Introduction
Rice (Oryza sativa L.), domesticated into subspecies Xian (indica) and Geng (japonica), is one of the top three cereal crops in China in terms of area and production (1-2). Rice blast, caused by the fungal pathogen Magnaporthe oryzae (Mo; syn. Pyricularia oryzae), is the most concerned and worried disease in both Southern Xian and Northern Geng cropping areas (2-5). Like other diseases, it is believed that genetic resistance is the most critical and promising approach to control rice blast, but the effectiveness and durability of resistance depends on population dynamics in each region (3,6-8).
The gene-for-gene principal, in which the pathogen carries a gene for avirulence (Avr gene) that corresponds to each resistance gene (R gene) in the cognate host, plays a pivotal role in shaping population structures of plant pathogens (2,5,8-12). The AvrPii of Mo, which was isolated through association genetic approach (AB498874; 13), was one of the most attractive Avr genes being widely investigated in various populations. Xing et al. (14) characterized 182 isolates of Mo, which were collected from Hunan, China, by rice monogenic lines-based pathogenicity assay and PCR amplification with gene-specific primer sets. The results showed that among 22.5% of Pii-avirulent isolates, only 2.5% gained AvrPii-amplicons. The distribution and mutation of AvrPii in Yunnan, China, was also studied (3). Out of 454 isolates tested, 291 were Pii-virulent. Among them, 24 Pii-virulent isolates gained AvrPii-specific amplicons, suggesting that the AvrPii carried by the 24 isolates has lost avirulence function through smaller variations such as base substitution. Conversely, 105 out of the 163 were Pii-avirulent isolates had no AvrPii-specific amplicon, indicating that two pairs of AvrPii-specific primers used in the investigation might not be suitable for these isolates, and/or avirulent performance of such isolates against the Pii-carrier conveyed by another gene pair of Avr/R genes. In the characterization of 77 isolates collected from Thailand, only 46 isolates successfully achieved coding sequence (CDS) amplification by AvrPii-specific primer pair. The CDS of the AvrPii gene from the 46 isolates displayed three different haplotypes with 11 nucleotide polymorphic sites, including one major haplotype (44 out of 46 isolates) and two minor haplotypes (one a single isolate each; 11). The four Avr genes, AvrPita1, AvrPik, AvrPiz-t, AvrPia, and AvrPii, were surveyed in a Mo population consisting of 258 isolates collected during 1975-2009 from Arkansas state, the United States. PCR products were obtained from 232 isolates, and the remaining 26 isolates without any amplicon might be due to absence of the Avr genes tested, and/or to less matching of Avr gene-specific primer sets used in the study. It was surprised that only one isolate carried AvrPii, compared to 225 for AvrPiz-t, and 174 for AvrPita1 (15). Altogether, there was a certain numerical gap between a higher rate of Pii-avirulent isolates and a lower rate of AvrPii-specific amplicons in the Mo population tested.
Our aim in this study was to investigate whether the main reason for the numerical gap was a higher proportion of sequence-distinct haplotypes in the AvrPii family that failed to be amplified by primers designed from the reference isolate? If as hypothesized, how was the AvrPii family featured with divergent haplotypes in the Mo population?
Materials and Methods
Discovery of a new haplotype AvrPii-C
Ten isolates (CHL22, 346, 584, 590, 624, EHL0314, 0317, 0319, 0329, 0445) that were avirulent on a rice monogenic line carrying
Pii, IRBLi-F5, were selected from various
Mo populations in China and were subjected to amplification of both coding sequence (CDS) and the full-length (FL) sequence of
AvrPii via the regular polymerase chain reaction (PCR) system, with the key primer pairs AvrPii-J/C-CDS_F/R, and AvrPii-J-FL_F/R, respectively. Both primer pairs were designed based on the known reference sequence of
AvrPii aforementioned. Because a half of the ten isolates tested did not produce any amplicon with the FL primer pair, the alternative approach called a thermal asymmetric interlaced PCR (Tail-PCR) system was employed for creating specific sequences of both 5’ and 3’ regions of the representative isolate, CHL346, both might be distinct from the reference sequence (
Figure S1). After three rounds of Tail-PCR, the certain amplicons derived from 5’ and 3’ regions were subjected to sequencing. Then, the key primer pair, AvrPii-C-FL_F/R, was devised based on both sequences for re-amplifying the target gene of the ten isolates via the regular PCR system. The full-length sequences of the ten isolates were compared at Multalin website (
http://multalin.toulouse.inra.fr/multalin/). Information on the primers and PCR programs were shown in
Table S1.
2.1. Functional validation of both haplotypes
To construct transgenic vectors of both haplotypes, the full-length sequences of
AvrPii-C (as that was firstly discovered from the Chinese isolates in the present study) and
AvrPii-J (as it was firstly discovered from the Japanese isolate Ina168; 13), respectively, were re-amplified using Q5 High-Fidelity DNA polymerase (Tsingke, Nanjing, China) based on the genomic DNA of isolates CHL346 and CHL22. The amplicons were inserted into pGEM-T (Promega, Dalian, China) to generate two constructs pGEM-T_
AvrPii-C and _
AvrPii-J. Following their amplification from the pGEM-T templates with the key primer pairs,
AvrPii-C-GT_F/R (GT, genetic transformation), and
AvrPii-J-GT_F/R, the transgene sequences were introduced into the binary vector pBHT2-AscI with an
Asc I recognition site (
Table S1). Before their transformation into a virulent recipient isolate CHL357, each construct was validated by sequencing. At least five independent hygromycin-resistant transformants per construct were carried forward for testing their functionality onto the
Pii carrier, IRBLi-F5, and their specificity onto four monogenic lines carrying individual resistance genes, i.e., IRBL5-M (
Pi5), IRBLk-Ka (
Pik), IRBLb-B (
Pib), and IRBLta-CP1 (
Pita) (16). Phenotype of each isolate was scored as A (0 reaction: avirulent), MA (1 to 2 reaction: moderately avirulent), MV (3 to 4 reaction: moderately virulent), and V (5 reaction: virulent) through the routine spraying inoculation procedure as described previously (2,5,17).
