1. Introduction
The history of the SARS-CoV-2 pandemic has been associated with an unprecedented level of global genomic surveillance. This surveillance has been fundamental for the identification and monitoring of virus variants as they have continued to emerge. In November 2021, WHO designated Omicron (B.1.1.529) a variant of concern [
1]. Omicron became the dominant SARS-CoV-2 variant globally due to increased transmissibility and the ability to evade both natural and vaccine-induced immunity [
2,
3]. Omicron has distinct sub-lineages, one of which is BA.2. Sub-lineage BA.2 has continued to evolve, and its phylogenetic “offspring” include the sub-lineage BA.4 and BA.5.2.1 which were first described in South Africa in early 2022. The sub-lineages BA.4 and BA.5.2.1 have a spike protein sequence comparable to BA.2 but have additional mutations resulting in several amino acid changes; a deletion 69-70, and substitutions L452R, F486V, and R493Q [
4,
5]. Sub-lineage BA.4 used in this study has an additional non-defining amino acid substitution in the spike (S640F). Other mutations result in substitutions in nucleoprotein (N:P151S) and Orf7b (L11F). BA.4 has a deletion not found in other sub-lineages in Orf1a resulting in the loss of three amino acids; Nsp1: Δ141-143 [
4]. Evidence suggests that existing COVID-19 ancestral spike-based vaccinations may be less efficacious at producing neutralizing antibodies to BA.4 and BA.5.2.1 [
6]. This study sought to investigate potential differences in pathogenicity of these sub-lineages.
2. Materials and Methods
Viruses. The GISAID ID or source of the isolation swabs/ viruses used in this study are as follows: Ancestral (EPI_ISL_406844, [
7]), BA.1 (EPI_ISL_7400555), BA.2 (not available), BA.2.12.1 (Gavin Screaton, University of Oxford), BA.4 (EPI_ISL_13157810), BA.5.2.1 (EPI_ISL_12810908), BQ.1.22 (EPI_ISL_15581064), XBB.1.1 (EPI_ISL_15682231). Viruses were isolated and propagated on Vero/hSLAM (ECACC 04091501, European Collection of Authenticated Cell Cultures (ECACC) UKHSA, Porton Down, UK) with quality control checks and whole genome sequencing performed as previously described [
8]. Sequence data of the virus banks used in this study are available in
Supplementary Data File S1.
Animals. Twenty-four healthy, Golden Syrian hamsters (Mesocricetus auratus), aged 7-12 weeks, were obtained from a UK Home Office accredited supplier (Envigo RMS UK). Animals were housed individually at Advisory Committee on Dangerous Pathogens (ACDP) containment level 3. Cages met with the UK Home Office Code of Practice for the Housing and Care of Animals Bred, Supplied or Used for Scientific Procedures (December 2014). Access to food and water was ad libitum and environmental enrichment was provided. All experimental work was conducted under the authority of a UK Home Office project license that had been subject to local ethical review at UKHSA Porton Down by the Animal Welfare and Ethical Review Body (AWERB) as required by the Home Office Animals (Scientific Procedures) Act 1986.
Study Design. Hamsters (n=6 per group, equal male: female ratio) were randomly assigned to groups to minimise bias using the RAND() function in Excel. A biothermal identifier chip (Plexx IPTT-300 temperature transponder) was inserted subcutaneously into each animal under sedation. Prior to infection, animals were sedated with isoflurane. Virus was delivered by intranasal instillation (200 μL total, 100 μL per nare) diluted in phosphate buffered saline (PBS). Groups 1 and 2 were infected with SARS-CoV-2 Omicron BA.4 for a target dose of 1.0E+04 (FFU). Groups 3 and 4 were infected identically, but with the sub-variant BA.5.2.1. Groups 1 and 3 were taken for necropsy 7 days after infection and groups 2 and 4 taken at day 28. Hamsters were throat swabbed at day -2, 2, 4, 6, 8, 10, 14 and 21 post infection, as well directly before necropsy (in-life). Blood samples were collected at baseline and necropsy to measure the humoral immune response.
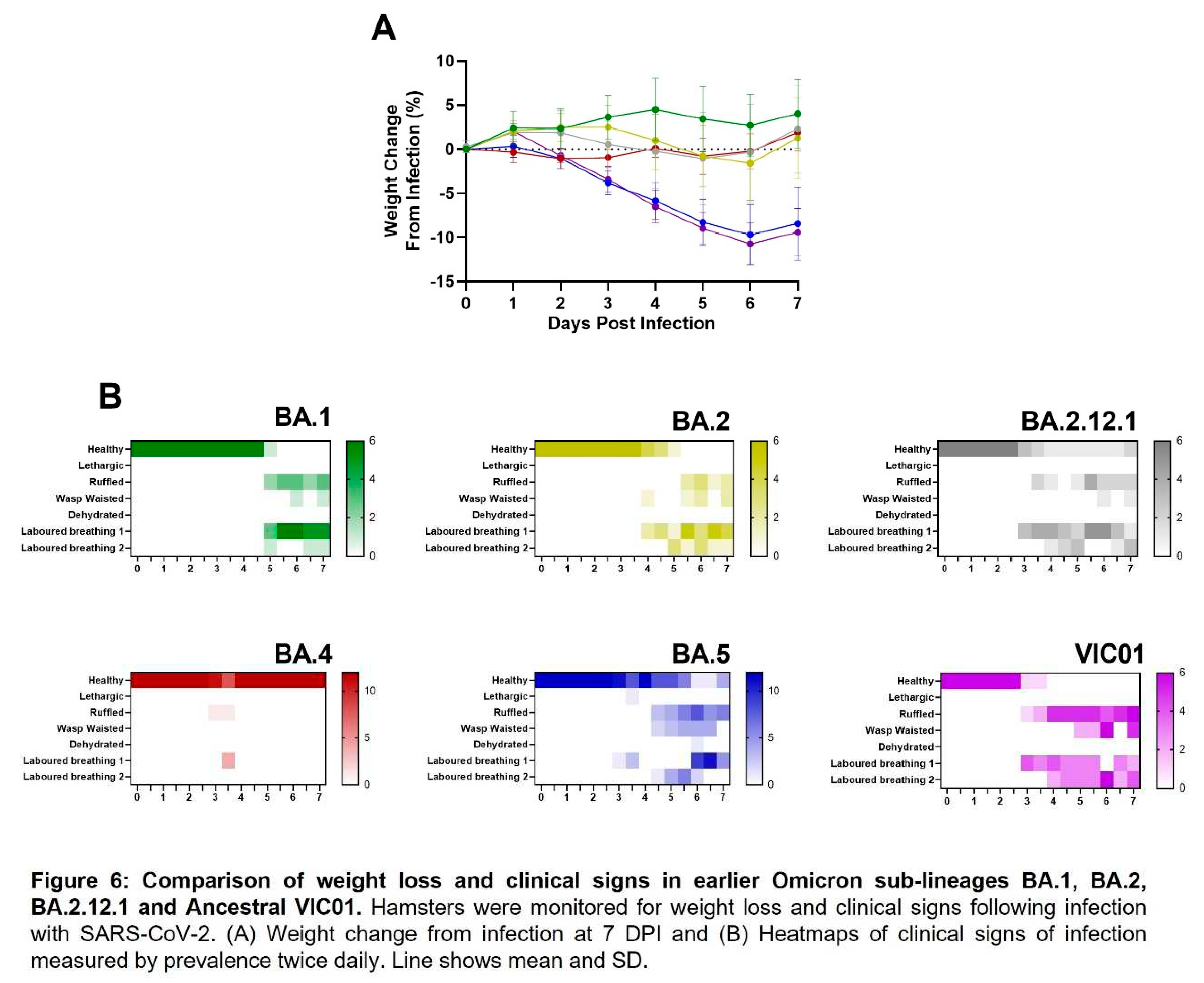
Clinical observations. Hamsters were monitored twice daily (approximately 8 h apart) to record temperature via biothermal Identichip and clinical signs of disease. Clinical signs of disease were assigned based upon the following criteria: healthy, lethargy, behavioural change, sunken eyes, ruffled, wasp waited, dehydrated, arched, coughing, laboured breathing 1 - occasional catch or skip in breathing rate and laboured breathing 2 - abdominal effort with breathing difficulties. The prevalence of each sign produced a weighted score at each time point. Clinical signs not observed have not been presented. Animals were weighed at the same time of each day until scheduled necropsy.
