Submitted:
20 April 2023
Posted:
21 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Technical system for the development of new drugs and personalized medicine
2.1. Research and development of personalized medicine based on artificial intelligence technology
2.2. Approaches with multidimensional omics data
2.3. Study on high-throughput targets of chemical proteomics technology
2.4. Computer aided drug discovery system for the development of personalized medicine
3. Anti-tumor personalized medicine
3.1. ALK inhibitors
3.2. EGFR inhibitors
4. Pharmacogenetics of the anti-cancer natural products
5. The future challenges of personalized therapy
6. Conclusions and future perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflict of Interest
References
- Su, Miao, Zhe Zhang, Li Zhou, Chao Han, Canhua Huang, and Edouard C Nice. "Proteomics, Personalized Medicine and Cancer." Cancers 13, no. 11 (2021): 2512. [CrossRef]
- Chen, Rui, and Michael Snyder. "Promise of Personalized Omics to Precision Medicine." Wiley Interdisciplinary Reviews: Systems Biology and Medicine 5, no. 1 (2013): 73-82. [CrossRef]
- Carrasco-Ramiro, Fernando, Ramón Peiró-Pastor, and Begoña Aguado. "Human Genomics Projects and Precision Medicine." Gene therapy 24, no. 9 (2017): 551-61. [CrossRef]
- Jain, Kewal K. "Personalized Medicine." Current opinion in molecular therapeutics 4, no. 6 (2002): 548-58.
- Goetz, L. H., and N. J. Schork. "Personalized Medicine: Motivation, Challenges, and Progress." Fertil Steril 109, no. 6 (2018): 952-63. [CrossRef]
- Najjar, S., and K. H. Allison. "Updates on Breast Biomarkers." Virchows Arch 480, no. 1 (2022): 163-76. [CrossRef]
- Cecchin, E., and G. Stocco. "Pharmacogenomics and Personalized Medicine." Genes (Basel) 11, no. 6 (2020). [CrossRef]
- Chang, C. J., C. B. Chen, S. I. Hung, C. Ji, and W. H. Chung. "Pharmacogenetic Testing for Prevention of Severe Cutaneous Adverse Drug Reactions." Front Pharmacol 11 (2020): 969. [CrossRef]
- Zastrozhin, M. S., A. S. Sorokin, T. V. Agibalova, E. A. Grishina, AР Antonenko, I. N. Rozochkin, D. V. Duzhev, V. Y. Skryabin, T. E. Galaktionova, I. V. Barna, A. V. Orlova, A. D. Aguzarov, L. M. Savchenko, E. A. Bryun, and D. A. Sychev. "Using a Personalized Clinical Decision Support System for Bromdihydrochlorphenylbenzodiazepine Dosing in Patients with Anxiety Disorders Based on the Pharmacogenomic Markers." Hum Psychopharmacol 33, no. 6 (2018): e2677. [CrossRef]
- Workman, P. "Pharmacogenomics in Cancer Drug Discovery and Development: Inhibitors of the Hsp90 Molecular Chaperone." Cancer Detect Prev 26, no. 6 (2002): 405-10. [CrossRef]
- Letai, A., P. Bhola, and A. L. Welm. "Functional Precision Oncology: Testing Tumors with Drugs to Identify Vulnerabilities and Novel Combinations." Cancer Cell 40, no. 1 (2022): 26-35. [CrossRef]
- Shirley, M. "Bruton Tyrosine Kinase Inhibitors in B-Cell Malignancies: Their Use and Differential Features." Target Oncol 17, no. 1 (2022): 69-84. [CrossRef]
- Zhong, L., Y. Li, L. Xiong, W. Wang, M. Wu, T. Yuan, W. Yang, C. Tian, Z. Miao, T. Wang, and S. Yang. "Small Molecules in Targeted Cancer Therapy: Advances, Challenges, and Future Perspectives." Signal Transduct Target Ther 6, no. 1 (2021): 201. [CrossRef]
- Pushpakom, S., F. Iorio, P. A. Eyers, K. J. Escott, S. Hopper, A. Wells, A. Doig, T. Guilliams, J. Latimer, C. McNamee, A. Norris, P. Sanseau, D. Cavalla, and M. Pirmohamed. "Drug Repurposing: Progress, Challenges and Recommendations." Nat Rev Drug Discov 18, no. 1 (2019): 41-58. [CrossRef]
- Grover, A., and P. C. Sharma. "Development and Use of Molecular Markers: Past and Present." Crit Rev Biotechnol 36, no. 2 (2016): 290-302. [CrossRef]
- Nakagawa, K., E. B. Garon, T. Seto, M. Nishio, S. Ponce Aix, L. Paz-Ares, C. H. Chiu, K. Park, S. Novello, E. Nadal, F. Imamura, K. Yoh, J. Y. Shih, K. H. Au, D. Moro-Sibilot, S. Enatsu, A. Zimmermann, B. Frimodt-Moller, C. Visseren-Grul, and M. Reck. "Ramucirumab Plus Erlotinib in Patients with Untreated, Egfr-Mutated, Advanced Non-Small-Cell Lung Cancer (Relay): A Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial." Lancet Oncol 20, no. 12 (2019): 1655-69. [CrossRef]
- Casak, S. J., I. Fashoyin-Aje, S. J. Lemery, L. Zhang, R. Jin, H. Li, L. Zhao, H. Zhao, H. Zhang, H. Chen, K. He, M. Dougherty, R. Novak, S. Kennett, S. Khasar, W. Helms, P. Keegan, and R. Pazdur. "Fda Approval Summary: Ramucirumab for Gastric Cancer." Clin Cancer Res 21, no. 15 (2015): 3372-6. [CrossRef]
- Domingos, S., V. André, S. Quaresma, I. C. Martins, M. F. Minas da Piedade, and M. T. Duarte. "New Forms of Old Drugs: Improving without Changing." J Pharm Pharmacol 67, no. 6 (2015): 830-46. [CrossRef]
- Vlahopoulos, S., E. Critselis, I. F. Voutsas, S. A. Perez, M. Moschovi, C. N. Baxevanis, and G. P. Chrousos. "New Use for Old Drugs? Prospective Targets of Chloroquines in Cancer Therapy." Curr Drug Targets 15, no. 9 (2014): 843-51. [CrossRef]
- Joyce, C. R. "New Drugs for Mental Diseases? New Diseases for Old Drugs?" J R Soc Med 80, no. 7 (1987): 406-9. [CrossRef]
- Cortez-Maya, S., A. Moreno-Herrera, I. Palos, and G. Rivera. "Old Antiprotozoal Drugs: Are They Still Viable Options for Parasitic Infections or New Options for Other Diseases?" Curr Med Chem 27, no. 32 (2020): 5403-28. [CrossRef]