2.2. Construction and characterization of haplotype-chimeric mutant
Because multiple variations were scattered in the whole sequences of both haplotypes (
Figure S2), the haplotype-chimeric mutants were constructed via domain-swapping approach (
Figure S3). To create the two haplotype-chimeric mutants in 5’ and 3’ regions, two chimeric templets were amplified with respective primer pairs with outside one being haplotype-specific (pure color), and inside one being haplotype-chimeric (chimeric color); then the haplotype-chimeric mutants were generated by overlap-PCR with both haplotype specific primers. To create three haplotype-chimeric mutants targeted in CDS region, self-ligation-PCR was conducted with a set of laddering chimeric primers (
Table S1 and
Figure S3).
Seven haplotype-chimeric mutants (Mai1-7) with two donor and one recipient isolates were phenotyped as aforementioned. Then the repeated and typical phenotype of each mutant was quantified based on
Mo growth in the leaves of IRBLi-F5 by estimating the ratio between the copy number of a
Mo sequence (
Po2) and the rice
Ubi gene (GenBank accession D12629) with qPCR system as described previously (8). To establish relationships (if any) between phenotypes and protein structures of the haplotype-chimeric mutants, two models were employed to draw their 3D-structures (I-TASSER,
https://zhanggroup.org/I-TASSER/; SWISS-MODEL,
https://swissmodel.expasy.org/interactive) (
Figure S4).
Interactions of the wild types and three CDS derived haplotype-chimeric mutants with the paired Pii gene were tested in the yeast two-hybrid (Y2H) system (Clontech, Dalian, China), in which the mature and the full-length AvrPii proteins with the five versions, i.e., FL, CC (coiled coil), NBS (nucleotide binding site), LRR (leucine rich repeat), CtNL (carbon terminal non-LRR sequence), of the paired Pii gene in reciprocal bait/prey combinations (
Figure S5). The detailed procedure was adopted from the previous studies (7,18).
2.3. Shape of population structure
Six
Mo populations ((Guangdong (GD), Hunan (HN), and Guizhou (GZ) from southern; and Liaoning (LN), Jilin (JL), and Heilongjiang (HLJ) from northern China)), each consisting of 60 rice-derived isolates, were selected based on regionally representative rice cultivars and sites sampled (
Table S2). An additional set of 17 weed-derived
Mo isolates was used as outgroup for checking the origins of haplotypes. A total of 377 isolates were phenotyped on the
Pii carrier as aforementioned. The isolates were genotyped with four markers, each two, a shorter fragment (MK1) tending to CDS, and a longer one (MK2) to 5’ region for the individual haplotypes, by the key PCR primer pairs, AvrPii-C-MK1_F/R, AvrPii-C-MK2_F/R, AvrPii-J-MK1_F/R, and AvrPii-J-MK2_F/R. Each marker was given a genotype code of either 0 (absence of amplicon) or 1 (presence of amplicon). Then, the genotype of each isolate was simply scored as 0 (absence of
AvrPii), 1-C (one copy of
AvrPii-C), 1-J (one copy of
AvrPii-J), and 1-C_J (two copies of both
AvrPii-C and
AvrPii-J), when genotypes of each two markers for the individual haplotype were consistent. Taken together, parameter of each combination of phenotype/genotype was counted out for each population for shaping its population structure.
2.4. Resequencing and evolution analysis
All the isolates with amplicons including weed-derived
Mo isolates were re-amplified by the key primer pairs, AvrPii-C-RS_F/R (RS, resequencing), and/or AvrPii-J-RS_F/R. Particularly, an isolate belonging to genotype 1-C/J might gain two amplicons by both key primer pairs. Then all the amplicons carrying the FL sequences were subjected to resequencing, directly. The
AvrPii allelic sequences were aligned with the known reference sequence of isolate Ina168 using DnaSP v6.12.03 software (
http://www.ub.edu/dnasp/), and their CDSs were subjected to evolutionary analysis with the following parameters. Segregating site (
S) was derived from pair comparison among isolates tested. Nucleotide diversity (
π) was estimated by Nei’s function with the Jukes and Cantor correction (19-20). Selection on the family was evaluated by the Fu and Li’s
D* and
F* parameters (21), and the average rate of non-synonymous (
Ka) and synonymous (
Ks) substitutions (
Ka/
Ks) was assessed as described by Nei and Gojobori (22). Then, three kinds of key selection forces were briefly defined, the positive selection (also defined as Darwinian selection or directional selection) is featured with key evolutionary parameters,
D < 0 and/or
Ka/
Ks (
dN/
dS) >1, that leads to fixation of target allele that increases the fitness of individuals; the balancing selection (disruptive selection or diversified selection) is featured with key parameter,
D > 0, that favors diversity; and the purifying selection (negative or stabilizing selection) is featured with key parameter,
Ka/
Ks ≤ 1, that eliminates deleterious mutations (23-25).
For deeply dissecting evolutionary features of the AvrPii family, six Mo natural populations were recombined into four comparable groups, i.e., all AvrPii group, haplotype AvrPii-J group, haplotype AvrPii-C group, and a regional group. The target genes were further divided into the entire coding region, non-signal peptide region (mature protein), and signal peptide region, if necessary. Additionally, 17 weed-derived isolates served as an outgroup for evolutionary comparison, if any.