Necropsy Procedures. Hamsters were given an anaesthetic overdose (sodium pentabarbitone Dolelethal, Vetquinol UK Ltd, 140 mg/kg) via intraperitoneal injection and exsanguination was by cardiac puncture. A necropsy was performed immediately after confirmation of death.
RNA Extraction. Throat swabs were inactivated in AVL (Qiagen, UK) and ethanol, and RNA was isolated. Downstream extraction was performed using the BioSprint™96 One-For-All Vet kit (Indical, UK) and Kingfisher Flex platform as per manufacturer’s instructions.
Quantification of Viral RNA by RT-qPCR. Reverse transcription-quantitative polymerase chain reaction (RT-qPCR) targeting a region of the SARS-CoV-2 nucleocapsid (N) gene was performed as previously described [
9]. Positive swab and fluid samples detected below the limit of quantification (LLOQ) of 12,857 copies mL, were assigned the value of 5 copies/μL, this equates to 6,429 copies/mL, whilst undetected samples were assigned the value of < 2.3 copies/μL, equivalent to the assay’s lower limit of detection (LLOD) which equates to 2,957 copies/mL.
Focus forming assay (FFA). The viral titre of infection material and throat swabs (TS) were determined by focus forming assay on Vero/E6 cells [ECACC 85020206] as previously described [
9]. Cell lines were obtained from the European Collection of Authenticated Cell Cultures (ECACC) UKHSA, Porton Down, UK.
SARS-CoV-2 focus reduction neutralization test. Test sera were heat-inactivated at 56°C for 30 minutes to destroy any complement activity. Neutralization and staining were assessed as previously described [
10], with the following modifications for Omicron sub-lineages: Monolayers were incubated with overlay media containing 1% CMC (Sigma, UK) after virus/antibody removal for 22 (BA.5.2.1) or 26 hours (BA.4, BQ.1.22, XBB.1.1) at 37°C. After cells were fixed, and prior to the removal of residual endogenous peroxidase activity with 0.3% hydrogen peroxide (Sigma, UK), cell monolayers were permeabilized with 0.2% Triton-X-100/PBS. Cells were incubated for 1 hour with a primary rabbit polyclonal antibody against SARS-CoV-2 anti-nucleocapsid, diluted 1:1000 (Sinobiological, 40588-T62) before resumption of the previously described protocol [
10]. Both primary and secondary antibody were diluted in 0.2% Triton-X-100/PBS.
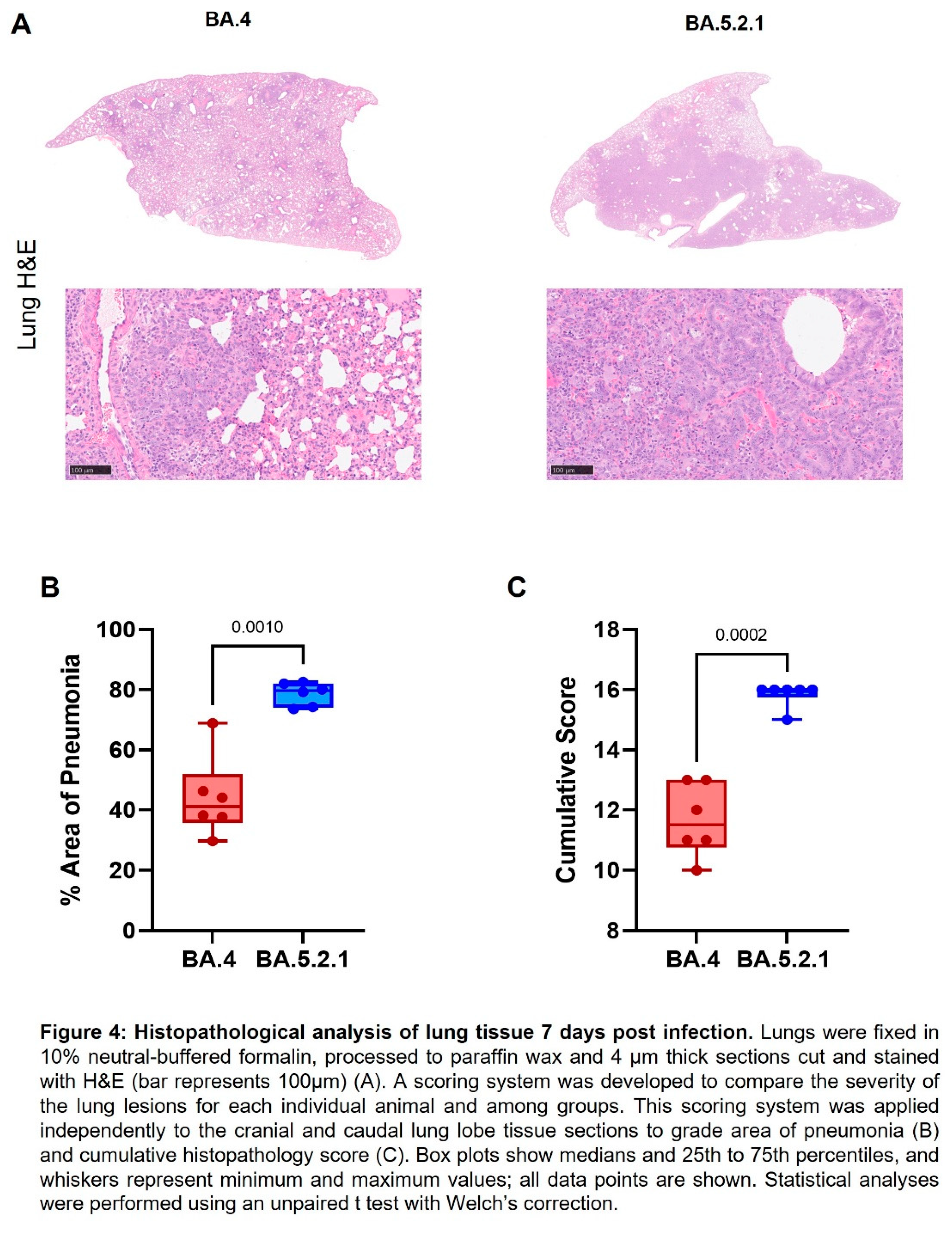
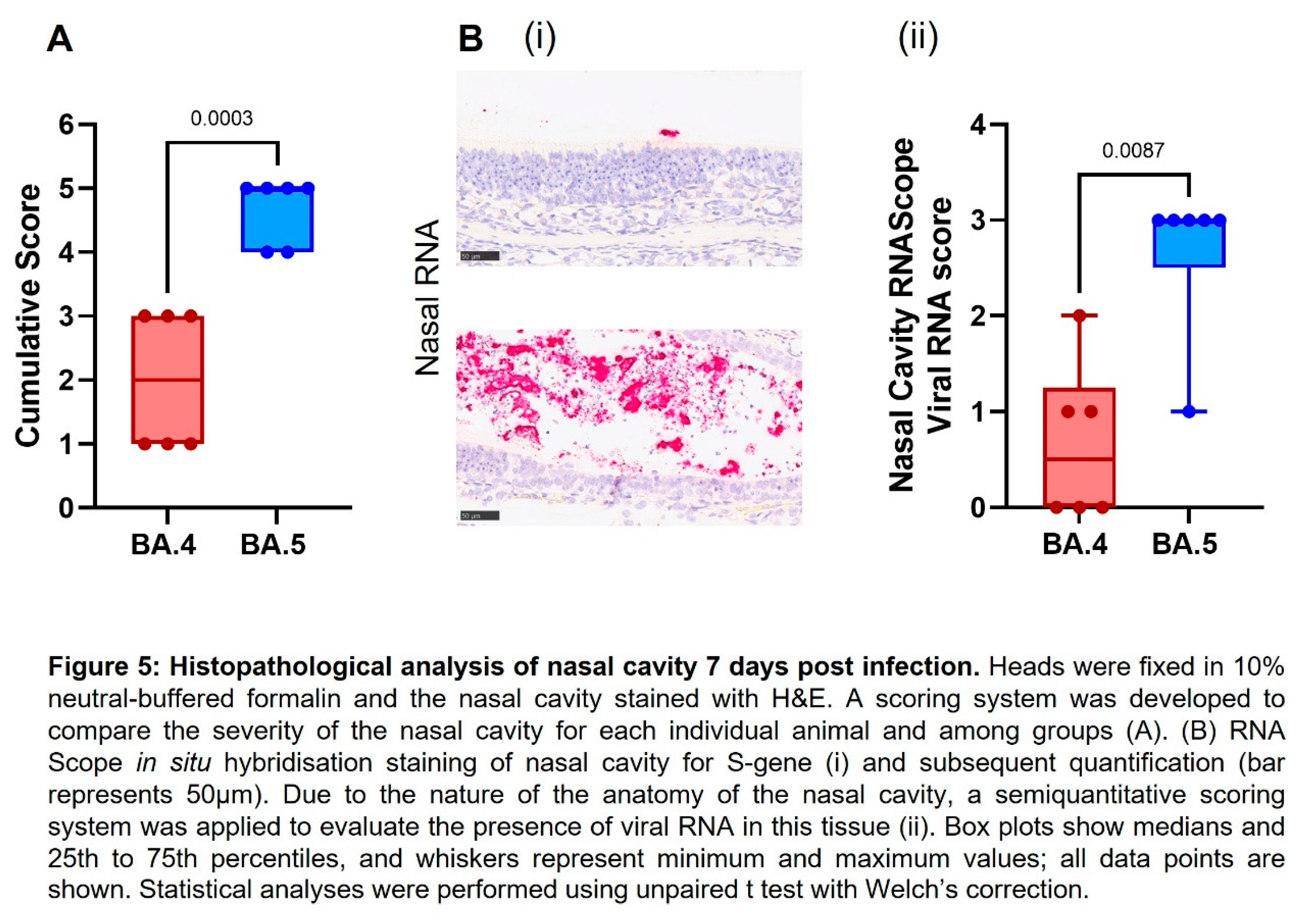
Histopathology. The following samples from each animal were fixed in 10% neutral-buffered formalin, processed to paraffin wax and 4 µm thick sections cut and stained with hematoxylin and eosin (H&E); nasal cavity and respiratory tract (left lung lobe). Tissue sections were scanned using a Hamamatsu S360 scanner and viewed by ndp.view2 software (Hamamatsu, U12388-01). Tissue sections were evaluated subjectively by a veterinary pathologist blinded to treatment and group details and the slides were randomized prior to examination to prevent bias. A scoring system previously reported by our group was applied independently to the lung tissue sections and nasal cavity [