- Arbel, Y., W. Abuzeid, R. S. Rosenson, A. Weisman, and M. E. Farkouh. "Old Drugs for New Indications in Cardiovascular Medicine." Cardiovasc Drugs Ther 32, no. 2 (2018): 223-32. [CrossRef]
- Kaiser, D., and E. Oetjen. "Something Old, Something New and Something Very Old: Drugs for Treating Type 2 Diabetes." Br J Pharmacol 171, no. 12 (2014): 2940-50. [CrossRef]
- Janiaud, P., S. Serghiou, and J. P. A. Ioannidis. "New Clinical Trial Designs in the Era of Precision Medicine: An Overview of Definitions, Strengths, Weaknesses, and Current Use in Oncology." Cancer Treat Rev 73 (2019): 20-30. [CrossRef]
- Di Sanzo, M., L. Cipolloni, M. Borro, R. La Russa, A. Santurro, M. Scopetti, M. Simmaco, and P. Frati. "Clinical Applications of Personalized Medicine: A New Paradigm and Challenge." Curr Pharm Biotechnol 18, no. 3 (2017): 194-203. [CrossRef]
- Jørgensen, J. T. "Oncology Drug-Companion Diagnostic Combinations." Cancer Treat Res Commun 29 (2021): 100492. [CrossRef]
- Zwart, H., L. Landeweerd, and P. Lemmens. "Continental Philosophical Perspectives on Life Sciences and Emerging Technologies." Life Sci Soc Policy 12, no. 1 (2016): 8. [CrossRef]
- Hartl, D., V. de Luca, A. Kostikova, J. Laramie, S. Kennedy, E. Ferrero, R. Siegel, M. Fink, S. Ahmed, J. Millholland, A. Schuhmacher, M. Hinder, L. Piali, and A. Roth. "Translational Precision Medicine: An Industry Perspective." J Transl Med 19, no. 1 (2021): 245. [CrossRef]
- Lian, M. A., W. Ke-Bin, and W. U. Sheng-Xian. "[Research and Development of New Traditional Chinese Medicine Drugs for Certain Syndromes Based on "Theoretical Innovation"]." Zhongguo Zhong Yao Za Zhi 45, no. 20 (2020): 5048-56. [CrossRef]
- Jiang, Z., X. Zhou, R. Li, J. J. Michal, S. Zhang, M. V. Dodson, Z. Zhang, and R. M. Harland. "Whole Transcriptome Analysis with Sequencing: Methods, Challenges and Potential Solutions." Cell Mol Life Sci 72, no. 18 (2015): 3425-39. [CrossRef]
- Sherman, R. M., and S. L. Salzberg. "Pan-Genomics in the Human Genome Era." Nat Rev Genet 21, no. 4 (2020): 243-54. [CrossRef]
- Wang, Y., and W. Yang. "Proteome-Scale Analysis of Protein S-Acylation Comes of Age." J Proteome Res 20, no. 1 (2021): 14-26. [CrossRef]
- Balashova, E. E., O. P. Trifonova, D. L. Maslov, S. R. Lichtenberg, P. G. Lokhov, and A. I. Archakov. "[Metabolome Profiling in the Study of Aging Processes]." Biomed Khim 68, no. 5 (2022): 321-38. [CrossRef]
- Li, X., J. Ma, L. Leng, M. Han, M. Li, F. He, and Y. Zhu. "Mogcn: A Multi-Omics Integration Method Based on Graph Convolutional Network for Cancer Subtype Analysis." Front Genet 13 (2022): 806842. [CrossRef]
- Farber, G., K. E. Boczar, C. C. Wiefels, J. G. E. Zelt, E. C. Guler, R. A. deKemp, R. S. Beanlands, and B. H. Rotstein. "The Future of Cardiac Molecular Imaging." Semin Nucl Med 50, no. 4 (2020): 367-85. [CrossRef]
- Bagchi, A. "Latest Trends in Structure Based Drug Design with Protein Targets." Adv Protein Chem Struct Biol 121 (2020): 1-23. [CrossRef]
- Joshi, R. R., and V. V. Samant. "Bayesian Data Mining of Protein Domains Gives an Efficient Predictive Algorithm and New Insight." J Mol Model 13, no. 1 (2007): 275-82. [CrossRef]
- Ramesh, A. N., C. Kambhampati, J. R. Monson, and P. J. Drew. "Artificial Intelligence in Medicine." Ann R Coll Surg Engl 86, no. 5 (2004): 334-8.
- Carrera-Rivera, A., W. Ochoa, F. Larrinaga, and G. Lasa. "How-to Conduct a Systematic Literature Review: A Quick Guide for Computer Science Research." MethodsX 9 (2022): 101895. [CrossRef]
- Rasteau, S., D. Ernenwein, C. Savoldelli, and P. Bouletreau. "Artificial Intelligence for Oral and Maxillo-Facial Surgery: A Narrative Review." J Stomatol Oral Maxillofac Surg 123, no. 3 (2022): 276-82. [CrossRef]
- Wang, A., X. Xiu, S. Liu, Q. Qian, and S. Wu. "Characteristics of Artificial Intelligence Clinical Trials in the Field of Healthcare: A Cross-Sectional Study on Clinicaltrials.Gov." Int J Environ Res Public Health 19, no. 20 (2022). [CrossRef]
- Wang, L., and C. A. Alexander. "Big Data Analytics in Medical Engineering and Healthcare: Methods, Advances and Challenges." J Med Eng Technol 44, no. 6 (2020): 267-83. [CrossRef]
- Dallas, S., C. Sensenhauser, A. Batheja, M. Singer, M. Markowska, C. Zakszewski, R. N. Mamidi, M. McMillia, C. Han, H. Zhou, and J. Silva. "De-Risking Bio-Therapeutics for Possible Drug Interactions Using Cryopreserved Human Hepatocytes." Curr Drug Metab 13, no. 7 (2012): 923-9. [CrossRef]
- Lee, Y. W., J. W. Choi, and E. H. Shin. "Machine Learning Model for Predicting Malaria Using Clinical Information." Comput Biol Med 129 (2021): 104151. [CrossRef]
- Chan, S., V. Reddy, B. Myers, Q. Thibodeaux, N. Brownstone, and W. Liao. "Machine Learning in Dermatology: Current Applications, Opportunities, and Limitations." Dermatol Ther (Heidelb) 10, no. 3 (2020): 365-86. [CrossRef]
- Jiang, Y., M. Yang, S. Wang, X. Li, and Y. Sun. "Emerging Role of Deep Learning-Based Artificial Intelligence in Tumor Pathology." Cancer Commun (Lond) 40, no. 4 (2020): 154-66. [CrossRef]
