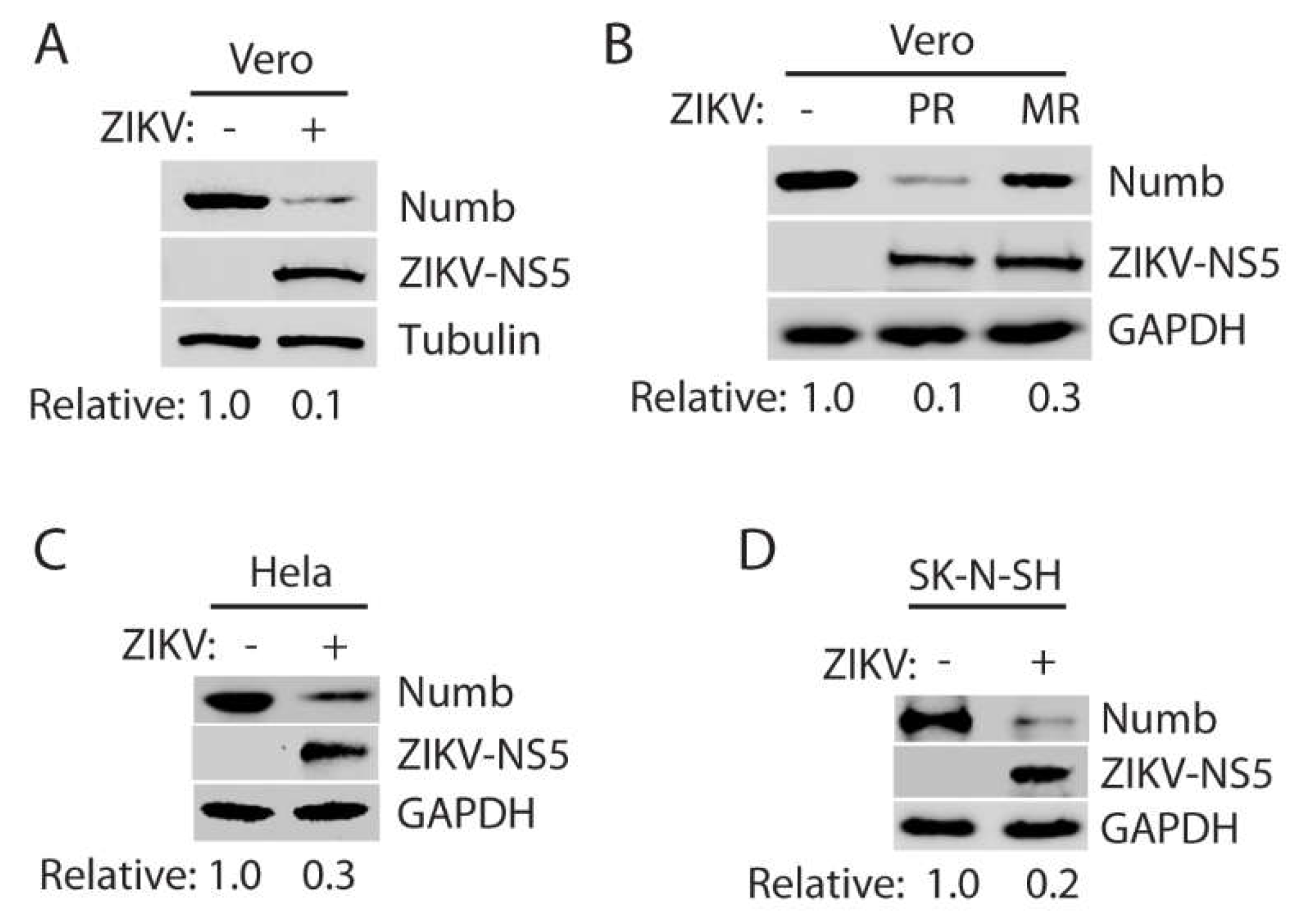
3.1. ZIKV infection reduces the Numb protein level.
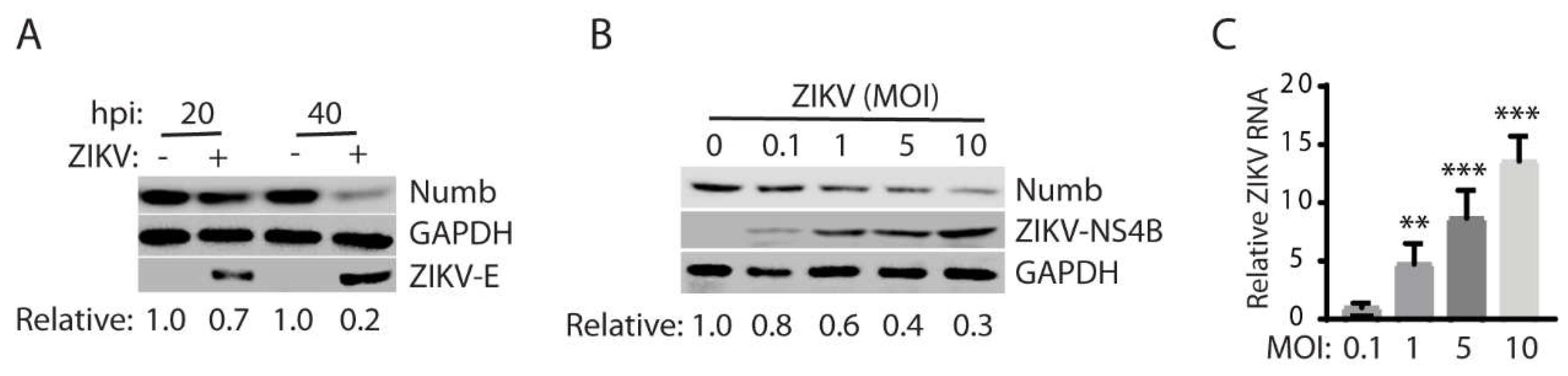
3.2. ZIKV reduces the Numb protein level in a temporal and dose-dependent manner.
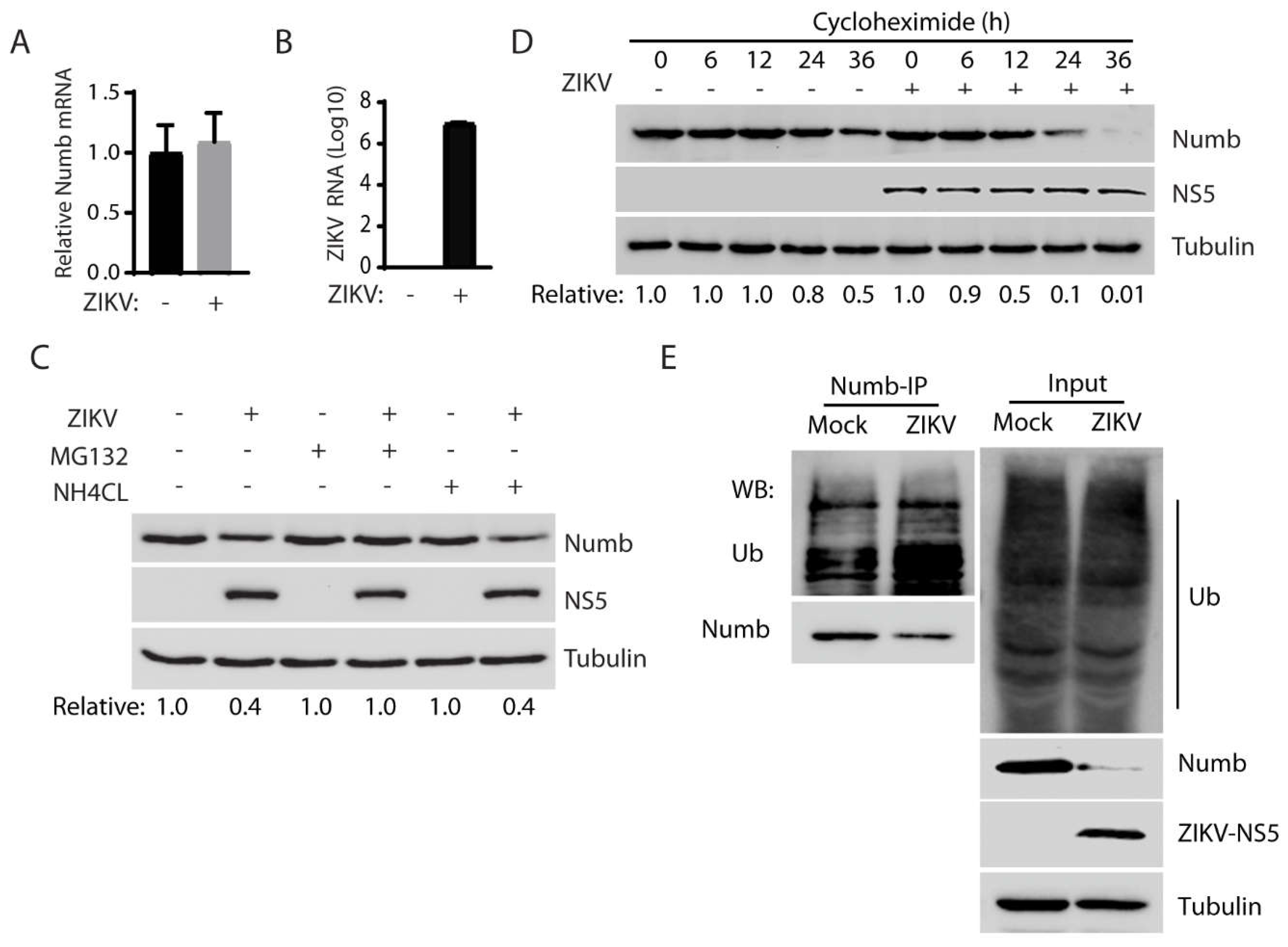
3.3. ZIKV reduces the Numb protein via the ubiquitin-proteasome pathway.
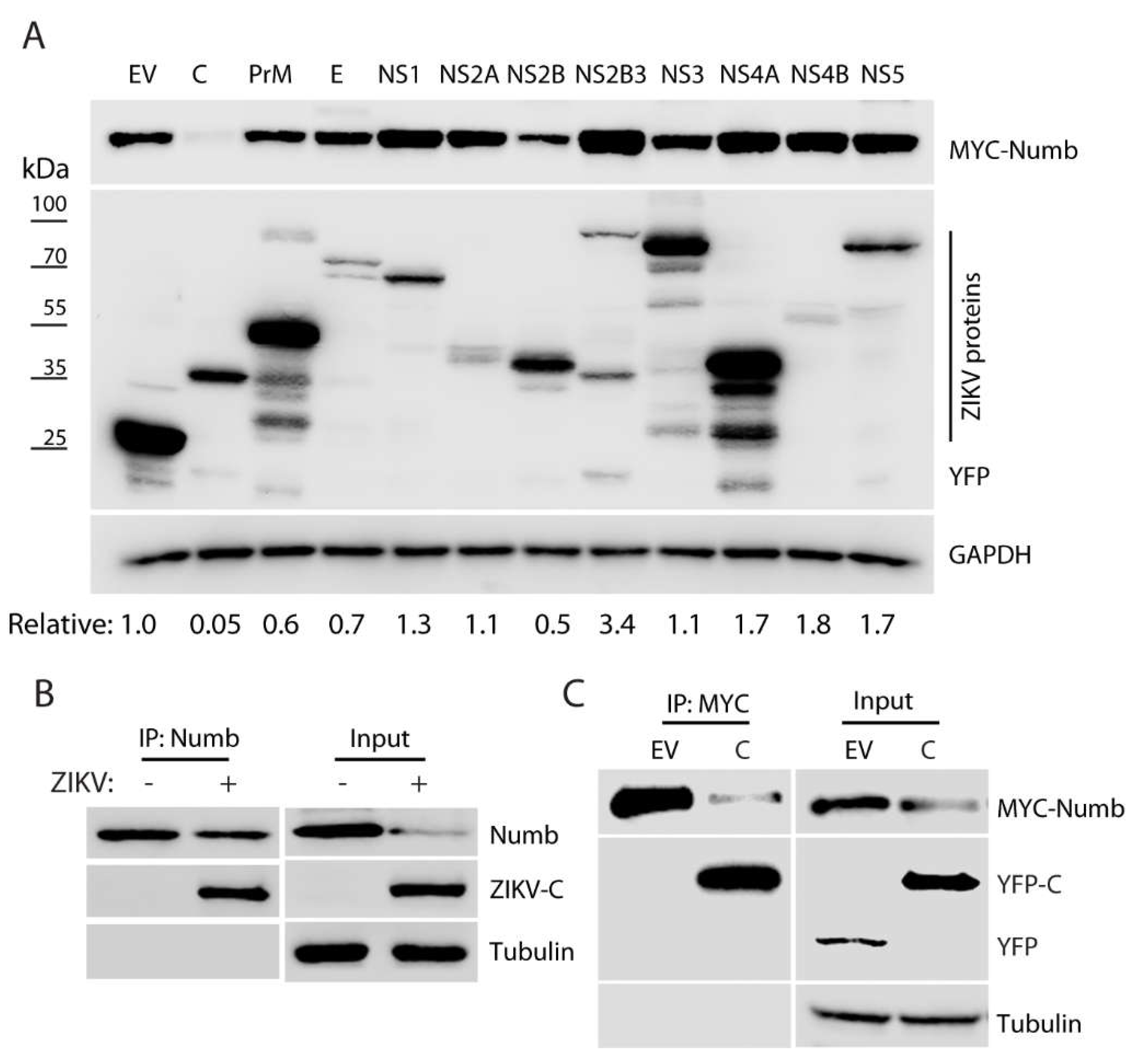
3.4. ZIKV capsid protein induces the Numb reduction.
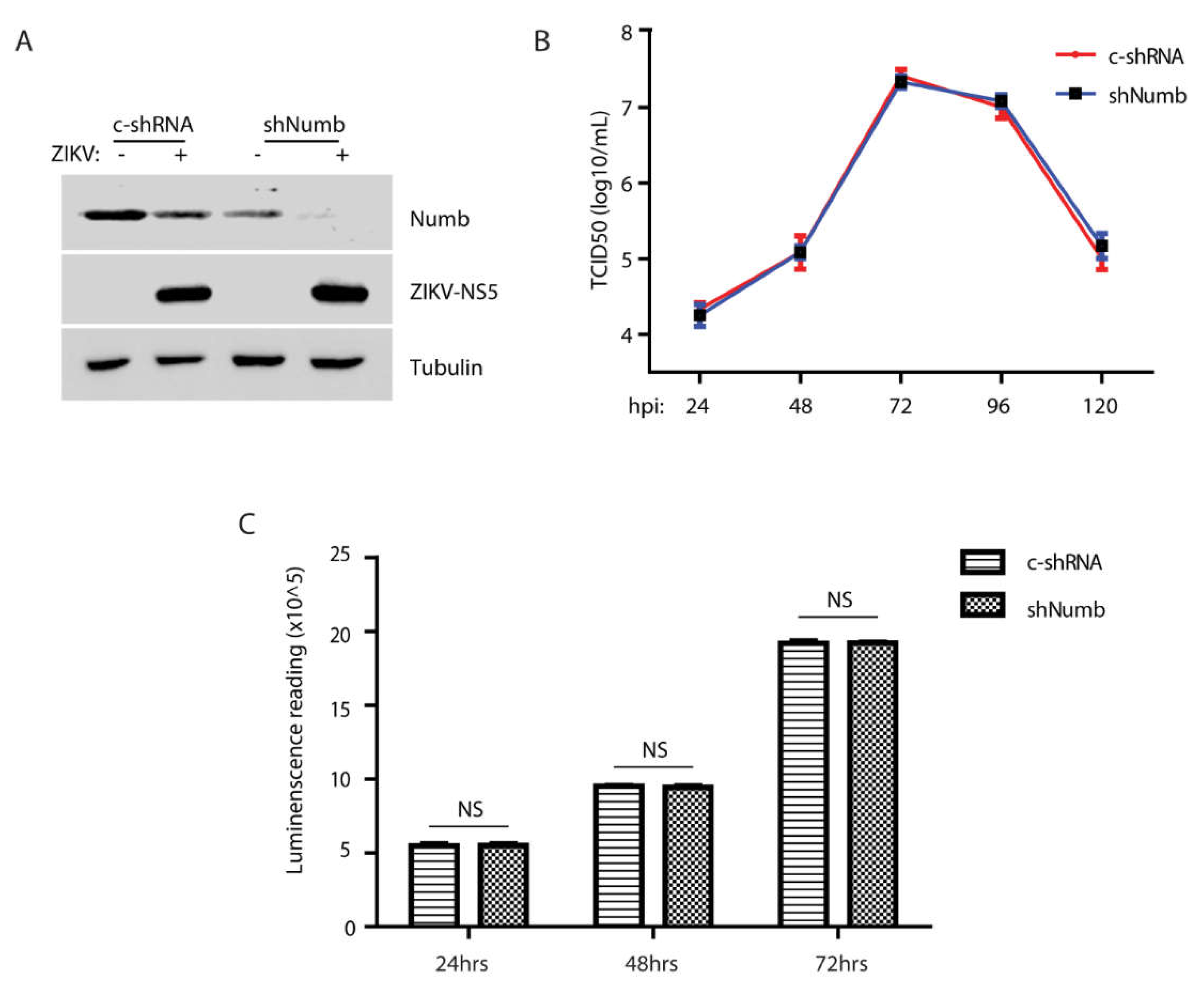
3.5. The Numb knockdown has minimal effect on ZIKV replication.
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dick, G.W. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg 1952, 46, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Dick, G.W.; Kitchen, S.F.; Haddow, A.J. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg 1952, 46, 509–520. [Google Scholar] [CrossRef]
- Duffy, M.R.; Chen, T.H.; Hancock, W.T.; Powers, A.M.; Kool, J.L.; Lanciotti, R.S.; Pretrick, M.; Marfel, M.; Holzbauer, S.; Dubray, C.; et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 2009, 360, 2536–2543. [Google Scholar] [CrossRef] [PubMed]
- Campos, G.S.; Bandeira, A.C.; Sardi, S.I. Zika Virus Outbreak, Bahia, Brazil. Emerg Infect Dis 2015, 21, 1885–1886. [Google Scholar] [CrossRef] [PubMed]
- Brooks, T.; Roy-Burman, A.; Tuholske, C.; Busch, M.P.; Bakkour, S.; Stone, M.; Linnen, J.M.; Gao, K.; Coleman, J.; Bloch, E.M. Real-Time Evolution of Zika Virus Disease Outbreak, Roatan, Honduras. Emerg Infect Dis 2017, 23, 1360–1363. [Google Scholar] [CrossRef] [PubMed]
- Bonilla-Soto, L.A. Zika Virus in the Americas: An Environmental Health Perspective. P R Health Sci J 2018, 37, S5–S14. [Google Scholar] [PubMed]