11]. The severity of pulmonary microscopic lesion was also evaluated using digital image analysis (Nikon-NIS Br) to calculate the percentage area of pneumonia in H&E-stained lung tissue sections.
Statistics. Normal distribution of the numeric data was evaluated, and appropriate parametric or non-parametric statistical tests applied. Weight percentage change data was analysed by Tukey-corrected pairwise multiple comparison ANOVA. Parametric statistical analyses were selected as the data were expected to conform to a log-normal distribution (for qPCR results) or a normal distribution (for weights and antibody titres) based on historical observations of data from similar hamster challenge studies. Histopathological results were analysed by Mann-Whitney’s U test. Paired t-tests were used to analyse variant differences in convalescent hamster sera.
3. Results
Study design. Hamsters (n=12 per group, with equal numbers of males and females) were infected intranasally with either BA.4 or BA.5.2.1 to achieve a target dose of 1.0E+04 focus forming units (FFU) in a total volume of 200 μL (100 μL per nare) per hamster. Inocula were back titrated by focus forming assay (FFA) [
9] on the day of infection to confirm doses. Throat swabs (TS) were taken at -2, 2, 4, 6, 8, 10, 14, 21 and 28-days post infection (DPI). Six hamsters infected with BA.4 and six infected with BA.5.2.1 were culled on day seven post infection. Remaining hamsters were culled at day 28 post infection. Viral loads in the lung were determined at day 7 and day 28 post infection. Pathology was assessed in the nasal turbinates and lung. Sera was taken at baseline and cull (either 7- or 28-days post infection) for analysis of humoral response.
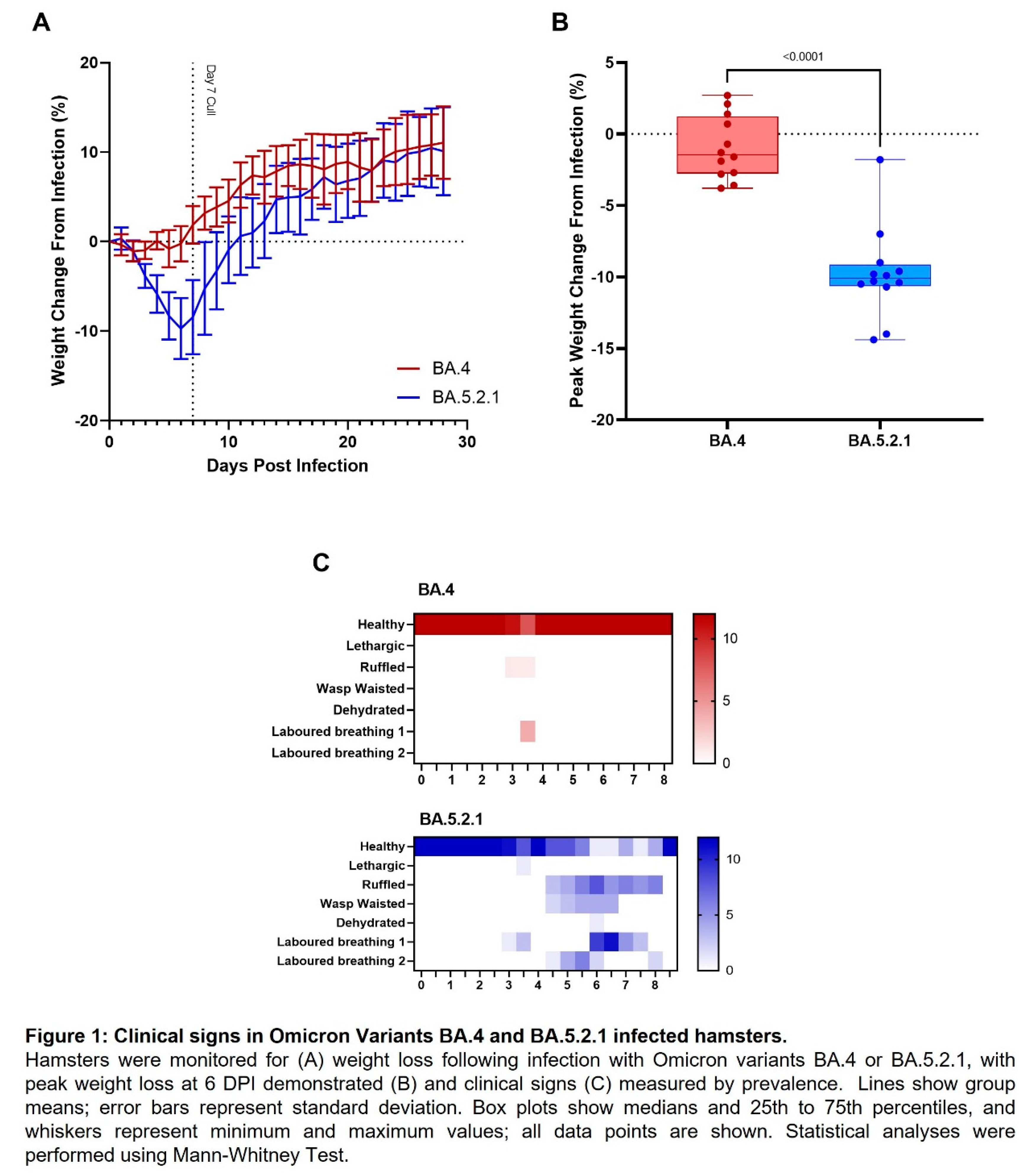
BA.5 infected animals exhibit weight loss. In our Golden Syrian Hamster model of SARS-CoV-2 infection [
12] and others [
13], weight loss is typically one of the first clinical signs of infection observed (from 1 – 2 days post infection). Hamsters were weighed daily, and a percentage weight change calculated from the day of infection. Animals that were infected with BA.4 ceased to gain weight in the first few days following infection (Figure 1A, red line), however, from the first day following infection onwards, the animals infected with BA.5.2.1 demonstrated prolonged weight loss until 7 days post infection (Figure 1A, blue line). Peak weight loss (seen at 6 DPI) was significantly greater (p<0.0001) in the BA.5.2.1 infected hamsters when compared with that seen in the BA.4 infected animals (Figure 1B). As other clinical signs of infection subsided, BA.5.2.1 infected animals began to regain weight and were back to their pre-infection weights by 11 days post infection. From that timepoint, their weights were comparable to the weights observed in the BA.4 infected hamsters.