- Dong, L., W. He, R. Zhang, Z. Ge, Y. X. Wang, J. Zhou, J. Xu, L. Shao, Q. Wang, Y. Yan, Y. Xie, L. Fang, H. Wang, Y. Wang, X. Zhu, J. Wang, C. Zhang, H. Wang, Y. Wang, R. Chen, Q. Wan, J. Yang, W. Zhou, H. Li, X. Yao, Z. Yang, J. Xiong, X. Wang, Y. Huang, Y. Chen, Z. Wang, C. Rong, J. Gao, H. Zhang, S. Wu, J. B. Jonas, and W. B. Wei. "Artificial Intelligence for Screening of Multiple Retinal and Optic Nerve Diseases." JAMA Netw Open 5, no. 5 (2022): e229960. [CrossRef]
- Yu, K. H., A. L. Beam, and I. S. Kohane. "Artificial Intelligence in Healthcare." Nat Biomed Eng 2, no. 10 (2018): 719-31. [CrossRef]
- Ji, Y., S. Liu, X. Hong, Y. Lu, X. Wu, K. Li, K. Li, and Y. Liu. "Advances in Artificial Intelligence Applications for Ocular Surface Diseases Diagnosis." Front Cell Dev Biol 10 (2022): 1107689. [CrossRef]
- Song, Q., and X. Li. "[Application and Development of Voice Analysis and Endoscopic Technology Combined with Artificial Intelligence in the Diagnosis and Treatment of Throat Disease]." Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi 36, no. 8 (2022): 647-50. [CrossRef]
- Ye, Y., Z. Zhang, Y. Liu, L. Diao, and L. Han. "A Multi-Omics Perspective of Quantitative Trait Loci in Precision Medicine." Trends Genet 36, no. 5 (2020): 318-36. [CrossRef]
- Chong, J., D. S. Wishart, and J. Xia. "Using Metaboanalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis." Curr Protoc Bioinformatics 68, no. 1 (2019): e86. [CrossRef]
- Nygaard, H. B. "Targeting Fyn Kinase in Alzheimer's Disease." Biol Psychiatry 83, no. 4 (2018): 369-76. [CrossRef]
- Wang, M., A. Li, M. Sekiya, N. D. Beckmann, X. Quan, N. Schrode, M. B. Fernando, A. Yu, L. Zhu, J. Cao, L. Lyu, E. Horgusluoglu, Q. Wang, L. Guo, Y. S. Wang, R. Neff, W. M. Song, E. Wang, Q. Shen, X. Zhou, C. Ming, S. M. Ho, S. Vatansever, HÜ Kaniskan, J. Jin, M. M. Zhou, K. Ando, L. Ho, P. A. Slesinger, Z. Yue, J. Zhu, P. Katsel, S. Gandy, M. E. Ehrlich, V. Fossati, S. Noggle, D. Cai, V. Haroutunian, K. M. Iijima, E. Schadt, K. J. Brennand, and B. Zhang. "Transformative Network Modeling of Multi-Omics Data Reveals Detailed Circuits, Key Regulators, and Potential Therapeutics for Alzheimer's Disease." Neuron 109, no. 2 (2021): 257-72.e14. [CrossRef]
- Parker, G., and H. Brotchie. "Do the Old Psychostimulant Drugs Have a Role in Managing Treatment-Resistant Depression?" Acta Psychiatr Scand 121, no. 4 (2010): 308-14. [CrossRef]
- Whirl-Carrillo, M., E. M. McDonagh, J. M. Hebert, L. Gong, K. Sangkuhl, C. F. Thorn, R. B. Altman, and T. E. Klein. "Pharmacogenomics Knowledge for Personalized Medicine." Clin Pharmacol Ther 92, no. 4 (2012): 414-7. [CrossRef]
- Qu, J., F. S. Kalyani, L. Liu, T. Cheng, and L. Chen. "Tumor Organoids: Synergistic Applications, Current Challenges, and Future Prospects in Cancer Therapy." Cancer Commun (Lond) 41, no. 12 (2021): 1331-53. [CrossRef]
- Park, M., D. Kim, K. Moon, and T. Park. "Integrative Analysis of Multi-Omics Data Based on Blockwise Sparse Principal Components." Int J Mol Sci 21, no. 21 (2020). [CrossRef]
- Ding, Z., N. Wang, N. Ji, and Z. S. Chen. "Proteomics Technologies for Cancer Liquid Biopsies." Mol Cancer 21, no. 1 (2022): 53. [CrossRef]
- Battaglini, D., L. Al-Husinat, A. G. Normando, A. P. Leme, K. Franchini, M. Morales, P. Pelosi, and P. R. Rocco. "Personalized Medicine Using Omics Approaches in Acute Respiratory Distress Syndrome to Identify Biological Phenotypes." Respir Res 23, no. 1 (2022): 318. [CrossRef]
- Spradlin, J. N., E. Zhang, and D. K. Nomura. "Reimagining Druggability Using Chemoproteomic Platforms." Acc Chem Res 54, no. 7 (2021): 1801-13. [CrossRef]
- Kanduc, D. "The Role of Proteomics in Defining Autoimmunity." Expert Rev Proteomics 18, no. 3 (2021): 177-84. [CrossRef]
- Elyada, E., M. Bolisetty, P. Laise, W. F. Flynn, E. T. Courtois, R. A. Burkhart, J. A. Teinor, P. Belleau, G. Biffi, M. S. Lucito, S. Sivajothi, T. D. Armstrong, D. D. Engle, K. H. Yu, Y. Hao, C. L. Wolfgang, Y. Park, J. Preall, E. M. Jaffee, A. Califano, P. Robson, and D. A. Tuveson. "Cross-Species Single-Cell Analysis of Pancreatic Ductal Adenocarcinoma Reveals Antigen-Presenting Cancer-Associated Fibroblasts." Cancer Discov 9, no. 8 (2019): 1102-23. [CrossRef]
- Pang, Z., M. A. Schafroth, D. Ogasawara, Y. Wang, V. Nudell, N. K. Lal, D. Yang, K. Wang, D. M. Herbst, J. Ha, C. Guijas, J. L. Blankman, B. F. Cravatt, and L. Ye. "In Situ Identification of Cellular Drug Targets in Mammalian Tissue." Cell 185, no. 10 (2022): 1793-805.e17. [CrossRef]
- Ambure, P., and K. Roy. "Cadd Modeling of Multi-Target Drugs against Alzheimer'S Disease." Curr Drug Targets 18, no. 5 (2017): 522-33. [CrossRef]
- Lomenick, B., R. Hao, N. Jonai, R. M. Chin, M. Aghajan, S. Warburton, J. Wang, R. P. Wu, F. Gomez, J. A. Loo, J. A. Wohlschlegel, T. M. Vondriska, J. Pelletier, H. R. Herschman, J. Clardy, C. F. Clarke, and J. Huang. "Target Identification Using Drug Affinity Responsive Target Stability (Darts)." Proc Natl Acad Sci U S A 106, no. 51 (2009): 21984-9. [CrossRef]
- Li, G., P. Lin, K. Wang, C. C. Gu, and S. Kusari. "Artificial Intelligence-Guided Discovery of Anticancer Lead Compounds from Plants and Associated Microorganisms." Trends Cancer 8, no. 1 (2022): 65-80. [CrossRef]