Results
3.1. Haplotypic features of AvrPii
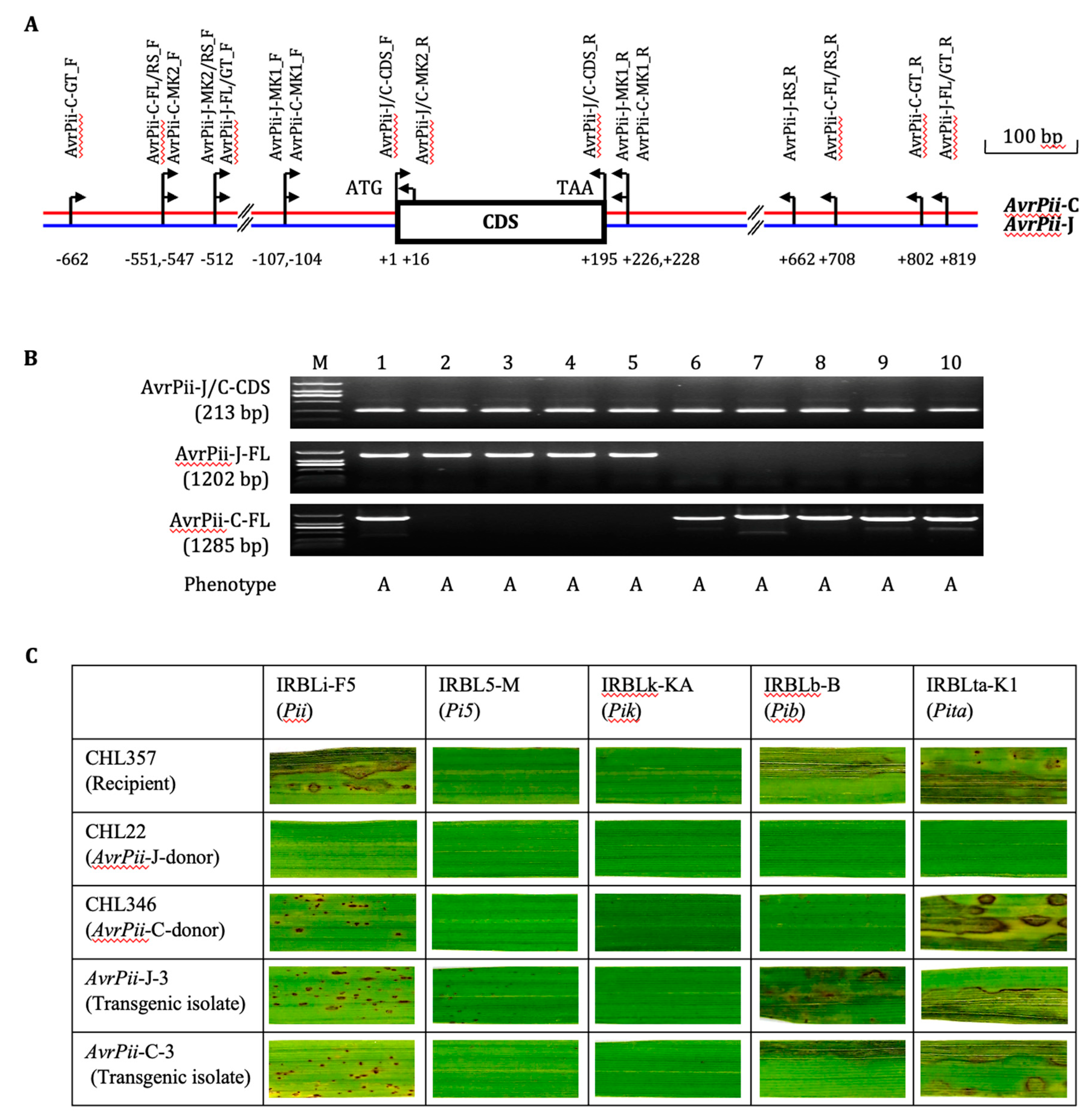
All the ten avirulent isolates selected from various Chinese
Mo populations were amplified the expective CDS products with the key primer pair, AvrPii-J/C-CDS_F/R, whereas only the first-five isolates were with the expected FL amplicons by the key primer pair, AvrPii-J-FL_F/R, indicating that sequences of the second primer pair might be less matched with those of the second-five isolates (
Figure 1A, B). The key primer pair, AvrPii-C-FL_F/R, which was devised based on the specific sequences generated from both 5’ and 3’ regions of the representative isolates CHL346 via Tail-PCR system (
Figure S1), was employed to re-amplify the ten isolates. The reverse result was observed, i.e., the first-five isolates were without any amplicon, except for #1, CHL22, and the second-five one with the expectative products. The reasonable explanation for genotype of the exceptional isolate CHL22 is that it carries two copies of
AvrPii, i.e.,
AvrPii-J and
AvrPii-C, which resembles some isolates shown in
Table S2.
Thus, the key primer pairs, AvrPii-J-RS_F/R, and AvrPii-C-RS_F/R, respectively, were used for amplifying the first- and second-five isolates for the haplotypic amplicon-based sequencing. As expected, there were extensive variations in both 5’ and 3’ regions, and intermediate ones in CDS region between the two haplotypes (two typical sequence difference as shown in
Figure S1-2). The target gene from the first-five isolates was temporally defined as
AvrPii-J, and that from the second-five isolates as
AvrPii-C. Transgenic progenies derived from both haplotypes expressed a consistent functionality and specificity on five monogenic rice lines carrying
Pii and other four individual resistance genes (
Figure 1C). Noticeably, both haplotypes conveyed moderately avirulent to the
Pii carrier.
Altogether, the new haplotype, AvrPii-C, is a novel and functional one in the AvrPii family.
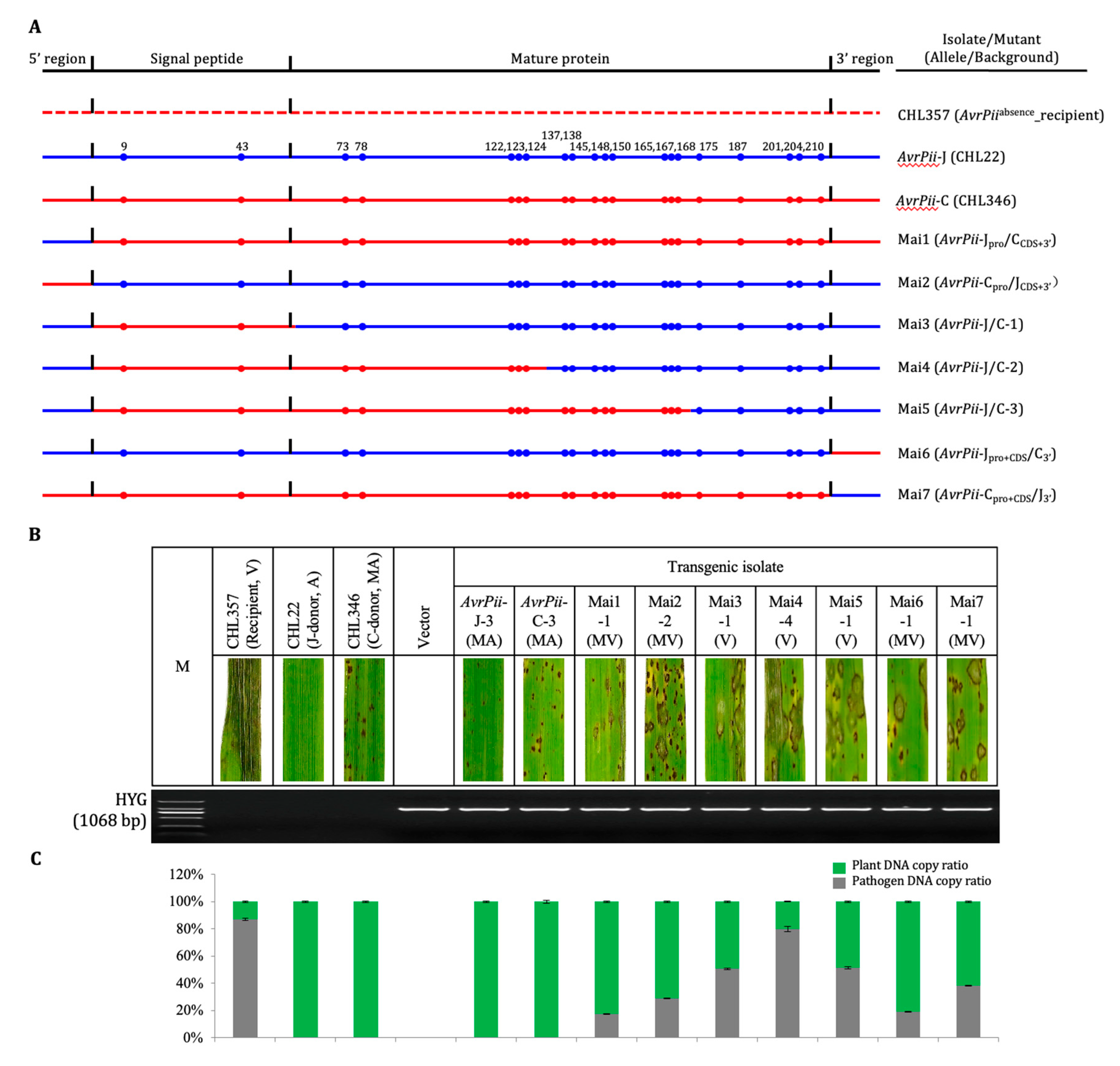
3.2. Performance of haplotype-chimeric mutant
Compared to moderately avirulent performance of transgenic isolates derived from both wild haplotypes, reactions of each two haplotype-chimeric mutants derived from both 5’ (Mai1-2) and 3’ (Mai6-7) were quantified as moderately virulent (
Figure 2). It suggests that both 5’ and 3’ regions were specific to the individual wild haplotypes because replacement of either 5’ or 3’ regions led to decline expression of their avirulence against the
Pii carrier (
Figure S3;
Table S3). While the performance of three haplotype-chimeric mutants, which were derived from CDS regions (Mai3-5), were almost changed from moderately avirulent to virulent. It indicates that the integrity of CDS was crucial to express functionality of the individual haplotypes. Correspondingly, the protein structures of CDS-derived chimeric mutants were distinct from both wild type ones, i.e., Mai1-2 and 6-7 (
Figure S4).
Furthermore, there was not any positive interaction in Bait/Prey reciprocal combinations between four mature proteins (AvrPii-J
WT20-70, AvrPii-J/C-2
20-70, AvrPii-J/C-3
20-70, and AvrPii-C
WT20-70) and a series of domains of the paired genes of Pii (