BA.5 infected animals exhibit increased clinical signs. As required by our UK Home Office project license, clinical signs of infection were monitored and recorded twice a day following challenge. Except for 3 days post infection, few clinical signs were seen in BA.4 infected hamsters; laboured breathing 1 was observed in 4/12 and ruffled fur in 1/12 at day 3 post-infection at one monitoring point only (Figure 1C, red heatmap). In contrast, multiple clinical signs were seen in all hamsters infected with BA.5.2.1 over several, sequential days (Figure 1C, blue heatmap); these were most frequently; ruffled fur, wasp waisted appearance and laboured breathing (where a score of 1 is ‘evident’ and 2 is ‘pronounced’).
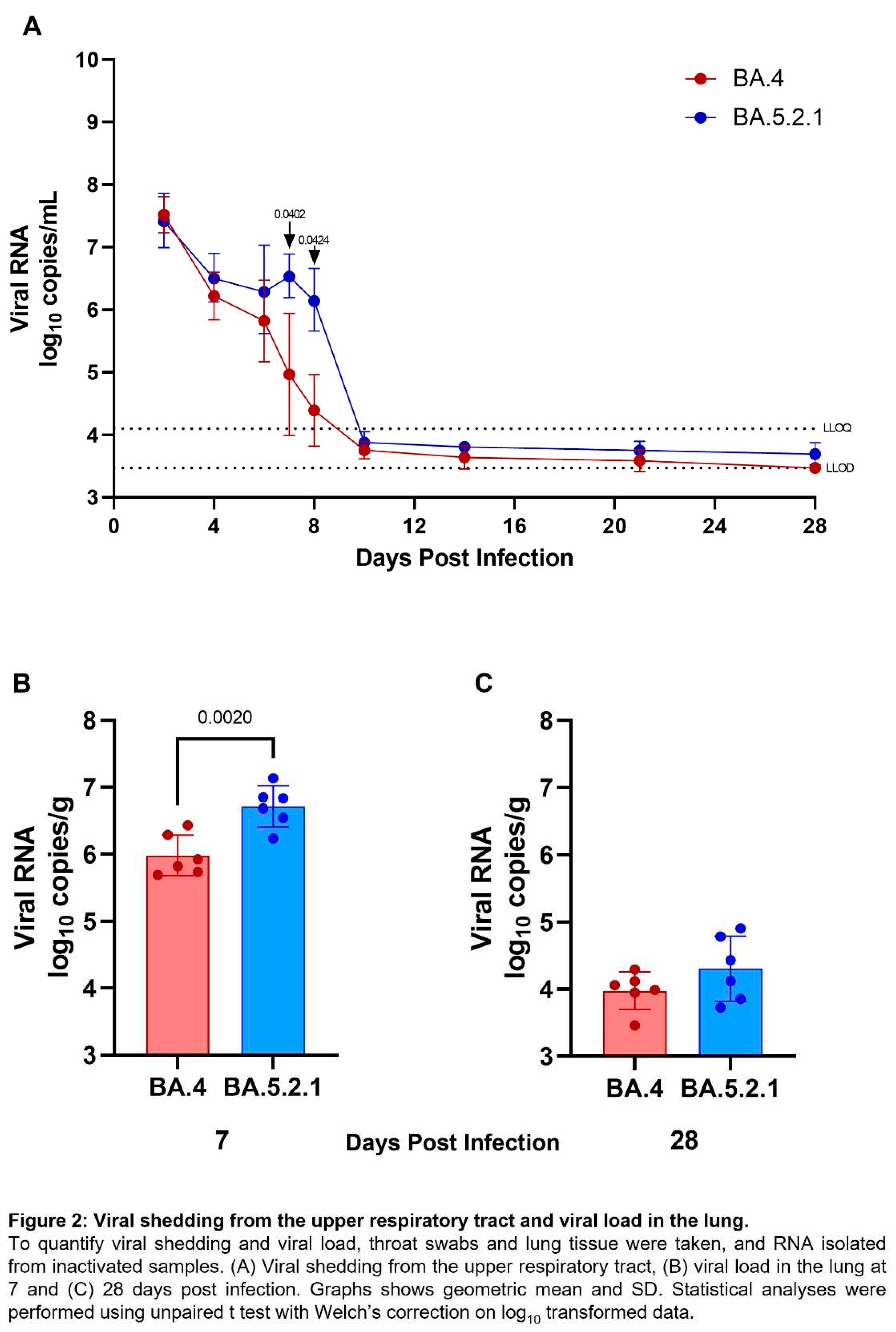
Viral shedding and lung viral loads are reduced in BA.4 infected hamsters. Viral shedding was determined by RT-qPCR targeting a region of the SARS-CoV-2 nucleocapsid (N) gene. At 2- and 4-days post infection, similar levels of viral shedding were seen in all hamsters, regardless of which Omicron virus was administered by the intranasal route (Figure 2A). Assessment of live virus load by FFA in throat swabs at 2 days post infection also confirmed no difference in live viral titre (
Figure S1). From 6 days post infection onwards, reduced viral shedding was observed in BA.4 infected hamsters. The difference in shedding from the upper respiratory tract from BA.4 and BA.5.2.1 infected hamsters was found to be significant at both 7- and 8-days post infection (p=0.0402 and 0.0424 respectively). From day 10 onwards levels of viral shedding fell below the lower levels of quantification and detection for all infected hamsters. At cull, in lung tissue from hamsters infected with BA.5.2.1 for 7 days, there was significantly more viral load compared to BA.4 infected hamsters (Figure 2B) (p=0.002); however, at cull 28 days post infection, there was no difference, and the viral load was observed to be below the lower limit of quantification (Figure 2C).