- Mlakar, J.; Korva, M.; Tul, N.; Popovic, M.; Poljsak-Prijatelj, M.; Mraz, J.; Kolenc, M.; Resman Rus, K.; Vesnaver Vipotnik, T.; Fabjan Vodusek, V.; et al. Zika Virus Associated with Microcephaly. N Engl J Med 2016, 374, 951–958. [Google Scholar] [CrossRef]
- Panchaud, A.; Stojanov, M.; Ammerdorffer, A.; Vouga, M.; Baud, D. Emerging Role of Zika Virus in Adverse Fetal and Neonatal Outcomes. Clin Microbiol Rev 2016, 29, 659–694. [Google Scholar] [CrossRef]
- Plourde, A.R.; Bloch, E.M. A Literature Review of Zika Virus. Emerg Infect Dis 2016, 22, 1185–1192. [Google Scholar] [CrossRef]
- Raposo-Amaral, C.E. Microcephaly: Consequence of the Zika Virus Global Outbreak. J Craniofac Surg 2016, 27, 1383–1384. [Google Scholar] [CrossRef]
- Crisanto-Lopez, I.E.; Jesus, P.L.; Lopez-Quecho, J.; Flores-Alonso, J.C. Congenital Zika syndrome. Bol Med Hosp Infant Mex 2023, 80, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Garcez, P.P.; Loiola, E.C.; Madeiro da Costa, R.; Higa, L.M.; Trindade, P.; Delvecchio, R.; Nascimento, J.M.; Brindeiro, R.; Tanuri, A.; Rehen, S.K. Zika virus impairs growth in human neurospheres and brain organoids. Science 2016, 352, 816–818. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Herrlinger, S.; Yang, S.L.; Lai, F.; Moore, J.M.; Brindley, M.A.; Chen, J.F. Zika virus infection disrupts neurovascular development and results in postnatal microcephaly with brain damage. Development 2016, 143, 4127–4136. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.S.; Sampaio, G.L.; Pereira, C.S.; Campos, G.S.; Sardi, S.I.; Freitas, L.A.; Figueira, C.P.; Paredes, B.D.; Nonaka, C.K.; Azevedo, C.M.; et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci Rep 2016, 6, 39775. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Maj, T.; Kryczek, I.; Li, W.; Wu, K.; Zhao, L.; Wei, S.; Crespo, J.; Wan, S.; Vatan, L.; et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol 2016, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Abraham, R.; Shim, B.S.; Choe, H.; Page, D.T. Zika virus infection during the period of maximal brain growth causes microcephaly and corticospinal neuron apoptosis in wild type mice. Sci Rep 2016, 6, 34793. [Google Scholar] [CrossRef] [PubMed]
- Lazear, H.M.; Diamond, M.S. Zika Virus: New Clinical Syndromes and Its Emergence in the Western Hemisphere. J Virol 2016, 90, 4864–4875. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Ling, L.; Zhang, Z.; Marin-Lopez, A. Current Advances in Zika Vaccine Development. Vaccines (Basel) 2022, 10. [Google Scholar] [CrossRef]
- Uemura, T.; Shepherd, S.; Ackerman, L.; Jan, L.Y.; Jan, Y.N. numb, a gene required in determination of cell fate during sensory organ formation in Drosophila embryos. Cell 1989, 58, 349–360. [Google Scholar] [CrossRef]
- Flores, A.N.; McDermott, N.; Meunier, A.; Marignol, L. NUMB inhibition of NOTCH signalling as a therapeutic target in prostate cancer. Nat Rev Urol 2014, 11, 499–507. [Google Scholar] [CrossRef]
- Zilian, O.; Saner, C.; Hagedorn, L.; Lee, H.Y.; Sauberli, E.; Suter, U.; Sommer, L.; Aguet, M. Multiple roles of mouse Numb in tuning developmental cell fates. Curr Biol 2001, 11, 494–501. [Google Scholar] [CrossRef] [PubMed]
- Ciechanover, A. Proteolysis: from the lysosome to ubiquitin and the proteasome. Nat Rev Mol Cell Biol 2005, 6, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Jackson, M.P.; Hewitt, E.W. Cellular proteostasis: degradation of misfolded proteins by lysosomes. Essays Biochem 2016, 60, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Yang, L.; Chang, P.; Yang, S.; Lin, S.; Tang, Q.; Wang, X.; Zhang, Y.J. Zika virus NS2A protein induces the degradation of KPNA2 (karyopherin subunit alpha 2) via chaperone-mediated autophagy. Autophagy 2020, 16, 2238–2251. [Google Scholar] [CrossRef]