Figure S5A, B). The results repeated those reported in the previous studies (3,26). While there were three positive reactions only in Prey/Bait combinations between five full-length proteins AvrPii (Prey) and a series of domains of the paired genes of Pii (Bait), i.e., a weaker interaction between AvrPii-J/C-3
FL and Pii-1
CC, and a stronger and weaker interactions, respectively, between AvrPii-C
WTFL and Pii-1
CC and Pii-2
LRR (
Figure S5C, D). These results indicate that the younger prey of the AvrPii-C, particularly its latter part, could be interacted with the cognate bait of the Pii, during their long-term co-evolution process.
3.3. Population structure of the AvrPii family
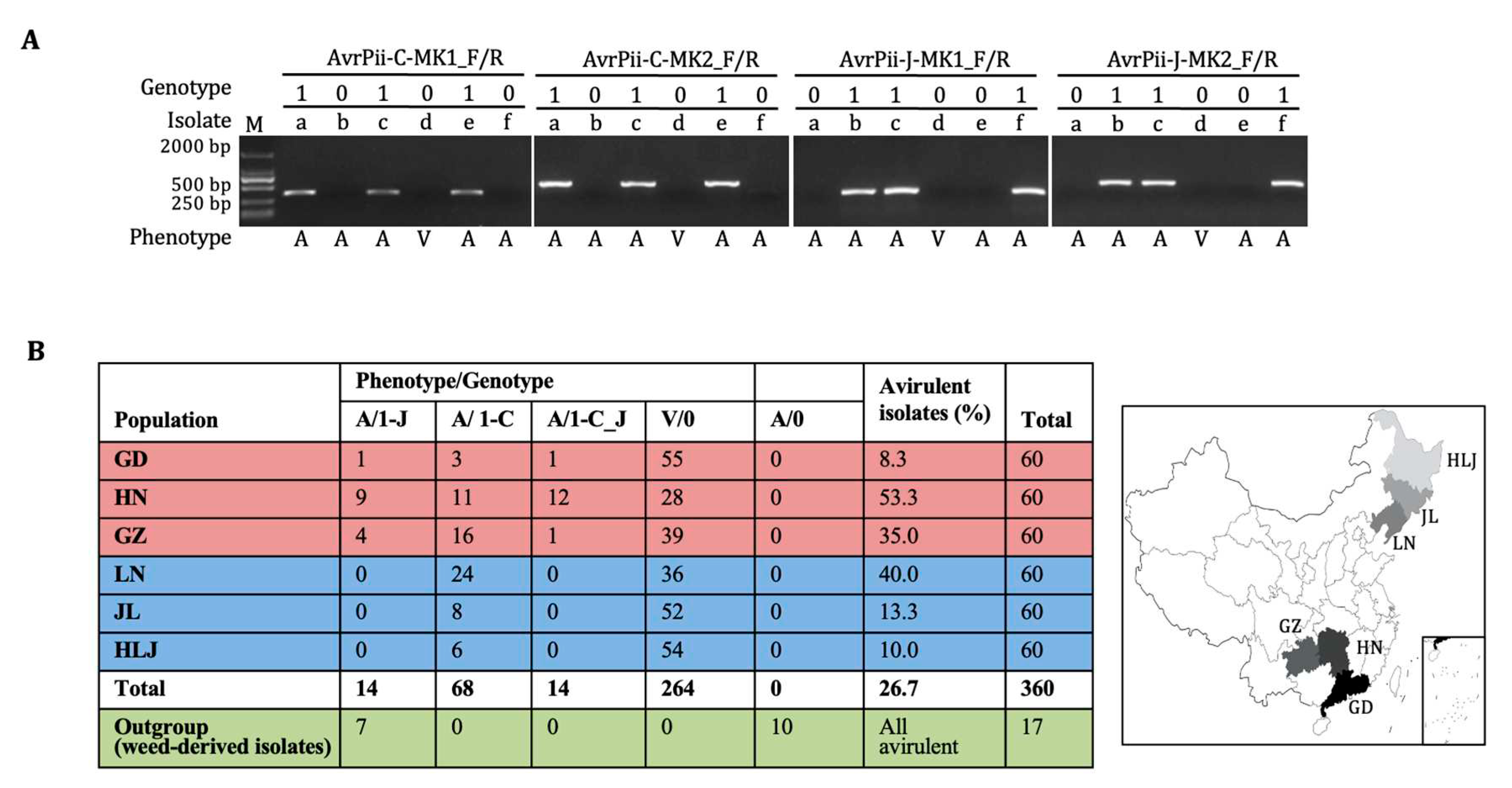
As for
Pii-based phenotyping, frequencies of avirulent isolates ranged from 8.3% (GD) to 53.3% (HN), indicating that the
AvrPii family was less avirulent and diverse in both southern and northern regions in China (
Figure 3;
Table S2). As for the two haplotype-specific marker-based genotyping, the results derived from the shorter fragment markers (MK1) were completely repeated by that from the longer one (MK2), thereby simply giving only four genotypes, 0, 1-C, 1-J, and 1-C_J, among six
Mo populations derived from rice (
Figure 1A; 3A;
Table S2). Intriguingly, combinations of phenotypes/genotypes were also only four, V/0, A/1-C, A/1-J, and A/1-C_J, because all the
Pii-virulent isolates restricted to genotype 0, whereas the
Pii-avirulent ones with the rest three. Furthermore, all the four combinations were detected in the three southern populations, and only two A/1-C and V/0 in all the northern ones, suggesting that genic diversity in southern region was higher than those in the northern one (
Figure 3B). Together, the
AvrPii family was rarer and less diverse in all the six
Mo populations, particularly in the northern region.
Regarding to outgroup, all 17 weed-derived isolates could not infect rice, and only seven were identified as A/1-J, indicating that the
AvrPii-J, which emerged before rice domestication, was older than
AvrPii-C (
Figure 3B;
Table S2).
3.4. Evolutionary and regional divergence of the AvrPii family
The genetic diversity of the
AvrPii family among the six populations was explored in some detail by re-sequencing all 110 amplicons derived from 96 isolates, of which 14 carrying two copies of both haplotypes (
Figure 3B;
Table S2). The CDS harbored 20 SNPs, of which eleven were non-synonymous (data not shown). The non-signal domain harbored 18 of the SNP sites, while the other two were within the signal peptide domain; as a result, the level of nucleotide diversity in the former domain (0.045) was higher than in the latter (0.013) (
Table 1). As to the individual haplotypes, only one segregating site was found in
AvrPii-C group and none in
AvrPii-J one, indicating that there were extremely less diverse in both haplotypes. Regarding the regional groups, both haplotypes were identified in the southern group, thereby raising 20 segregating sites for reaching a higher level (0.045) of the
AvrPii diversity. On the contrary, only one haplotype,
AvrPii-C, was placed in the northern one without any segregating site. Again, it was clearly suggested that diversity of the