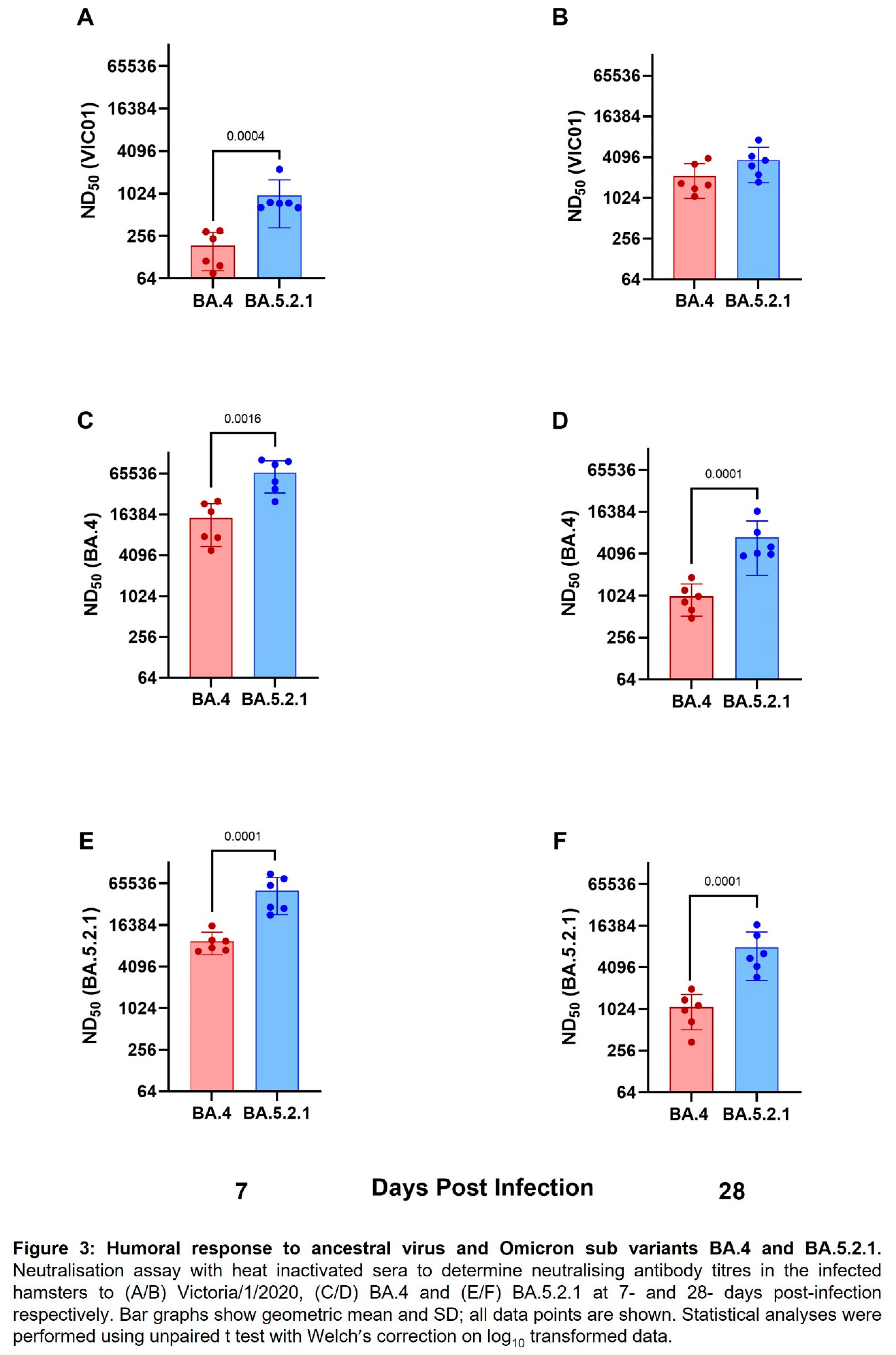
Convalescent hamsters infected with BA.4 have lower neutralising antibody titres against ancestral wildtype virus and Omicron variants. Geometric neutralising antibody titres (ND50) in hamster sera were determined in samples taken at 7-and 28- days post infection against ancestral virus (Australia/VIC01/2020, VIC01), BA.4 and BA.5.2.1. Neutralising antibody responses were detected in all hamsters against ancestral virus and both Omicron sub-variants regardless of challenge virus; however, hamsters infected with BA.5.2.1 had higher titres against ancestral, BA.4 and BA.5.2.1 at both timepoints. Geometric mean neutralising titres are shown in
Table 1. Figures 3A and 3B illustrate neutralising antibodies to VIC01, Figures 3C and 3D to BA.4 and Figures 3E and 3F to BA.5.2.1. Live virus neutralising antibody titres against BA.4 and BA.5.2.1 fell between 7- and 28-days post infection in contrast to the increase over the same time-period for the titres detected against the ancestral virus. We also noted that hamsters infected with BA.5.2.1 elicited higher live virus neutralising antibody titres against BA.4 than hamsters infected with BA.4.
Pathology. Histopathological examination showed multifocal areas of bronchointerstitial pneumonia in all hamsters at day 7 post infection (Figure 4A). However, the severity of microscopic lesions, the area of the lung showing pneumonia (Figure 4B) and the total histopathology score (Figure 4C) of the lung was significantly lower in BA.4 infected hamsters when compared with BA.5.2.1 infected hamsters. At day 28 post infection, hamsters showed less severe lung lesions, with type II pneumocyte hyperplasia and low grade peribronchiolar and perivascular cuffing as the main observed changes in the lung. Viral RNA (detected by in situ hybridisation of the -S-gene region) was seen in the lung of only four hamsters (two infected with BA.4, two infected with BA.5.2.1) at day 7 post infection (data not shown). No viral RNA was detected in any hamsters at day 28 post infection in the lung.
Cell necrosis was observed in the respiratory and olfactory epithelium associated to inflammatory exudates within the nasal cavity laminae and the presence of viral RNA at day 7 post infection, showing significantly less pathology (Figure 5A) and viral RNA Figure 5B) in the epithelial cells and exudates in the BA.4 infected hamsters compared to the BA.5.2.1 infected hamsters. Only minimal microscopic changes and no viral RNA were observed in the nasal cavity at day 28 post infection (data not shown).
Comparison to earlier Omicron variants. We have also performed equivalent studies with the earlier Omicron sub-lineages BA.1, BA.2 and BA.2.12.1 in parallel with ancestral virus. Weight loss was only seen in hamster infected with BA.5.2.1 and ancestral virus, lack of weight gain in the days following infection was observed with BA.1, BA.2, BA.2.12.1 and BA.4 (Figure 6A). Clinical signs of infection were observed for all other Omicron variants except for hamsters infected with BA.4. The incidence of clinical signs in BA.1, BA.2 and BA.2.12.1 infected hamsters was typically observed around 24 hours after the onset in BA.5.2.1 or ancestrally infected hamsters (Figure 6). Viral shedding from the upper respiratory tract was approximately ten-fold lower in hamsters infected with BA.1, BA.2 and BA.2.12.1 when compared to ancestral and BA.5.2.1 isolates for the 7 days following infection (
Figure S2A), however there was no statistical difference in viral load in the lung between Omicron variants, only between ancestral virus and BA.1, BA.2.12.1 and BA.4, where P = 0.0244, 0.0102 and 0.0007 respectively (
Figure S2B). This was also true for viral RNA (detected by in situ hybridisation with the -S-gene), although this lack of statistical difference is likely due to an outlier in the BA.5.2.1 infected hamsters (
Figure S3A). Furthermore, histopathological analyses demonstrated that areas of the lung showing microscopic lesions were significantly higher in BA.5.2.1 and ancestrally infected hamsters as well as BA.2 infected hamsters (
Figure S3B, C, D and E). Collectively, data show that BA.5.2.1 suggests a return to ancestral virus pathology in the hamster model relative to the previously circulating Omicron variants whilst BA.4 shows a notable lack of clinical signs despite only a few genetic changes.
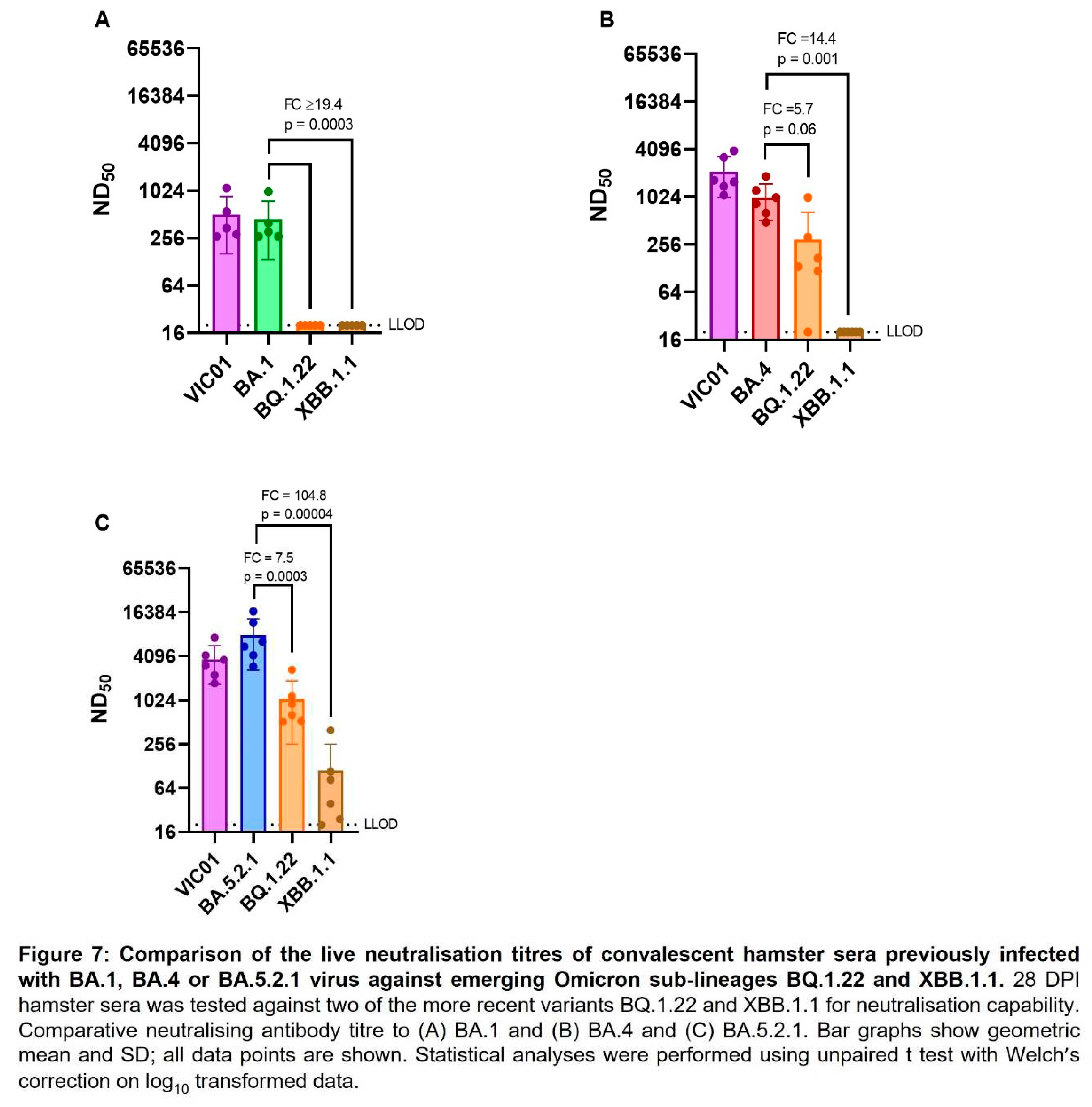
Neutralising antibody titres against emerging variants. As SARS-CoV-2 continues to evolve and new variants emerge, we have assessed live virus neutralising antibody levels in BA.1, BA.4 and BA.5.2.1 convalescent hamster sera (28 DPI) against more recent variants BQ.1.22 and XBB.1.1. The geometric mean neutralising antibody titres of each group of hamsters are shown in
Table 2. BA.1 convalescent sera demonstrated a ≥18-fold reduction in neutralisation titre when we compared the homologous titre against BA.1 (373) with those detected against BQ.1.22 and XBB.1.1 (≤20). Both BA.4 and BA.5.2.1 convalescent sera also demonstrated a reduction in neutralising titres to the emerging sub-lineages, but BA.5.2.1 infection appeared to induce better neutralising antibody titres against all viruses tested. Figure 7 illustrates these differences in neutralising antibody titres by comparing homologous titres with ancestral and Omicron sub-lineages.