- Liu, X.H.; Yao, S.; Levine, A.C.; Kirschenbaum, A.; Pan, J.; Wu, Y.; Qin, W.; Collier, L.; Bauman, W.A.; Cardozo, C.P. Nandrolone, an anabolic steroid, stabilizes Numb protein through inhibition of mdm2 in C2C12 myoblasts. J Androl 2012, 33, 1216–1223. [Google Scholar] [CrossRef]
- Tan, T.Y.; Fibriansah, G.; Kostyuchenko, V.A.; Ng, T.S.; Lim, X.X.; Zhang, S.; Lim, X.N.; Wang, J.; Shi, J.; Morais, M.C.; et al. Capsid protein structure in Zika virus reveals the flavivirus assembly process. Nat Commun 2020, 11, 895. [Google Scholar] [CrossRef]
- Farelo, M.A.; Korrou-Karava, D.; Brooks, K.F.; Russell, T.A.; Maringer, K.; Mayerhofer, P.U. Dengue and Zika Virus Capsid Proteins Contain a Common PEX19-Binding Motif. Viruses 2022, 14. [Google Scholar] [CrossRef]
- Fontaine, K.A.; Leon, K.E.; Khalid, M.M.; Tomar, S.; Jimenez-Morales, D.; Dunlap, M.; Kaye, J.A.; Shah, P.S.; Finkbeiner, S.; Krogan, N.J.; et al. The Cellular N.M.D. Pathway Restricts Zika Virus Infection and Is Targeted by the Viral Capsid Protein. MBio 2018, 9. [Google Scholar] [CrossRef]
- Gestuveo, R.J.; Royle, J.; Donald, C.L.; Lamont, D.J.; Hutchinson, E.C.; Merits, A.; Kohl, A.; Varjak, M. Analysis of Zika virus capsid-Aedes aegypti mosquito interactome reveals proviral host factors critical for establishing infection. Nat Commun 2021, 12, 2766. [Google Scholar] [CrossRef]
- Hou, S.; Kumar, A.; Xu, Z.; Airo, A.M.; Stryapunina, I.; Wong, C.P.; Branton, W.; Tchesnokov, E.; Gotte, M.; Power, C.; et al. Zika Virus Hijacks Stress Granule Proteins and Modulates the Host Stress Response. J Virol 2017, 91. [Google Scholar] [CrossRef]