AvrPii in the southern group was largely higher than that in the northern one.
In all
AvrPii group, the first set of selection-related parameters
D* and
F* in both entire (
D*=1.74;
F*=2.65) and non-signal peptide region (
D*=1.68;
F*=2.59) were positive and higher than those in signal peptide region (
D*=0.68;
F*=1.11), indicating that the target locus has been experienced a stronger balancing selection in both regions and slighter one in signal peptide region (
Table 1). In the haplotype groups, only slight balancing selection pressure (
D*=0.51;
F*=0.79) was detected within the haplotype
AvrPii-C group, and none in haplotype
AvrPii-J one. As to the regional group, there has been a stronger balancing selection pressure (
D*=1.72;
F*=3.04) on the
AvrPii family in the southern group, but none in the northern one. As to the second set of selection-related parameters (
Ka,
Ks, and
Ka/
Ks),
Ka/
Ks in both all
AvrPii and the southern groups were less than 0.57, indicating that the
AvrPii family has been imposed by a stronger purifying selection (negative selection) in both groups. While a slight positive selection (
Ka>
Ks) was detected within the haplotype
AvrPii-C group. Intriguingly, neither positive nor purifying selection pressures was detected in haplotype
AvrPii-J, the regional north, and out groups (
Table 1).
4. DISCUSSION
4.1. The lesson learned from case study on population structures of the AvrPii family
Having identified the novel haplotype,
AvrPii-C, dynamic population structures of the
AvrPii family were finely dissected among six
Mo populations, each three from southern and northern regions of China (
Figure 1-3). Whereas the large numerical gaps between higher rates of
Pii-avirulent isolates and/or lower rates of presence of
AvrPii were detected in various
Mo populations previously (3,11,14-15). Comparing to lower rate of presence of
AvrPii being detected, the higher rate of
Pii-avirulent isolates was partly due to
Pii independent that was conveyed by another gene pair of
Avr/
R gene in such combinations (3). It was, however, certainly conceivable that failing in detecting the novel haplotype
AvrPii-C in such
Pii-avirulent isolates as reported in the current study, was the major reason for creating the numerical gaps and/or the lower rates of presence of
AvrPii. When revisiting all PCR primer sequences used in the previous studies (3,11,14-15), it was revealed that all such primers were designed from the known reference sequence of the
AvrPii-J (AB498874 that resembles to CHL22 as shown in
Figure S1, 2), those were less matching enough for amplifying the
AvrPii-C as shown in
Figure 1. Because proportion of the
AvrPii-C was almost five times higher than that of the
AvrPii-J in the six
Mo populations tested in the present research (
Figure 3), in addition to the fact that higher resistance frequencies of the
Pii carrier, IRBLi-F5 were detected in Japan (27), Bangladesh (28), Kenya (29), and Vietnam (30), it was conceivable that the numerical gaps and/or extremely lower proportions of presence of
AvrPii in various populations were largely due to missing the novel haplotype,
AvrPii-C (3,11,14-15).