4. Discussion
Genetic mutation and viral recombination have resulted in an “alphabet soup” of SARS-CoV-2 variants. The rapid emergence of some of these new variants constitutes a continued challenge to the success of the global vaccination programme and controls directed at preventing continued transmission and associated morbidity and mortality.
In this study we have demonstrated that there are two distinct disease profiles in hamsters after intranasal infection with either Omicron sub-variants BA.4 or BA.5.2.1 despite these viruses having almost identical spike protein. Hamsters infected intranasally with BA.5.2.1 demonstrated significant weight loss (-9.8±3.2%) by 7 DPI and multiple clinical signs of infection – comparable to those seen in animals infected with ancestral virus (Australia/VIC01/2020) [
9]. In contrast, hamsters infected with an equivalent virus dose of BA.4 maintained weight and displayed none of the clinical signs that have so far been seen in all the virulence studies we have conducted to date. Despite these clinical differences, viral shedding from the upper respiratory tract was similar for both Omicron variants in the days immediately following infection.
We note this outcome is different to the recent publication by Uraki
et al (2022), who describe no discernible differences between BA.4 and BA.5, where most notably there is no weight loss in hamsters infected with BA.5 [
14]. Whilst we use an infection inoculum volume of 200 µL [
12], Uraki
et al used an infection inoculum of 30 µL. We postulate the differences observed between the results we present and those of Uraki
et al could be due to the difference in IN inoculum volume or the additional changes in BA.5.2.1 used in this study. Our previously published data [
12] demonstrate SARS-CoV-2 disease severity in the Golden Syrian hamster model of infection is related to the volume of IN inoculum..
There are several possible explanations for the differences in pathogenicity observed in hamsters infected with either BA.4 or BA.5.2.1. Our hypothesis is that the deletion in the SARS-CoV-2 viral non-structural protein 1 (Nsp1) is the most likely. A nine-nucleotide deletion specific to BA.4 was identified in ORF1a (∆686-694) leading to the loss of three amino acids (aa 141-143) in Nsp1.
In SARS-CoV-2, Nsp1 is likely to play many different key roles in the host cell [
15]. Other previously published studies have reported that Nsp1 inhibits host protein translation and disrupts the mRNA export machinery to inhibit host gene expression [
16,
17,
18]. In addition, Nsp1 of SARS-CoV was shown to cause the decay of host mRNA [
19] and down-regulate type I IFN response (IFN-α and IFN-β) [
20,
21].
Lin
et al [
20] tracked the molecular evolution and clinical features of SARS-CoV-2-infected patients in China and showed that ∆500-532 in Nsp1 correlated with lower viral load, less severe symptoms of infection, and lower serum IFN-β. They also showed that IFN-I responses were significantly reduced in Calu-3 cells infected with deletion mutation viruses isolated from clinical samples or engineered using reverse genetics [
20].
Fisher
et al (2022), have performed mutagenesis studies to demonstrate that Nsp1 is a major immune evasion factor in SARS-CoV-2 [
22]. They generated a mutant SARS-CoV-2 with an amino acid deletion (aa 155-156) by reverse genetics, and infected Vero E6 (which do not produce type I interferon) [
23] and Calu-3 (which generate intact interferon response) cells. They showed comparable viral titres in Vero-E6 cells, but the mutant viral titres were lower in the Calu-3 cells compared to wildtype virus. They also demonstrated a stronger induction of interferon-stimulated genes in Calu-3 cells infected with mutant virus to support Nsp1’s role in down-regulating the interferon response [
22].
Deletions in related Coronaviruses have been shown to play an important part in their virulence [
18,
24,
25]. Given the role of Nsp1 as a major pathogenicity factor, it is a potential target for drug or vaccine design. Vaccine design could be in the form of recombinant virus, with mutated Nsp1 incapable of down-regulating the interferon response, or by a live-attenuated virus. Liu
et al (2022), have shown that a single intranasal delivery of an attenuated SARS-CoV-2, which included a pair of mutations in Nsp1, induced both mucosal and systemic IgA and IgG mediated protection in the Golden Syrian hamster COVID-19 model [
3].
There is still much to be learned about the different activities of Nsp1 in different SARS-CoV-2 variants of concern. This study supports previous observations that deletions in Nsp1 of coronaviruses are attenuating, as demonstrated by a lack of weight loss and clinical signs in hamsters infected with BA.4. While it is reasonable to assume that the deletion in BA.4 Nsp1 is responsible for the observed attenuation in this study, a confirmatory study in which the deletion in BA.4 was genetically introduced into BA.5.2.1 and a hamster challenge study performed would test this hypothesis conclusively. Further work to support this hypothesis may lead to a better understanding of the drivers of virulence and molecular targets for anti-viral therapy and for the rational design of future mucosal, live attenuated vaccines.
Supplementary Materials
The following supporting information can be downloaded at the website of this paper posted on Preprints.org. Supplementary Data File 1: Sequence data of the virus banks; Figure S1: Assessment of live viral titre in throat swabs 2 DPI; Figure S2: Assessment of viral shedding from respiratory tract and viral load in the lung against earlier Omicron sub-lineages and ancestral virus VIC01, Figure S3: Comparison histopathological analysis of nasal cavity and lung 7 days post infection to previous Omicron sub-lineages and ancestral virus.
Author Contributions
Conceptualization, E.R.D., K.A.R., O.T.C., Y.H., and S.G.P.F.; software, K.R.B.; formal analysis, E.R.D., F.J.S. and A.H.; investigation, E.R.D., N.S.C., O.T.C., S.B., M.C., A.C., E.S.C., A.H., D.N. and T.M.W.; writing—original draft preparation, E.R.D.; writing—review and editing, E.R.D., K.A.R., K.R.B., F.J.S., Y.H. and S.G.P.F.; visualization, E.R.D.; supervision, K.A.R., Y.H., S.G.P.F.; project administration, E.R.D., K.A.R. and Y.H.; funding acquisition, Y.H. and S.G.P.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the Coalition for Epidemic Preparedness Innovations’ (CEPI) Agility Program.