- Zeng, J.; Dong, S.; Luo, Z.; Xie, X.; Fu, B.; Li, P.; Liu, C.; Yang, X.; Chen, Y.; Wang, X.; et al. The Zika Virus Capsid Disrupts Corticogenesis by Suppressing Dicer Activity and miRNA Biogenesis. Cell Stem Cell 2020, 27, 618–632. [Google Scholar] [CrossRef] [PubMed]
- Neveu, G.; Ziv-Av, A.; Barouch-Bentov, R.; Berkerman, E.; Mulholland, J.; Einav, S. AP-2-associated protein kinase 1 and cyclin G-associated kinase regulate hepatitis C virus entry and are potential drug targets. J Virol 2015, 89, 4387–4404. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Kawaguchi, K.; Honda, M.; Hashimoto, S.; Shirasaki, T.; Okada, H.; Orita, N.; Shimakami, T.; Yamashita, T.; Sakai, Y.; et al. Notch signaling facilitates hepatitis B virus covalently closed circular DNA transcription via cAMP response element-binding protein with E3 ubiquitin ligase-modulation. Sci Rep 2019, 9, 1621. [Google Scholar] [CrossRef] [PubMed]
- Frise, E.; Knoblich, J.A.; Younger-Shepherd, S.; Jan, L.Y.; Jan, Y.N. The Drosophila Numb protein inhibits signaling of the Notch receptor during cell-cell interaction in sensory organ lineage. Proc Natl Acad Sci U S A 1996, 93, 11925–11932. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Feder, J.N.; Jiang, M.M.; Jan, L.Y.; Jan, Y.N. Asymmetric localization of a mammalian numb homolog during mouse cortical neurogenesis. Neuron 1996, 17, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Bray, S.J. Notch signalling in context. Nat Rev Mol Cell Biol 2016, 17, 722–735. [Google Scholar] [CrossRef]
- McGill, M.A.; McGlade, C.J. Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J Biol Chem 2003, 278, 23196–23203. [Google Scholar] [CrossRef]
- Jiang, J.; Hui, C.C. Hedgehog signaling in development and cancer. Dev Cell 2008, 15, 801–812. [Google Scholar] [CrossRef]