Considering haplotype divergence is one of the most important mechanisms underpinning
Avr gene evolution in various plant pathosystems (3,7,15,31-38), the lesion learned from case studies on population structures of the
AvrPii family is that much attention should be focused on haplotype divergence of target gene when there were certain numerical gaps, or extremely lower proportions of presence of the target gene as reported in the previous investigations (3,11,14-15). Briefly, the population structure of individual target gene(s) is better to dissect by both phenotypic and genotypic datasets so that the reasons for the numerical gap and/or extremely lower proportion of target gene could be easily found. If done, the specific sequence for unknown haplotype, which is distinct from the known one, should be created via the Tail-PCR system in a case of less enough reference sequences available, and/or through gene similarity analysis using reference sequence of the known one for inquiring whether there was/were candidate distinct haplotype(s) in public database (as shown in
Figure S1-2). Then primer sets for the novel haplotype, if any, should be re-devised based the haplotype-specific sequence for genotyping and resequencing (as shown in
Figure 1, 3). Furthermore, the novel haplotype(s) might be functional validated via transformation test, if possible and necessary.
4.2. The lesson learned from evolutionary story of the AvrPii family
Accumulating evidence from population genetic study on various
Avr genes demonstrated that the gene-for-gene principal has played a pivotal role in featuring their population structures (3,7-9,11,34,39-40). As per the definitions of three kinds of the key selections, stronger balancing and purifying selection forces exerted to the Chinese populations, and only positive selection pressure to the Thai population, when datasets available in both Lu et al. 2019 (3) and Sirisathaworn et al. 2017 (11) were re-assayed by the same approaches employed in the current study (
Table S4). As pathogen
Avr genes have generally evolved in response to positive selection (7-9,11,39-41), the unique population structures of the
AvrPii family found in China have significant implications for understanding how the
AvrPii family has kept an artful balance and purify among its members (haplotypes) those keenly interact with the cognate
R gene,
Pii, under gene-for-gene relationships.
Furthermore, several intriguing points, which were extracted from five comparable groups in the current study, should be noteworthy. First, it could be clearly deemed that the haplotype
AvrPii-C was a mutant being emerged after rice domestication, and the haplotype
AvrPii-J as wild type being emerged before rice domestication (
Figure 3;
Table S2). Second, the population structures between haplotypes
AvrPii-C and
AvrPii-J groups were slightly different, the former one being received weaker balancing and positive selection forces thereby scattering in all the six
Mo populations across China, and the latter one being received almost no selection pressure thereby being restricted in the southern region (
Table 1;
Figure 3). Third, the population structures between southern and northern groups were distinct. The former one has experienced both stronger balancing and purifying selection pressure thereby keeping all the four combinations of phenotypes/genotypes (A/1-J, A/1-C, A/1-C_J, and V/0), and the latter one has received almost no selection pressure thereby being restricted only two (A/1-C and V/0), which led to raising certainly higher genetic diversity in the southern region. Fourth, higher frequencies of avirulent isolates were detected in southern HN (53.3%) and GZ (35.0%), and in northern LN (40.0%), but over three-fold lower ones in their neighboring regions, i.e., the southern GD (8.3%), and the northern JL (13.3 %) and HLJ (10.0%) (
Figure 3). That, in turn, indicated that the cognate
R gene,
Pii, could be continuously served as basic and critical resistance resource in HN, GZ, and LN populations, even though it was recognized as one of the extremely rarer and weaker
R genes in the Chinese rice breeding programs (2,5,12,42-44). Again, it was providing further weight to the idea that the
AvrPii family was a smarter one to keep a sophisticated balance and purify not only its members but also its living places.
It is worthy learning more marvelous lessons from evolution stories of the AvrPii family featured by its balanced, purified, and directed haplotypes in global Mo populations including trans-species ones.
5. Conclusions
The study has identified a sequence-distinct and functional haplotype AvrPii-C, compared to the known one, AvrPii-J. It was believed that failing in amplifying of AvrPii-C was the major reason for causing the numerical gaps between higher proportions of the Pii-avirulent isolates and/or extremely lower proportions of presence of AvrPii in various populations previously reported. The unique population structure of the AvrPii family was shaped by its balanced, purified, and directed haplotypes in the Chinese populations. The AvrPii-C was recognized as a younger one that emerged after rice domestication. The cognate resistance gene Pii could be continuously served as a basic and critical resistance resource in Hunan, Guizhou, and Liaoning provinces, China.
6. Patent
Qinghua Pan, Xing Wang, Cheng Li, Jianqiang Wen, Yu Zhang, Ling Wang. Identification and applications of the avirulence gene AvrPii-C of Magnaporthe oryzae (ZL201811273480.3, Granted on August 14, 2020)
Supplementary Materials
Figure S1: Discovery of a novel haplotype AvrPii-C in Chinese populations of M. oryzae; Figure S2: Alignment of the full-length sequences of both haplotypes, AvrPii-J and AvrPii-C; Figure S3: Constructure of haplotype-chimeric mutants by PCR-based domain swapping from AvrPii-J to AvrPii-C; Figure S4: The 3D-structures of a set of five AvrPii proteins; Figure S5: Interactions between AvrPii and Pii proteins in the Y2H system; Table S1. PCR primers and programs used in the current study; Table S2. Genotypes and phenotypes of 377 isolates of M. oryzae characterized in the current study; Table S3. The full-length sequences of seven haplotype-chimeric mutants of the AvrPii family; Table S4. Comparison of evolutionary parameters of the AvrPii family among three Mo populations, of which parameters of the two populations previously reported were re-computed by the same approaches used in the current study.
Author Contributions
Conceptualization, Q.P.; methodology, X.W., W.W., J.W. (Jinyan Wang), J.W. (Jianqiang Wen), Y.Z., C.L., S.Z.; investigation, X.W., W.W., J.W. (Jinyan Wang), J.W. (Jianqiang Wen), C.L., S.Z., Y.Z., Y.Y., W.L., Z.Z.; formal analysis, X.W., W.W., J.W. (Jinyan Wang), J.W. (Jianqiang Wen); validation, X.W., J.W. (Jinyan Wang), W.W., J.Z., Q.P.; resource, Y.Z., Y.Y., W.L., Z.Z.; data curation, X.W., J.W. (Jinyan Wang); funding acquisition, W.L., Q.P.; project administration, W.L.; writing-original draft, X.W., W.W., Y.Z., Q.P.; writing-review & editing, J.Z., Q.P. All authors have read and agreed to the published version of the manuscript.