Institutional Review Board Statement
All experimental work was conducted under the authority and in compliance with a UK Home Office approved project license that had been subject to local ethical review at UKHSA Porton Down by the Animal Welfare and Ethical Review Body (AWERB) as required by the Home Office Animals (Scientific Procedures) Act 1986.
Data Availability Statement
Not applicable
Acknowledgments
The authors gratefully acknowledge the support from the Biological Investigations Group at the UK Health Security Agency, Porton Down, United Kingdom. We also thank the Doherty Institute, Melbourne, Australia for the generous supply of SARS-CoV-2 Victoria/01/2020. The views expressed in this paper are those of the authors and not necessarily those of the funding body. This work was funded by the Coalition for Epidemic Preparedness Innovations’ (CEPI) Agility Program. The authors are grateful for the critical review of the manuscript by Amy C. Shurtleff.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript or in the decision to publish the results.
References
- Cele, S.; Jackson, L.; Khoury, D. S.; Khan, K.; Moyo-Gwete, T.; Tegally, H.; San, J. E.; Cromer, D.; Scheepers, C.; Amoako, D.; Karim, F.; Bernstein, M.; Lustig, G.; Archary, D.; Smith, M.; Ganga, Y.; Jule, Z.; Reedoy, K.; Hwa, S. H.; Giandhari, J.; Blackburn, J. M.; Gosnell, B. I.; Abdool Karim, S. S.; Hanekom, W.; Ngs, S. A.; Team, C.-K.; von Gottberg, A.; Bhiman, J.; Lessells, R. J.; Moosa, M. S.; Davenport, M. P.; de Oliveira, T.; Moore, P. L.; Sigal, A., SARS-CoV-2 Omicron has extensive but incomplete escape of Pfizer BNT162b2 elicited neutralization and requires ACE2 for infection. medRxiv 2021. [CrossRef]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Toffa, S.; Rickeard, T.; Gallagher, E.; Gower, C.; Kall, M.; Groves, N.; O'Connell, A. M.; Simons, D.; Blomquist, P. B.; Zaidi, A.; Nash, S.; Iwani Binti Abdul Aziz, N.; Thelwall, S.; Dabrera, G.; Myers, R.; Amirthalingam, G.; Gharbia, S.; Barrett, J. C.; Elson, R.; Ladhani, S. N.; Ferguson, N.; Zambon, M.; Campbell, C. N. J.; Brown, K.; Hopkins, S.; Chand, M.; Ramsay, M.; Lopez Bernal, J., Covid-19 Vaccine Effectiveness against the Omicron (B.1.1.529) Variant. N Engl J Med 2022, 386, (16), 1532-1546. [CrossRef]
- Liu, L.; Iketani, S.; Guo, Y.; Chan, J. F.; Wang, M.; Liu, L.; Luo, Y.; Chu, H.; Huang, Y.; Nair, M. S.; Yu, J.; Chik, K. K.; Yuen, T. T.; Yoon, C.; To, K. K.; Chen, H.; Yin, M. T.; Sobieszczyk, M. E.; Huang, Y.; Wang, H. H.; Sheng, Z.; Yuen, K. Y.; Ho, D. D., Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature 2022, 602, (7898), 676-681. [CrossRef]
- Control., E. C. f. D. P. a. Epidemiological update: SARS-CoV-2 Omicron sub-lineages BA.4 and BA.5. https://www.ecdc.europa.eu/en/news-events/epidemiological-update-sars-cov-2-omicron-sub-lineages-ba4-and-ba5 (18 October),.
- Tegally, H.; Moir, M.; Everatt, J.; Giovanetti, M.; Scheepers, C.; Wilkinson, E.; Subramoney, K.; Makatini, Z.; Moyo, S.; Amoako, D. G.; Baxter, C.; Althaus, C. L.; Anyaneji, U. J.; Kekana, D.; Viana, R.; Giandhari, J.; Lessells, R. J.; Maponga, T.; Maruapula, D.; Choga, W.; Matshaba, M.; Mbulawa, M. B.; Msomi, N.; consortium, N.-S.; Naidoo, Y.; Pillay, S.; Sanko, T. J.; San, J. E.; Scott, L.; Singh, L.; Magini, N. A.; Smith-Lawrence, P.; Stevens, W.; Dor, G.; Tshiabuila, D.; Wolter, N.; Preiser, W.; Treurnicht, F. K.; Venter, M.; Chiloane, G.; McIntyre, C.; O'Toole, A.; Ruis, C.; Peacock, T. P.; Roemer, C.; Kosakovsky Pond, S. L.; Williamson, C.; Pybus, O. G.; Bhiman, J. N.; Glass, A.; Martin, D. P.; Jackson, B.; Rambaut, A.; Laguda-Akingba, O.; Gaseitsiwe, S.; von Gottberg, A.; de Oliveira, T., Emergence of SARS-CoV-2 Omicron lineages BA.4 and BA.5 in South Africa. Nat Med 2022, 28, (9), 1785-1790. [CrossRef]
- Qu, P.; Faraone, J.; Evans, J. P.; Zou, X.; Zheng, Y. M.; Carlin, C.; Bednash, J. S.; Lozanski, G.; Mallampalli, R. K.; Saif, L. J.; Oltz, E. M.; Mohler, P. J.; Gumina, R. J.; Liu, S. L., Neutralization of the SARS-CoV-2 Omicron BA.4/5 and BA.2.12.1 Subvariants. N Engl J Med 2022, 386, (26), 2526-2528. [CrossRef]
- Caly, L.; Druce, J.; Roberts, J.; Bond, K.; Tran, T.; Kostecki, R.; Yoga, Y.; Naughton, W.; Taiaroa, G.; Seemann, T.; Schultz, M. B.; Howden, B. P.; Korman, T. M.; Lewin, S. R.; Williamson, D. A.; Catton, M. G., Isolation and rapid sharing of the 2019 novel coronavirus (SARS-CoV-2) from the first patient diagnosed with COVID-19 in Australia. Med J Aust 2020, 212, (10), 459-462. [CrossRef]
- Coombes, N. S.; Bewley, K. R.; Le Duff, Y.; Hurley, M.; Smith, L. J.; Weldon, T. M.; Osman, K.; Pullan, S.; Berry, N.; Hallis, B.; Charlton, S.; Hall, Y.; Funnell, S. G. P., Assessment of the Biological Impact of SARS-CoV-2 Genetic Variation Using an Authentic Virus Neutralisation Assay with Convalescent Plasma, Vaccinee Sera, and Standard Reagents. Viruses 2023, 15, (3). [CrossRef]
- Ryan, K. A.; Bewley, K. R.; Fotheringham, S. A.; Slack, G. S.; Brown, P.; Hall, Y.; Wand, N. I.; Marriott, A. C.; Cavell, B. E.; Tree, J. A., Dose-dependent response to infection with SARS-CoV-2 in the ferret model and evidence of protective immunity. Nature communications 2021, 12, (1), 1-13. [CrossRef]
- Bewley, K. R.; Coombes, N. S.; Gagnon, L.; McInroy, L.; Baker, N.; Shaik, I.; St-Jean, J. R.; St-Amant, N.; Buttigieg, K. R.; Humphries, H. E.; Godwin, K. J.; Brunt, E.; Allen, L.; Leung, S.; Brown, P. J.; Penn, E. J.; Thomas, K.; Kulnis, G.; Hallis, B.; Carroll, M.; Funnell, S.; Charlton, S., Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat Protoc 2021, 16, (6), 3114-3140. [CrossRef]
- Dowall, S.; Salguero, F. J.; Wiblin, N.; Fotheringham, S.; Hatch, G.; Parks, S.; Gowan, K.; Harris, D.; Carnell, O.; Fell, R.; Watson, R.; Graham, V.; Gooch, K.; Hall, Y.; Mizen, S.; Hewson, R., Development of a Hamster Natural Transmission Model of SARS-CoV-2 Infection. Viruses 2021, 13, (11). [CrossRef]