- Di Marcotullio, L.; Ferretti, E.; Greco, A.; De Smaele, E.; Po, A.; Sico, M.A.; Alimandi, M.; Giannini, G.; Maroder, M.; Screpanti, I.; et al. Numb is a suppressor of Hedgehog signalling and targets Gli1 for Itch-dependent ubiquitination. Nat Cell Biol 2006, 8, 1415–1423. [Google Scholar] [CrossRef]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of Snail-dominant colorectal cancer stem cells. Nat Cell Biol 2014, 16, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Colaluca, I.N.; Tosoni, D.; Nuciforo, P.; Senic-Matuglia, F.; Galimberti, V.; Viale, G.; Pece, S.; Di Fiore, P.P. NUMB controls p53 tumour suppressor activity. Nature 2008, 451, 76–80. [Google Scholar] [CrossRef]
- Juven-Gershon, T.; Shifman, O.; Unger, T.; Elkeles, A.; Haupt, Y.; Oren, M. The Mdm2 oncoprotein interacts with the cell fate regulator Numb. Mol Cell Biol 1998, 18, 3974–3982. [Google Scholar] [CrossRef] [PubMed]
- Pece, S.; Serresi, M.; Santolini, E.; Capra, M.; Hulleman, E.; Galimberti, V.; Zurrida, S.; Maisonneuve, P.; Viale, G.; Di Fiore, P.P. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J Cell Biol 2004, 167, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Sheng, W.; Dong, M.; Chen, C.; Li, Y.; Liu, Q.; Dong, Q. Musashi2 promotes the development and progression of pancreatic cancer by down-regulating Numb protein. Oncotarget 2017, 8, 14359–14373. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.Y.; Seok, J.; Kang, G.H.; Lim, K.M.; Cho, S.G. The role of NUMB/NUMB isoforms in cancer stem cells. BMB Rep 2021, 54, 335–343. [Google Scholar] [CrossRef]
- Imai, T.; Tokunaga, A.; Yoshida, T.; Hashimoto, M.; Mikoshiba, K.; Weinmaster, G.; Nakafuku, M.; Okano, H. The neural RNA-binding protein Musashi1 translationally regulates mammalian numb gene expression by interacting with its mRNA. Mol Cell Biol 2001, 21, 3888–3900. [Google Scholar] [CrossRef] [PubMed]