- Wong, M., A. Badri, C. Gasparis, G. Belfort, and M. Koffas. "Modular Optimization in Metabolic Engineering." Crit Rev Biochem Mol Biol 56, no. 6 (2021): 587-602. [CrossRef]
- Gomeni, R., M. Bani, C. D'Angeli, M. Corsi, and A. Bye. "Computer-Assisted Drug Development (Cadd): An Emerging Technology for Designing First-Time-in-Man and Proof-of-Concept Studies from Preclinical Experiments." Eur J Pharm Sci 13, no. 3 (2001): 261-70. [CrossRef]
- Vintonyak, V. V., A. P. Antonchick, D. Rauh, and H. Waldmann. "The Therapeutic Potential of Phosphatase Inhibitors." Curr Opin Chem Biol 13, no. 3 (2009): 272-83. [CrossRef]
- Avram, S., M. Mernea, C. Limban, F. Borcan, and C. Chifiriuc. "Potential Therapeutic Approaches to Alzheimer's Disease by Bioinformatics, Cheminformatics and Predicted Adme-Tox Tools." Curr Neuropharmacol 18, no. 8 (2020): 696-719. [CrossRef]
- Akalin, P. K. "Introduction to Bioinformatics." Mol Nutr Food Res 50, no. 7 (2006): 610-9. [CrossRef]
- Wishart, D. S. "Introduction to Cheminformatics." Curr Protoc Bioinformatics Chapter 14 (2007): Unit 14.1. [CrossRef]
- Irwin, J. J., G. Gaskins, T. Sterling, M. M. Mysinger, and M. J. Keiser. "Predicted Biological Activity of Purchasable Chemical Space." J Chem Inf Model 58, no. 1 (2018): 148-64. [CrossRef]
- Mitusińska, K., A. Raczyńska, M. Bzówka, W. Bagrowska, and A. Góra. "Applications of Water Molecules for Analysis of Macromolecule Properties." Comput Struct Biotechnol J 18 (2020): 355-65. [CrossRef]
- Zobdeh, F., A. Ben Kraiem, M. M. Attwood, V. N. Chubarev, V. V. Tarasov, H. B. Schiöth, and J. Mwinyi. "Pharmacological Treatment of Migraine: Drug Classes, Mechanisms of Action, Clinical Trials and New Treatments." Br J Pharmacol 178, no. 23 (2021): 4588-607. [CrossRef]
- Pinzi, L., and G. Rastelli. "Molecular Docking: Shifting Paradigms in Drug Discovery." Int J Mol Sci 20, no. 18 (2019). [CrossRef]
- Reddy, K. K., R. S. Rathore, P. Srujana, R. R. Burri, C. R. Reddy, M. Sumakanth, P. Reddanna, and M. R. Reddy. "Performance Evaluation of Docking Programs- Glide, Gold, Autodock & Surflexdock, Using Free Energy Perturbation Reference Data: A Case Study of Fructose-1, 6-Bisphosphatase-Amp Analogs." Mini Rev Med Chem 20, no. 12 (2020): 1179-87. [CrossRef]
- Collier, T. A., T. J. Piggot, and J. R. Allison. "Molecular Dynamics Simulation of Proteins." Methods Mol Biol 2073 (2020): 311-27. [CrossRef]
- Wang, L., J. Chambers, and R. Abel. "Protein-Ligand Binding Free Energy Calculations with Fep." Methods Mol Biol 2022 (2019): 201-32.
- Gupta, R., D. Srivastava, M. Sahu, S. Tiwari, R. K. Ambasta, and P. Kumar. "Artificial Intelligence to Deep Learning: Machine Intelligence Approach for Drug Discovery." Mol Divers 25, no. 3 (2021): 1315-60. [CrossRef]
- Hessler, G., and K. H. Baringhaus. "Artificial Intelligence in Drug Design." Molecules 23, no. 10 (2018). [CrossRef]
- Mueller, R., E. S. Dawson, J. Meiler, A. L. Rodriguez, B. A. Chauder, B. S. Bates, A. S. Felts, J. P. Lamb, U. N. Menon, S. B. Jadhav, A. S. Kane, C. K. Jones, K. J. Gregory, C. M. Niswender, P. J. Conn, C. M. Olsen, D. G. Winder, K. A. Emmitte, and C. W. Lindsley. "Discovery of 2-(2-Benzoxazoyl Amino)-4-Aryl-5-Cyanopyrimidine as Negative Allosteric Modulators (Nams) of Metabotropic Glutamate Receptor 5 (Mglu₅): From an Artificial Neural Network Virtual Screen to an in Vivo Tool Compound." ChemMedChem 7, no. 3 (2012): 406-14. [CrossRef]
- Ijjaali, I., C. Barrere, J. Nargeot, F. Petitet, and E. Bourinet. "Ligand-Based Virtual Screening to Identify New T-Type Calcium Channel Blockers." Channels (Austin) 1, no. 4 (2007): 300-4. [CrossRef]
- Dembic, Z. "Antitumor Drugs and Their Targets." Molecules 25, no. 23 (2020). [CrossRef]
- Jain, K. K. "Personalized Immuno-Oncology." Med Princ Pract 30, no. 1 (2021): 1-16. [CrossRef]
- Du, X., Y. Shao, H. F. Qin, Y. H. Tai, and H. J. Gao. "Alk-Rearrangement in Non-Small-Cell Lung Cancer (Nsclc)." Thorac Cancer 9, no. 4 (2018): 423-30. [CrossRef]
- Golding, B., A. Luu, R. Jones, and A. M. Viloria-Petit. "The Function and Therapeutic Targeting of Anaplastic Lymphoma Kinase (Alk) in Non-Small Cell Lung Cancer (Nsclc)." Mol Cancer 17, no. 1 (2018): 52. [CrossRef]
- Lin, J. J., G. J. Riely, and A. T. Shaw. "Targeting Alk: Precision Medicine Takes on Drug Resistance." Cancer Discov 7, no. 2 (2017): 137-55. [CrossRef]
- Camidge, D. R., H. R. Kim, M. J. Ahn, J. C. Yang, J. Y. Han, J. S. Lee, M. J. Hochmair, J. Y. Li, G. C. Chang, K. H. Lee, C. Gridelli, A. Delmonte, R. Garcia Campelo, D. W. Kim, A. Bearz, F. Griesinger, A. Morabito, E. Felip, R. Califano, S. Ghosh, A. Spira, S. N. Gettinger, M. Tiseo, N. Gupta, J. Haney, D. Kerstein, and S. Popat. "Brigatinib Versus Crizotinib in Alk-Positive Non-Small-Cell Lung Cancer." N Engl J Med 379, no. 21 (2018): 2027-39. [CrossRef]
- Horn, L., Z. Wang, G. Wu, E. Poddubskaya, T. Mok, M. Reck, H. Wakelee, A. A. Chiappori, D. H. Lee, V. Breder, S. Orlov, I. Cicin, Y. Cheng, Y. Liu, Y. Fan, J. G. Whisenant, Y. Zhou, V. Oertel, K. Harrow, C. Liang, L. Mao, G. Selvaggi, and Y. L. Wu. "Ensartinib Vs Crizotinib for Patients with Anaplastic Lymphoma Kinase-Positive Non-Small Cell Lung Cancer: A Randomized Clinical Trial." JAMA Oncol 7, no. 11 (2021): 1617-25. [CrossRef]