Funding
The project was supported by the Key Areas Research and Development Programs of Guangdong Province, China (2022B0202060005), the National Natural Science Foundation (31870137), and the Ministry of Science and Technology, China (2015ZX08001-002, 2016YDF0100601).
Data Availability Statement
Not applicable.
Acknowledgments
We are thankful to J. Yuan (Guizhou Academy of Agricultural Sciences), Z. Liu (Shenyang Agricultural University) for providing the rice blast samples. We also thank Mr. Jasper Pan (Rutgers University, USA) for critical reading of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Cheng, S. Retrospect and prospect of rice breeding in China. Pages 1-23 in: Rice Breeding and Cultivar Genealogies in China during 1986 to 2005. J. Wan, ed. China Agr. Press, Beijing, 2010.
- Huang, Z.; Wang, J.; Zhang, Y.; Yao, Y.; Huang, L.; Yang, X.; Wang, L.; Pan, Q.; Yao, Y.; Huang, L.; Yang, X.; Wang, L.; Pan, Q. Dynamics of race structures of Pyricularia oryzae populations across 18 seasons in Guangdong province, China. Plant Dis. 2021, 105, 144–148. [Google Scholar] [CrossRef]
- Lu, L.; Wang, Q.; Jia, Y.; Bi, Y.; Li, C.; Fan, H.; Li, J. Selection and mutation of the avirulence gene AvrPii of the rice blast fungus Magnaporthe oryzae. Plant Pathol. 2019, 68, 127–134. [Google Scholar] [CrossRef]
- Peng, Z.; Li, L.; Wu, S.; Chen, X.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Sun, P.; Deng, H.; Xing, J. Frequencies and variation of Magnaporthe oryzae avirulence genes in Hunan province, China. Plant Dis. 2021, 105, 3829–3834. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, J.; Yao, Y.; Jin, X.; Correll, J.; Wang, L.; Pan, Q. Dynamics of race structures of the rice blast pathogen populations in Heilongjiang province, China from 2006 through 2015. Plant Dis. 2019, 103, 2759–2763. [Google Scholar] [CrossRef] [PubMed]
- Chai, R.; Wang, J.; Wang, X.; Wen, J.; Liang, Z.; Ye, X.; Zhang, Y.; Yao, Y.; Zhang, J.; Zhang, Y.; et al. The Pid family has been diverged into Xian and Geng type resistance genes against rice blast disease. Genes 2022, 13, 891. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, L.; Zhang, S.; Li, Z.; Zhang, Y.; Lin, F.; Pan, Q. Stepwise arms race between AvrPik and Pik alleles in the rice blast pathosystem. Mol. Plant-Microbe Interac. 2014, 27, 759–769. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, L.; Wu, W.; He, L.; Yang, X.; Pan, Q. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci. Rep. 2015, 5, 11642. [Google Scholar] [CrossRef]
- Chen, C.; Chen, M.; Hu, J.; Zhang, W.; Zhong, Z.; Jia, Y.; Allaux, L.; Fournier, E.; Tharreau, D.; Wang, G.; et al. Sequence variation and recognition specificity of the avriulence gene AvrPiz-t in Magnaporthe oryzae field populations. Fungal Genom. Biol. 2014, 4, 1. [Google Scholar]
- Flor, H. Current status of the gene for gene concept. Ann. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Sirisathaworn, T.; Srirat, T.; Longya, A.; Jantasuriyarat, C. Evaluation of mating type distribution and genetic diversity of three Magnaporthe oryzae avirulence genes, PWL2, AvrPii and AvrPiz-t, in Thailand rice blast isolates. Agriculture and Natural Resources 2017, 51, 7–14. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Q.; Yao, Y.; Zhao, Z.; Correll, J.; Wang, L.; Pan, Q. The race structure of the rice blast pathogen across southern and northeastern China. Rice 2017, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Siatoh, H.; Fujisawa, S.; Kanzaki, H.; Matsumura, H.; Yoshida, K.; Tosa, Y.; Chuma, I.; Takano, Y.; Win, J.; et al. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae. Plant Cell 2009, 21, 1573–1591. [Google Scholar] [CrossRef] [PubMed]
- Xing, J.; Jia, Y.; Peng, Z.; Shi, Y.; He, Q.; Shu, F.; Zhang, W.; Zhang, Z.; Deng, H. Characterization of molecular identity and pathogenicity of rice blast fungus in Hunan province of China. Plant Dis. 2017, 101, 557–561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jia, Y.; Wang, Y.; Sun, G. A rapid survey of avirulence genes in field isolates of Magnaporthe oryzae. Plant Dis. 2020, 104, 717–723. [Google Scholar] [CrossRef] [PubMed]
- Tsunematsu, H.; Yanoria, M.; Ebron, L.; Hayashi, N.; Ando, I.; Kata, H.; Imbe, T.; Khush, G. Development of monogenic lines of rice for rice blast resistance. Breed. Sci. 2000, 50, 229–234. [Google Scholar] [CrossRef]
- Pan, Q.; Wang, L.; Ikehashi, H.; Tanisaka, T. Identification of a new blast resistance gene in the Xian rice cultivar Kasalath using Japanese differential cultivars and isozyme markers. Phytopathology 1996, 86, 1071–1075. [Google Scholar] [CrossRef]
- Zhai, C.; Zhang, Y.; Yao, N.; Lin, F.; Liu, Z.; Dong, Z.; Wang, L.; Pan, Q. Function and interaction of the coupled genes responsible for Pik-h encoded rice blast resistance. Plos One 2014, 9, e98067. [Google Scholar] [CrossRef]
- Lynch, M.; Crease, T. The analysis of population survey data on DNA sequence variation. Mol. Biol. Evol. 1990, 7, 377–394. [Google Scholar]
- Nei, M. 1987. Molecular Evolutionary Genetics. Columbia Univ. Press, New York.