- Kuang, W.; Tan, J.; Duan, Y.; Duan, J.; Wang, W.; Jin, F.; Jin, Z.; Yuan, X.; Liu, Y. Cyclic stretch induced miR-146a upregulation delays C2C12 myogenic differentiation through inhibition of Numb. Biochem Biophys Res Commun 2009, 378, 259–263. [Google Scholar] [CrossRef]
- Nie, J.; McGill, M.A.; Dermer, M.; Dho, S.E.; Wolting, C.D.; McGlade, C.J. L.N.X. functions as a RING type E3 ubiquitin ligase that targets the cell fate determinant Numb for ubiquitin-dependent degradation. EMBO J 2002, 21, 93–102. [Google Scholar] [CrossRef]
- Susini, L.; Passer, B.J.; Amzallag-Elbaz, N.; Juven-Gershon, T.; Prieur, S.; Privat, N.; Tuynder, M.; Gendron, M.C.; Israel, A.; Amson, R.; et al. Siah-1 binds and regulates the function of Numb. Proc Natl Acad Sci U S A 2001, 98, 15067–15072. [Google Scholar] [CrossRef]
- Teng, Y.; Liu, S.; Guo, X.; Liu, S.; Jin, Y.; He, T.; Bi, D.; Zhang, P.; Lin, B.; An, X.; et al. An Integrative Analysis Reveals a Central Role of P53 Activation via MDM2 in Zika Virus Infection Induced Cell Death. Front Cell Infect Microbiol 2017, 7, 327. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.J.; Stein, D.A.; Fan, S.M.; Wang, K.Y.; Kroeker, A.D.; Meng, X.J.; Iversen, P.L.; Matson, D.O. Suppression of porcine reproductive and respiratory syndrome virus replication by morpholino antisense oligomers. Vet Microbiol 2006, 117, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat Commun 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, R.; Ma, Z.; Xiao, Y.; Nan, Y.; Wang, Y.; Lin, S.; Zhang, Y.J. Porcine Reproductive and Respiratory Syndrome Virus Antagonizes JAK/STAT3 Signaling via nsp5, Which Induces STAT3 Degradation. J Virol 2017, 91, e02087–02016. [Google Scholar] [CrossRef] [PubMed]