- Camidge, D. R., H. R. Kim, M. J. Ahn, J. C. H. Yang, J. Y. Han, M. J. Hochmair, K. H. Lee, A. Delmonte, M. R. García Campelo, D. W. Kim, F. Griesinger, E. Felip, R. Califano, A. Spira, S. N. Gettinger, M. Tiseo, H. M. Lin, N. Gupta, M. J. Hanley, Q. Ni, P. Zhang, and S. Popat. "Brigatinib Versus Crizotinib in Advanced Alk Inhibitor-Naive Alk-Positive Non-Small Cell Lung Cancer: Second Interim Analysis of the Phase Iii Alta-1l Trial." J Clin Oncol 38, no. 31 (2020): 3592-603. [CrossRef]
- Popat, S., G. Liu, S. Lu, G. Song, X. Ma, and J. C. Yang. "Brigatinib Vs Alectinib in Crizotinib-Resistant Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer (Alta-3)." Future Oncol 17, no. 32 (2021): 4237-47. [CrossRef]
- Taniguchi, K., H. Konishi, A. Yoshinaga, M. Tsugane, H. Takahashi, F. Nishisaka, Y. Shishido, and A. Asai. "Efficacy of Combination Treatment Using Yho-1701, an Orally Active Stat3 Inhibitor, with Molecular-Targeted Agents on Cancer Cell Lines." Sci Rep 11, no. 1 (2021): 6685. [CrossRef]
- Camidge, D. R., H. R. Kim, M. J. Ahn, J. C. H. Yang, J. Y. Han, M. J. Hochmair, K. H. Lee, A. Delmonte, M. R. Garcia Campelo, D. W. Kim, F. Griesinger, E. Felip, R. Califano, A. I. Spira, S. N. Gettinger, M. Tiseo, H. M. Lin, Y. Liu, F. Vranceanu, H. Niu, P. Zhang, and S. Popat. "Brigatinib Versus Crizotinib in Alk Inhibitor-Naive Advanced Alk-Positive Nsclc: Final Results of Phase 3 Alta-1l Trial." J Thorac Oncol 16, no. 12 (2021): 2091-108. [CrossRef]
- Cooper, A. J., L. V. Sequist, and J. J. Lin. "Third-Generation Egfr and Alk Inhibitors: Mechanisms of Resistance and Management." Nat Rev Clin Oncol 19, no. 8 (2022): 499-514. [CrossRef]
- Shaw, A. T., B. J. Solomon, B. Besse, T. M. Bauer, C. C. Lin, R. A. Soo, G. J. Riely, S. I. Ou, J. S. Clancy, S. Li, A. Abbattista, H. Thurm, M. Satouchi, D. R. Camidge, S. Kao, R. Chiari, S. M. Gadgeel, E. Felip, and J. F. Martini. "Alk Resistance Mutations and Efficacy of Lorlatinib in Advanced Anaplastic Lymphoma Kinase-Positive Non-Small-Cell Lung Cancer." J Clin Oncol 37, no. 16 (2019): 1370-79. [CrossRef]
- Baba, K., and Y. Goto. "Lorlatinib as a Treatment for Alk-Positive Lung Cancer." Future Oncol 18, no. 24 (2022): 2745-66. [CrossRef]
- Hyman, D. M., S. A. Piha-Paul, H. Won, J. Rodon, C. Saura, G. I. Shapiro, D. Juric, D. I. Quinn, V. Moreno, B. Doger, I. A. Mayer, V. Boni, E. Calvo, S. Loi, A. C. Lockhart, J. P. Erinjeri, M. Scaltriti, G. A. Ulaner, J. Patel, J. Tang, H. Beer, S. D. Selcuklu, A. J. Hanrahan, N. Bouvier, M. Melcer, R. Murali, A. M. Schram, L. M. Smyth, K. Jhaveri, B. T. Li, A. Drilon, J. J. Harding, G. Iyer, B. S. Taylor, M. F. Berger, R. E. Cutler, Jr., F. Xu, A. Butturini, L. D. Eli, G. Mann, C. Farrell, A. S. Lalani, R. P. Bryce, C. L. Arteaga, F. Meric-Bernstam, J. Baselga, and D. B. Solit. "Her Kinase Inhibition in Patients with Her2- and Her3-Mutant Cancers." Nature 554, no. 7691 (2018): 189-94. [CrossRef]
- Harrison, P. T., S. Vyse, and P. H. Huang. "Rare Epidermal Growth Factor Receptor (Egfr) Mutations in Non-Small Cell Lung Cancer." Semin Cancer Biol 61 (2020): 167-79. [CrossRef]
- Sigismund, S., D. Avanzato, and L. Lanzetti. "Emerging Functions of the Egfr in Cancer." Mol Oncol 12, no. 1 (2018): 3-20. [CrossRef]
- Voldborg, B. R., L. Damstrup, M. Spang-Thomsen, and H. S. Poulsen. "Epidermal Growth Factor Receptor (Egfr) and Egfr Mutations, Function and Possible Role in Clinical Trials." Ann Oncol 8, no. 12 (1997): 1197-206. [CrossRef]
- Wu, S. G., and J. Y. Shih. "Management of Acquired Resistance to Egfr Tki-Targeted Therapy in Advanced Non-Small Cell Lung Cancer." Mol Cancer 17, no. 1 (2018): 38. [CrossRef]
- Yang, Z., A. Hackshaw, Q. Feng, X. Fu, Y. Zhang, C. Mao, and J. Tang. "Comparison of Gefitinib, Erlotinib and Afatinib in Non-Small Cell Lung Cancer: A Meta-Analysis." Int J Cancer 140, no. 12 (2017): 2805-19. [CrossRef]
- Cheng, H., H. Liu, Q. Du, H. Zhang, X. Zhang, Y. Wang, J. Shao, F. Yang, B. Zhang, J. Shi, Y. Liu, N. Wu, S. Xu, Q. Wei, Y. Sun, Q. Zhai, and B. Yu. "Efficacy and Safety of Domestic and Imported Gefitinib in Patients with Advanced Non-Small Cell Lung Cancer." Ann Palliat Med 10, no. 1 (2021): 10-15. [CrossRef]
- Çoban, Ö, and Z. Değim. "Development of Nanocochleates Containing Erlotinib Hcl and Dexketoprofen Trometamol and Evaluation of in Vitro Characteristic Properties." Turk J Pharm Sci 15, no. 1 (2018): 16-21. [CrossRef]
- Lynch, T. J., D. W. Bell, R. Sordella, S. Gurubhagavatula, R. A. Okimoto, B. W. Brannigan, P. L. Harris, S. M. Haserlat, J. G. Supko, F. G. Haluska, D. N. Louis, D. C. Christiani, J. Settleman, and D. A. Haber. "Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib." N Engl J Med 350, no. 21 (2004): 2129-39. [CrossRef]
- Zhong, W. Z., Q. Wang, W. M. Mao, S. T. Xu, L. Wu, Y. C. Wei, Y. Y. Liu, C. Chen, Y. Cheng, R. Yin, F. Yang, S. X. Ren, X. F. Li, J. Li, C. Huang, Z. D. Liu, S. Xu, K. N. Chen, S. D. Xu, L. X. Liu, P. Yu, B. H. Wang, H. T. Ma, J. J. Yang, H. H. Yan, X. N. Yang, S. Y. Liu, Q. Zhou, and Y. L. Wu. "Gefitinib Versus Vinorelbine Plus Cisplatin as Adjuvant Treatment for Stage Ii-Iiia (N1-N2) Egfr-Mutant Nsclc: Final Overall Survival Analysis of Ctong1104 Phase Iii Trial." J Clin Oncol 39, no. 7 (2021): 713-22. [CrossRef]