- Fu, Y.; Li, W. Statistical tests of neutrality of mutations. Genetics 1993, 133, 693–709. [Google Scholar] [CrossRef]
- Nei, M.; Gojobori, T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 1986, 3, 418–426. [Google Scholar]
- Han, G. Origin and evolution of the plant immune system. New Phytologist 2019, 222, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Hurst, L. Genetics and the understanding of selection. Nature Rev. Genet. 2009, 10, 83–93. [Google Scholar] [CrossRef] [PubMed]
- de Vries, S.; Stukenbrock, E.; Rose, L. Rapid evolution in plant-microbe interactions-and evolutionary genomics perspective. New Phytologist 2020, 226, 1256–1262. [Google Scholar] [CrossRef]
- Fujisaki, K.; Abe, Y.; Ito, A.; Saitoh, H.; Yoshida, K.; Kanzaki, H.; Kanzaki, E.; Utsushi, H.; Yamashita, T.; Kamoun, S.; et al. Rice Exo70 interacts with a fungal effector, AvrPii, and is required for AvrPii-triggered immunity. Plant J. 2015, 83, 875–887. [Google Scholar] [CrossRef]
- Kawasaki-Tanaka, A.; Hayashi, N.; Yanagihara, S.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Japan. Plant Dis. 2016, 100, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Ali, M.; Monsur, M.; Kawasaki-Tanaka, A.; Hayashi, N.; Yanagihara, S.; Obara, M.; Mia, M.; Latif, M.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Bangladesh. Plant Dis. 2016, 100, 2025–2033. [Google Scholar] [CrossRef]
- Fukuta, Y.; Telebanco-Yanoria, M.; Hayashi, N.; Yanagihara, S.; Machungo, C.; Makihara, D. Pathogenicities of rice blast (Pyricularia oryzae Cavara) isolates from Kenya. Plant Dis. 2019, 103, 3188. [Google Scholar] [CrossRef]
- Nguyet, N.; Long, H.; Ngoc, N.; Nhai, N.; Thuy, N.; Hayashi, N.; Fukuta, Y. Diversity and distribution of rice blast (Pyricularia oryzae Cavara) races in Vietnam. Plant Dis. 2020, 104, 381–387. [Google Scholar] [CrossRef]
- Chuma, I.; Isobe, C.; Hotta, Y.; Ibaragi, K.; Futamata, N.; Kusaba, M.; Yoshida, K.; Terauchi, R.; Fujita, Y.; Nakayashiki, H. Multiple translocations of the AVR-Pita effector gene among chromosomes of the rice blast fungus Magnaporthe oryzae and related species. PLoS Pathog. 2011, 7, e1002147. [Google Scholar] [CrossRef]
- Dai, Y.; Jia, Y.; Correll, J.; Wang, X.; Wang, Y. Diversification and evolution of the avirulence gene AVR-Pita1 in field isolates of Magnaporthe oryzae. Fungal Genet. Biol. 2010, 47, 973–980. [Google Scholar] [CrossRef]
- Khang, C.; Park, S.; Lee, Y.; Valent, B.; Kang, S. Genome organization and evolution of the AVR-Pita avirulence gene family in the Magnaporthe grisea species complex. Mol. Plant-Microbe Interac. 2008, 21, 658–670. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Q.; Li, C.; Bi, Y.; Fu, X.; Wang, R. Novel haplotypes and networks of AvrPik alleles in Magnaporthe oryzae. BMC Plant Biol. 2019, 19, 204. [Google Scholar] [CrossRef] [PubMed]
- Longya, A.; Chaipanya, C.; Franceschetti, M.; Maidment, J.; Banfiled, M.; Jantasuriyarat, C. Gene duplication and mutation in the emergence of a novel aggressive allele of the AvrPik effector in the rice blast fungus. Mol. Plant-Microbe Interac. 2019, 32, 740–749. [Google Scholar] [CrossRef] [PubMed]
- Praz, C.; Bourras, S.; Zeng, F.; Sanchez-Martin, J.; Menardo, F.; Xue, M.; Yang, L.; Roffler, S.; Boni, R.; Herren, G.; et al. AvrPm2 encodes an RNase-like avirulence effector which is conserved in the two different specialized forms of wheat and rye powdery mildew fungus. New Phytologist 2017, 213, 1301–1314. [Google Scholar] [CrossRef]
- Takahashi, M.; Ashizawa, T.; Hirayae, K.; Moriwaki, J.; Sone, T.; Sonoda, R.; Noguchi, M.; Nagashima, S.; Ishikawa, K.; Arai, M. One of two paralogs of AvrPita1 is functional in Japanese rice blast isolates. Phytopathology 2010, 100, 612–618. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Liu, H.; Duan, G.; Huang, Y.; Liu, S.; Fang, Z.; Wu, E.; Shang, L.; Zhan, J. The Phytophthora infestans Avr2 effector escapes R2 recognition through effector discording. Mol. Plant-Microbe Interac. 2020, 33, 921–931. [Google Scholar] [CrossRef]
- Dodds, P.; Thrall, P. Recognition events and host-pathogen co-evolution in gene-for-gene resistance to flax rust. Funct. Plant Biol. 2009, 36, 395–408. [Google Scholar] [CrossRef]
- Schürch, S.; Linde, C.; Knogge, W.; Jackson, L.; McDonald, B. Molecular population genetic analysis differentiates two virulence mechanisms of the fungal avirulence gene NIP1. Mol. Plant-Microbe Interac. 2004, 17, 1114–1125. [Google Scholar] [CrossRef]
- Daugherty, M.; Malik, H. Rules of engagement: Molecular insights from host-virus arms races. Annu. Rev. Genet. 2012, 46, 677–700. [Google Scholar] [CrossRef]
- Lin, L. Identification of rice blast resistance genes in current cultivars in Liaoning and Heilongjiang provinces, China. Master Dissertation, South China Agri University, Guangzhou, China, June 2021. (In Chinese with English Abstract). [Google Scholar]
- Ye, X. Identifying the composition of rice blast resistance genes and mining new allelic genes of backbone rice cultivars in Guangdong province and Guangxi Zhuang autonomous region. Master Dissertation, South China Agri University, Guangzhou, China, June 2021. (In Chinese with English Abstract). [Google Scholar]
- Zeng, S. Identification of rice blast resistance genes in the key parental cultivars. Master Dissertation, South China Agri University, Guangzhou, China, June 2020. (In Chinese with English Abstract). [Google Scholar]
- China National Rice Research Institute. Regionalization of Rice Cropping in China. Zhejiang Sci. Tech. Press, Hangzhou, China, 1988 (In Chinese).
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).