- Patel, D.; Opriessnig, T.; Stein, D.A.; Halbur, P.G.; Meng, X.J.; Iversen, P.L.; Zhang, Y.J. Peptide-conjugated morpholino oligomers inhibit porcine reproductive and respiratory syndrome virus replication. Antiviral Res 2008, 77, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wang, R.; Yang, S.; Ma, Z.; Lin, S.; Nan, Y.; Li, Q.; Tang, Q.; Zhang, Y.J. Karyopherin Alpha 6 Is Required for Replication of Porcine Reproductive and Respiratory Syndrome Virus and Zika Virus. J Virol 2018, 92, e00072–00018. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Wang, K.Y.; Stein, D.A.; Patel, D.; Watkins, R.; Moulton, H.M.; Iversen, P.L.; Matson, D.O. Inhibition of replication and transcription activator and latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus by morpholino oligomers. Antiviral Res 2007, 73, 12–23. [Google Scholar] [CrossRef]
| Primera | Sequences (5' to 3')b | Target gene/vector |
| ZIKV-RR-F | AARTACACATACCARAACAAAGTGGT | NS5 |
| ZIKV-RR-R | TCCRCTCCCYCTYTGGTCTTG | NS5 |
| Numb-F1 | CGAATTCAACAAATTACGGCAAAGTTT | Numb |
| Numb-R1 | GCTCGAGTTAAAGTTCAATTTCAAACG | Numb |
| Numb-RR-F1 | GCTACCACCAGTCCCTTCTT | Numb |
| Numb-RR-R1 | GTGCCTGTAGGAACCTCTGT | Numb |
| shNUMB1-F | GATCCGGAATAAATATTATATATATTCAAGAGATATATATAATATTTATTCCTTTTTTG | shNumb |
| shNUMB1-R | AATTCAAAAAAGGAATAAATATTATATATATCTCTTGAATATATATAATATTTATTCCG | shNumb |
| shNUMB2-F | GATCCGCTCTATAGAGAATATATATTCAAGAGATATATATTCTCTATAGAGCTTTTTTG | shNumb |
| shNUMB2-R | AATTCAAAAAAGCTCTATAGAGAATATATATCTCTTGAATATATATTCTCTATAGAGCG | shNumb |
| shNUMB3-F | GATCCGAATAAATATTATATATAATTCAAGAGATTATATATAATATTTATTCTTTTTTG | shNumb |
| shNUMB3-R | AATTCAAAAAAGAATAAATATTATATATAATCTCTTGAATTATATATAATATTTATTCG | shNumb |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