- Fu, Y., Y. Zhang, Z. Lei, T. Liu, T. Cai, A. Wang, W. Du, Y. Zeng, J. Zhu, Z. Liu, and J. A. Huang. "Abnormally Activated Opn/Integrin Avβ3/Fak Signalling Is Responsible for Egfr-Tki Resistance in Egfr Mutant Non-Small-Cell Lung Cancer." J Hematol Oncol 13, no. 1 (2020): 169. [CrossRef]
- Harvey, R. D., V. R. Adams, T. Beardslee, and P. Medina. "Afatinib for the Treatment of Egfr Mutation-Positive Nsclc: A Review of Clinical Findings." J Oncol Pharm Pract 26, no. 6 (2020): 1461-74. [CrossRef]
- Passaro, A., T. Mok, S. Peters, S. Popat, M. J. Ahn, and F. de Marinis. "Recent Advances on the Role of Egfr Tyrosine Kinase Inhibitors in the Management of Nsclc with Uncommon, Non Exon 20 Insertions, Egfr Mutations." J Thorac Oncol 16, no. 5 (2021): 764-73. [CrossRef]
- Leonetti, A., S. Sharma, R. Minari, P. Perego, E. Giovannetti, and M. Tiseo. "Resistance Mechanisms to Osimertinib in Egfr-Mutated Non-Small Cell Lung Cancer." Br J Cancer 121, no. 9 (2019): 725-37. [CrossRef]
- Ramalingam, S. S., J. Vansteenkiste, D. Planchard, B. C. Cho, J. E. Gray, Y. Ohe, C. Zhou, T. Reungwetwattana, Y. Cheng, B. Chewaskulyong, R. Shah, M. Cobo, K. H. Lee, P. Cheema, M. Tiseo, T. John, M. C. Lin, F. Imamura, T. Kurata, A. Todd, R. Hodge, M. Saggese, Y. Rukazenkov, and J. C. Soria. "Overall Survival with Osimertinib in Untreated, Egfr-Mutated Advanced Nsclc." N Engl J Med 382, no. 1 (2020): 41-50. [CrossRef]
- Remon, J., C. E. Steuer, S. S. Ramalingam, and E. Felip. "Osimertinib and Other Third-Generation Egfr Tki in Egfr-Mutant Nsclc Patients." Ann Oncol 29, no. suppl_1 (2018): i20-i27. [CrossRef]
- Fan, J., and I. A. de Lannoy. "Pharmacokinetics." Biochem Pharmacol 87, no. 1 (2014): 93-120. [CrossRef]
- Evans, W. E., and H. L. McLeod. "Pharmacogenomics--Drug Disposition, Drug Targets, and Side Effects." N Engl J Med 348, no. 6 (2003): 538-49. [CrossRef]
- Mougey, E. B., J. E. Lang, X. Wen, and J. J. Lima. "Effect of Citrus Juice and Slco2b1 Genotype on the Pharmacokinetics of Montelukast." J Clin Pharmacol 51, no. 5 (2011): 751-60. [CrossRef]
- Harvey, A. L., R. Edrada-Ebel, and R. J. Quinn. "The Re-Emergence of Natural Products for Drug Discovery in the Genomics Era." Nat Rev Drug Discov 14, no. 2 (2015): 111-29. [CrossRef]
- Xu, L., Y. Li, Y. Dai, and J. Peng. "Natural Products for the Treatment of Type 2 Diabetes Mellitus: Pharmacology and Mechanisms." Pharmacol Res 130 (2018): 451-65. [CrossRef]
- Rodrigues, T., D. Reker, P. Schneider, and G. Schneider. "Counting on Natural Products for Drug Design." Nat Chem 8, no. 6 (2016): 531-41. [CrossRef]
- Liu, X., J. M. Simon, H. Xie, L. Hu, J. Wang, G. Zurlo, C. Fan, T. S. Ptacek, L. Herring, X. Tan, M. Li, A. S. Baldwin, W. Y. Kim, T. Wu, M. W. Kirschner, K. Gong, and Q. Zhang. "Genome-Wide Screening Identifies Sfmbt1 as an Oncogenic Driver in Cancer with Vhl Loss." Mol Cell 77, no. 6 (2020): 1294-306.e5. [CrossRef]
- Pan, H., C. Xue, B. J. Auerbach, J. Fan, A. C. Bashore, J. Cui, D. Y. Yang, S. B. Trignano, W. Liu, J. Shi, C. O. Ihuegbu, E. C. Bush, J. Worley, L. Vlahos, P. Laise, R. A. Solomon, E. S. Connolly, A. Califano, P. A. Sims, H. Zhang, M. Li, and M. P. Reilly. "Single-Cell Genomics Reveals a Novel Cell State During Smooth Muscle Cell Phenotypic Switching and Potential Therapeutic Targets for Atherosclerosis in Mouse and Human." Circulation 142, no. 21 (2020): 2060-75. [CrossRef]
- Warren, N. J. H., and A. Eastman. "Comparison of the Different Mechanisms of Cytotoxicity Induced by Checkpoint Kinase I Inhibitors When Used as Single Agents or in Combination with DNA Damage." Oncogene 39, no. 7 (2020): 1389-401. [CrossRef]
- Cuevas, C., and A. Francesch. "Development of Yondelis (Trabectedin, Et-743). A Semisynthetic Process Solves the Supply Problem." Nat Prod Rep 26, no. 3 (2009): 322-37. [CrossRef]
- Italiano, A., N. Touati, S. Litière, F. Collin, P. Pourquier, and A. Gronchi. "Prospective Assessment of the Predictive Value of the Brca1 Gene Status in Sarcoma Patients Treated with Trabectedin: An Updated Analysis of the Eortc 62091 Trial." Cancer Med 7, no. 5 (2018): 1575-77. [CrossRef]
- Monk, B. J., D. Lorusso, A. Italiano, S. B. Kaye, M. Aracil, A. Tanović, and M. D'Incalci. "Trabectedin as a Chemotherapy Option for Patients with Brca Deficiency." Cancer Treat Rev 50 (2016): 175-82. [CrossRef]
- Risdon, E. N., C. H. Chau, D. K. Price, O. Sartor, and W. D. Figg. "Parp Inhibitors and Prostate Cancer: To Infinity and Beyond Brca." Oncologist 26, no. 1 (2021): e115-e29. [CrossRef]
- Yoshida, K., and Y. Miki. "Role of Brca1 and Brca2 as Regulators of DNA Repair, Transcription, and Cell Cycle in Response to DNA Damage." Cancer Sci 95, no. 11 (2004): 866-71. [CrossRef]
- Evans, D. G. "Neurofibromatosis 2." In Genereviews(®), edited by M. P. Adam, G. M. Mirzaa, R. A. Pagon, S. E. Wallace, L. J. H. Bean, K. W. Gripp and A. Amemiya. Seattle (WA): University of Washington, Seattle Copyright © 1993-2023, University of Washington, Seattle. GeneReviews is a registered trademark of the University of Washington, Seattle. All rights reserved., 1993.