- Handley, A.; Ryan, K. A.; Davies, E. R.; Bewley, K. R.; Carnell, O. T.; Challis, A.; Coombes, N. S.; Fotheringham, S. A.; Gooch, K. E.; Charlton, M.; Harris, D. J.; Kennard, C.; Ngabo, D.; Weldon, T. M.; Salguero, F. J.; Funnell, S. G. P.; Hall, Y., SARS-CoV-2 Disease Severity in the Golden Syrian Hamster Model of Infection is Related to the Volume of Intranasal Inoculum. Viruses 2023, 15, (3). [CrossRef]
- Munoz-Fontela, C.; Dowling, W. E.; Funnell, S. G. P.; Gsell, P. S.; Riveros-Balta, A. X.; Albrecht, R. A.; Andersen, H.; Baric, R. S.; Carroll, M. W.; Cavaleri, M.; Qin, C.; Crozier, I.; Dallmeier, K.; de Waal, L.; de Wit, E.; Delang, L.; Dohm, E.; Duprex, W. P.; Falzarano, D.; Finch, C. L.; Frieman, M. B.; Graham, B. S.; Gralinski, L. E.; Guilfoyle, K.; Haagmans, B. L.; Hamilton, G. A.; Hartman, A. L.; Herfst, S.; Kaptein, S. J. F.; Klimstra, W. B.; Knezevic, I.; Krause, P. R.; Kuhn, J. H.; Le Grand, R.; Lewis, M. G.; Liu, W. C.; Maisonnasse, P.; McElroy, A. K.; Munster, V.; Oreshkova, N.; Rasmussen, A. L.; Rocha-Pereira, J.; Rockx, B.; Rodriguez, E.; Rogers, T. F.; Salguero, F. J.; Schotsaert, M.; Stittelaar, K. J.; Thibaut, H. J.; Tseng, C. T.; Vergara-Alert, J.; Beer, M.; Brasel, T.; Chan, J. F. W.; Garcia-Sastre, A.; Neyts, J.; Perlman, S.; Reed, D. S.; Richt, J. A.; Roy, C. J.; Segales, J.; Vasan, S. S.; Henao-Restrepo, A. M.; Barouch, D. H., Animal models for COVID-19. Nature 2020, 586, (7830), 509-515. [CrossRef]
- Uraki, R.; Halfmann, P. J.; Iida, S.; Yamayoshi, S.; Furusawa, Y.; Kiso, M.; Ito, M.; Iwatsuki-Horimoto, K.; Mine, S.; Kuroda, M.; Maemura, T.; Sakai-Tagawa, Y.; Ueki, H.; Li, R.; Liu, Y.; Larson, D.; Fukushi, S.; Watanabe, S.; Maeda, K.; Pekosz, A.; Kandeil, A.; Webby, R. J.; Wang, Z.; Imai, M.; Suzuki, T.; Kawaoka, Y., Characterization of SARS-CoV-2 Omicron BA.4 and BA.5 isolates in rodents. Nature 2022, 612, (7940), 540-545. [CrossRef]
- Kao, H. T.; Orry, A.; Palfreyman, M. G.; Porton, B., Synergistic interactions of repurposed drugs that inhibit Nsp1, a major virulence factor for COVID-19. Sci Rep 2022, 12, (1), 10174. [CrossRef]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; Straub, J. H.; Sturzel, C. M.; Frohlich, T.; Berninghausen, O.; Becker, T.; Kirchhoff, F.; Sparrer, K. M. J.; Beckmann, R., Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, (6508), 1249-1255. [CrossRef]
- Zhang, K.; Miorin, L.; Makio, T.; Dehghan, I.; Gao, S.; Xie, Y.; Zhong, H.; Esparza, M.; Kehrer, T.; Kumar, A.; Hobman, T. C.; Ptak, C.; Gao, B.; Minna, J. D.; Chen, Z.; Garcia-Sastre, A.; Ren, Y.; Wozniak, R. W.; Fontoura, B. M. A., Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci Adv 2021, 7, (6). [CrossRef]
- Zust, R.; Cervantes-Barragan, L.; Kuri, T.; Blakqori, G.; Weber, F.; Ludewig, B.; Thiel, V., Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog 2007, 3, (8), e109. [CrossRef]
- Kamitani, W.; Narayanan, K.; Huang, C.; Lokugamage, K.; Ikegami, T.; Ito, N.; Kubo, H.; Makino, S., Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc Natl Acad Sci U S A 2006, 103, (34), 12885-90. [CrossRef]
- Lin, J. W.; Tang, C.; Wei, H. C.; Du, B.; Chen, C.; Wang, M.; Zhou, Y.; Yu, M. X.; Cheng, L.; Kuivanen, S.; Ogando, N. S.; Levanov, L.; Zhao, Y.; Li, C. L.; Zhou, R.; Li, Z.; Zhang, Y.; Sun, K.; Wang, C.; Chen, L.; Xiao, X.; Zheng, X.; Chen, S. S.; Zhou, Z.; Yang, R.; Zhang, D.; Xu, M.; Song, J.; Wang, D.; Li, Y.; Lei, S.; Zeng, W.; Yang, Q.; He, P.; Zhang, Y.; Zhou, L.; Cao, L.; Luo, F.; Liu, H.; Wang, L.; Ye, F.; Zhang, M.; Li, M.; Fan, W.; Li, X.; Li, K.; Ke, B.; Xu, J.; Yang, H.; He, S.; Pan, M.; Yan, Y.; Zha, Y.; Jiang, L.; Yu, C.; Liu, Y.; Xu, Z.; Li, Q.; Jiang, Y.; Sun, J.; Hong, W.; Wei, H.; Lu, G.; Vapalahti, O.; Luo, Y.; Wei, Y.; Connor, T.; Tan, W.; Snijder, E. J.; Smura, T.; Li, W.; Geng, J.; Ying, B.; Chen, L., Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe 2021, 29, (3), 489-502 e8. [CrossRef]
- Narayanan, K.; Huang, C.; Lokugamage, K.; Kamitani, W.; Ikegami, T.; Tseng, C. T.; Makino, S., Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J Virol 2008, 82, (9), 4471-9. [CrossRef]
- Fisher, T.; Gluck, A.; Narayanan, K.; Kuroda, M.; Nachshon, A.; Hsu, J. C.; Halfmann, P. J.; Yahalom-Ronen, Y.; Tamir, H.; Finkel, Y.; Schwartz, M.; Weiss, S.; Tseng, C. K.; Israely, T.; Paran, N.; Kawaoka, Y.; Makino, S.; Stern-Ginossar, N., Parsing the role of NSP1 in SARS-CoV-2 infection. Cell Rep 2022, 39, (11), 110954. [CrossRef]
- Emeny, J. M.; Morgan, M. J., Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol 1979, 43, (1), 247-52. [CrossRef]
- Lokugamage, K. G.; Narayanan, K.; Huang, C.; Makino, S., Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J Virol 2012, 86, (24), 13598-608. [CrossRef]
- Shen, Z.; Wang, G.; Yang, Y.; Shi, J.; Fang, L.; Li, F.; Xiao, S.; Fu, Z. F.; Peng, G., A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J Biol Chem 2019, 294, (37), 13606-13618. [CrossRef]
Table 1.
Geometric mean of neutralising antibody titres from BA.4 and BA.5.2.1 infected hamsters at 7- and 28-days post infection to ancestral (VIC01), BA.4 and BA.5.2.1 viruses.
Table 1.
Geometric mean of neutralising antibody titres from BA.4 and BA.5.2.1 infected hamsters at 7- and 28-days post infection to ancestral (VIC01), BA.4 and BA.5.2.1 viruses.
| Days Post Infection |
Challenge Virus |
Mean Neutralising Antibody Titre (ND50) |
| VIC01 |
BA.4
|
BA.5
|
| 7 |
BA.4 |
187 |
14471 |
9446 |
| BA.5 |
967 |
67363 |
51209 |
| 28 |
BA.4 |
2144 |
1008 |
1084 |
| BA.5 |
3709 |
7007 |
7970 |
Table 2.
Geometric mean of neutralising antibody titres from BA.1, BA.4 and BA.5.2.1 convalescent hamster sera against BQ.1.22 and XBB.1.1.
Table 2.
Geometric mean of neutralising antibody titres from BA.1, BA.4 and BA.5.2.1 convalescent hamster sera against BQ.1.22 and XBB.1.1.
| Infection Virus |
|
Geometric mean Neutralising Antibody Titre (ND50) |
| BA.1 |
BA.4 |
BA.5.2.1 |
BQ.1.22 |
XBB.1.1 |
| BA.1 |
373 |
|
|
≤20 |
≤20 |
| BA.4 |
|
1008 |
|
295 |
≤20 |
| BA.5.2.1 |
|
|
7970 |
1078 |
112 |
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).