- van der Valk, M. J. M., D. E. Hilling, E. Bastiaannet, E. Meershoek-Klein Kranenbarg, G. L. Beets, N. L. Figueiredo, A. Habr-Gama, R. O. Perez, A. G. Renehan, and C. J. H. van de Velde. "Long-Term Outcomes of Clinical Complete Responders after Neoadjuvant Treatment for Rectal Cancer in the International Watch & Wait Database (Iwwd): An International Multicentre Registry Study." Lancet 391, no. 10139 (2018): 2537-45. [CrossRef]
- Skiles, J. L., C. Chiang, C. H. Li, S. Martin, E. L. Smith, G. Olbara, D. R. Jones, T. A. Vik, S. Mostert, F. Abbink, G. J. Kaspers, L. Li, F. Njuguna, T. J. Sajdyk, and J. L. Renbarger. "Cyp3a5 Genotype and Its Impact on Vincristine Pharmacokinetics and Development of Neuropathy in Kenyan Children with Cancer." Pediatr Blood Cancer 65, no. 3 (2018). [CrossRef]
- Maki, K., T. Harada, Y. Akiyama, H. Ogasawara, and F. Kishi. "Giant Cell Tumor of the Rib." Intern Med 46, no. 14 (2007): 1151-2. [CrossRef]
- Egbelakin, A., M. J. Ferguson, E. A. MacGill, A. S. Lehmann, A. R. Topletz, S. K. Quinney, L. Li, K. C. McCammack, S. D. Hall, and J. L. Renbarger. "Increased Risk of Vincristine Neurotoxicity Associated with Low Cyp3a5 Expression Genotype in Children with Acute Lymphoblastic Leukemia." Pediatr Blood Cancer 56, no. 3 (2011): 361-7. [CrossRef]
- Uittenboogaard, A., C. L. G. Neutel, J. C. F. Ket, F. Njuguna, A. D. R. Huitema, G. J. L. Kaspers, and M. E. van de Velde. "Pharmacogenomics of Vincristine-Induced Peripheral Neuropathy in Children with Cancer: A Systematic Review and Meta-Analysis." Cancers (Basel) 14, no. 3 (2022). [CrossRef]
- Losuwannarak, N., A. Maiuthed, N. Kitkumthorn, A. Leelahavanichkul, S. Roytrakul, and P. Chanvorachote. "Gigantol Targets Cancer Stem Cells and Destabilizes Tumors Via the Suppression of the Pi3k/Akt and Jak/Stat Pathways in Ectopic Lung Cancer Xenografts." Cancers (Basel) 11, no. 12 (2019). [CrossRef]
- Bhummaphan, N., and P. Chanvorachote. "Gigantol Suppresses Cancer Stem Cell-Like Phenotypes in Lung Cancer Cells." Evid Based Complement Alternat Med 2015 (2015): 836564. [CrossRef]
- Losuwannarak, N., S. Roytrakul, and P. Chanvorachote. "Gigantol Targets Myc for Ubiquitin-Proteasomal Degradation and Suppresses Lung Cancer Cell Growth." Cancer Genomics Proteomics 17, no. 6 (2020): 781-93. [CrossRef]
- Verweij, J., M. Clavel, and B. Chevalier. "Paclitaxel (Taxol) and Docetaxel (Taxotere): Not Simply Two of a Kind." Ann Oncol 5, no. 6 (1994): 495-505. [CrossRef]
- McCorkle, J. R., J. W. Gorski, J. Liu, M. B. Riggs, A. B. McDowell, N. Lin, C. Wang, F. R. Ueland, and J. M. Kolesar. "Lapatinib and Poziotinib Overcome Abcb1-Mediated Paclitaxel Resistance in Ovarian Cancer." PLoS One 16, no. 8 (2021): e0254205. [CrossRef]
- van Eerden, R. A. G., L. van Doorn, F. M. de Man, N. Heersche, M. Doukas, T. P. P. van den Bosch, E. Oomen-de Hoop, P. de Bruijn, S. Bins, E. Ibrahim, S. Nikkessen, L. E. Friberg, S. L. W. Koolen, M. C. W. Spaander, and R. H. J. Mathijssen. "Tissue Type Differences in Abcb1 Expression and Paclitaxel Tissue Pharmacokinetics in Patients with Esophageal Cancer." Front Pharmacol 12 (2021): 759146. [CrossRef]
- Bhummaphan, N., N. Petpiroon, O. Prakhongcheep, B. Sritularak, and P. Chanvorachote. "Lusianthridin Targeting of Lung Cancer Stem Cells Via Src-Stat3 Suppression." Phytomedicine 62 (2019): 152932. [CrossRef]
- Wu, Q., W. Qian, X. Sun, and S. Jiang. "Small-Molecule Inhibitors, Immune Checkpoint Inhibitors, and More: Fda-Approved Novel Therapeutic Drugs for Solid Tumors from 1991 to 2021." J Hematol Oncol 15, no. 1 (2022): 143. [CrossRef]
- Nam, A. S., R. Chaligne, and D. A. Landau. "Integrating Genetic and Non-Genetic Determinants of Cancer Evolution by Single-Cell Multi-Omics." Nat Rev Genet 22, no. 1 (2021): 3-18. [CrossRef]
- Jardim, D. L., A. Goodman, D. de Melo Gagliato, and R. Kurzrock. "The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker." Cancer Cell 39, no. 2 (2021): 154-73. [CrossRef]
- Pareek, C. S., R. Smoczynski, and A. Tretyn. "Sequencing Technologies and Genome Sequencing." J Appl Genet 52, no. 4 (2011): 413-35. [CrossRef]
- Reuter, J. A., D. V. Spacek, and M. P. Snyder. "High-Throughput Sequencing Technologies." Mol Cell 58, no. 4 (2015): 586-97. [CrossRef]
- Hwang, B., J. H. Lee, and D. Bang. "Single-Cell Rna Sequencing Technologies and Bioinformatics Pipelines." Exp Mol Med 50, no. 8 (2018): 1-14. [CrossRef]
- Ngamcherdtrakul, W., and W. Yantasee. "Sirna Therapeutics for Breast Cancer: Recent Efforts in Targeting Metastasis, Drug Resistance, and Immune Evasion." Transl Res 214 (2019): 105-20. [CrossRef]
- Mu, Q., J. Yu, L. A. McConnachie, J. C. Kraft, Y. Gao, G. K. Gulati, and R. J. Y. Ho. "Translation of Combination Nanodrugs into Nanomedicines: Lessons Learned and Future Outlook." J Drug Target 26, no. 5-6 (2018): 435-47. [CrossRef]
- Stuver, R., G. L. Shah, N. S. Korde, L. E. Roeker, A. R. Mato, C. L. Batlevi, D. J. Chung, S. Doddi, L. Falchi, B. Gyurkocza, A. Hamilton, Y. H. Lin, A. A. Jakubowski, E. Joffe, H. L. Landau, R. J. Lin, S. Mailankody, M. L. Palomba, J. H. Park, M. A. Perales, D. M. Ponce, L. V. Ramanathan, G. A. Salles, M. Scordo, S. K. Seo, U. A. Shah, E. M. Stein, D. Straus, S. Z. Usmani, J. W. Young, A. D. Zelenetz, A. Noy, and S. A. Vardhana. "Activity of Azd7442 (Tixagevimab-Cilgavimab) against Omicron Sars-Cov-2 in Patients with Hematologic Malignancies." Cancer Cell 40, no. 6 (2022): 590-91. [CrossRef]
- Cilento, M. E., Y. T. Ong, P. R. Tedbury, and S. G. Sarafianos. "Drug Interactions in Lenacapavir-Based Long-Acting Antiviral Combinations." Viruses 14, no. 6 (2022). [CrossRef]
- Liang, Z., Y. He, and X. Hu. "Cardio-Oncology: Mechanisms, Drug Combinations, and Reverse Cardio-Oncology." Int J Mol Sci 23, no. 18 (2022). [CrossRef]
- McDonald, E. S., A. S. Clark, J. Tchou, P. Zhang, and G. M. Freedman. "Clinical Diagnosis and Management of Breast Cancer." J Nucl Med 57 Suppl 1 (2016): 9s-16s. [CrossRef]
- Ho, D., S. R. Quake, E. R. B. McCabe, W. J. Chng, E. K. Chow, X. Ding, B. D. Gelb, G. S. Ginsburg, J. Hassenstab, C. M. Ho, W. C. Mobley, G. P. Nolan, S. T. Rosen, P. Tan, Y. Yen, and A. Zarrinpar. "Enabling Technologies for Personalized and Precision Medicine." Trends Biotechnol 38, no. 5 (2020): 497-518. [CrossRef]
- Litman, T. "Personalized Medicine-Concepts, Technologies, and Applications in Inflammatory Skin Diseases." Apmis 127, no. 5 (2019): 386-424. [CrossRef]
- Botella, P., and E. Rivero-Buceta. "Safe Approaches for Camptothecin Delivery: Structural Analogues and Nanomedicines." J Control Release 247 (2017): 28-54. [CrossRef]
- Thomford, N. E., D. A. Senthebane, A. Rowe, D. Munro, P. Seele, A. Maroyi, and K. Dzobo. "Natural Products for Drug Discovery in the 21st Century: Innovations for Novel Drug Discovery." Int J Mol Sci 19, no. 6 (2018). [CrossRef]
| Technology | Principle | Advantage | Disadvantage |
| AfBPP | Affinity of target proteins to active small molecules on stationary phases | 1. No bias; 2. Systematic study of total protein; 3. It can enrich the target and is suitable for identification of low abundance proteins. |
1. A detailed understanding of the structure-activity relationship of active molecules is required; 2. Chemical derivatization of active molecules is required; 3. Targets with low abundance and low affinity are easy to be washed off; 4. Probes usually cannot enter cells. |
| ABPP | The target protein forms a covalent bond with a covalent small molecule. | 1. No bias; 2. Systematic study of whole protein; 3. It can enrich the target and is suitable for identification of low abundance proteins; Grasp low affinity targets; 4. Probes usually get into cells. |
1. A thorough understanding of the structure-activity relationship of active molecules is required; 2. Chemical derivatization of active molecules is required; 3. Non-specific covalent binding is easy to occur. |
| TPP | The thermal stability of the target protein increases after binding with small molecules and it is not easy to precipitate | 1. No bias; 2. Systematic study of whole protein; 3. No derivations of small active molecules are required. |
1. Limited effect on extreme conditions, such as heat insensitivity or heat unstable proteins; 2. Further measures should be taken to reduce the complexity of samples so as to realize the identification of low abundance proteins. |
| DARTS | The stability of the target protein increases after binding with small molecules and is not easily degraded by enzymes | 1. No bias; 2. Systematic study of whole protein; 3. No derivations of small active molecules are required. |
1. The protein that is not sensitive to enzyme digestion has limited effect; 2. Further measures should be taken to reduce the complexity of samples so as to realize the identification of low abundance proteins. |
| Seq_ID | Medicine | Personalized tag | Approval time | Molecular formula | Mechanism of action | Disease |
| 1 | Crizotinib | ALK+ | 2014 | 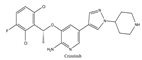 |
ALK inhibitor | Metastatic non-small cell lung cancer with ALK or ROS1 positive |
| 2 | Ceritinib | ALK+ | 2014 | 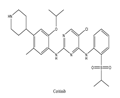 |
ALK inhibitor | Non-small-cell lung cancer |
| 3 | Alectinib | ALK+ | 2015 | 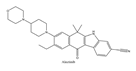 |
ALK inhibitor | Non-small-cell lung cancer |
| 4 | Brigatinib | ALK+ | 2017 | 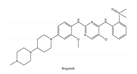 |
ALK inhibitor | Non-small-cell lung cancer |
| 5 | Lorlatinib | ALK+ is positive | 2018 | 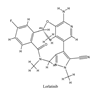 |
A dual-target inhibitoror of ALK/ROS1 | Non-small-cell lung cancer |
| 6 | Gefitinib | EGFR | 2003 | 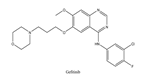 |
EGFR inhibitor | Non-small-cell lung cancer |
| 7 | Erlotinib | EGFR | 2004 | 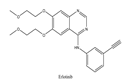 |
EGFR inhibitor | Non-small-cell lung cancer |
| 8 | Afatinib | EGFR | 2013 | 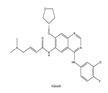 |
EGFR inhibitor | Non-small-cell lung cancer |
| 9 | Osimertinib | EGFR | 2015 | 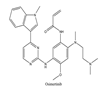 |
EGFR inhibitor | Non-small-cell lung cancer |
| Seq_ID | Natural products | Main Sources | Molecular formula | Related Gene | Disease |
| 1 | Trabectedin | Ecteinascidia turbinata | 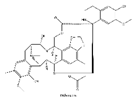 |
BRCA1, BRCA2 | Soft tissue sarcoma, Breast cancer |
| 2 | Vincristine | Catharanthus roseus | 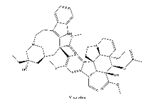 |
CYP3A enzymes, ABC transporters |
Leukemias, Lymphomas, Brain tumors, Solid tumors |
| 5 | Paclitaxel | Taxus baccata Linn | 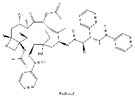 |
ABCB1 G2677T/A mutation | Ovarian cancer |
| 3 | Gigantol | Dendrobium draconis | 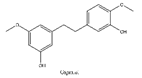 |
CD133, ALDH1A1 | Non-small-cell lung cancer |
| 6 | Chrysotoxine | Dendrobium pulchellum | 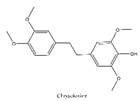 |
ABCG2 | Lung cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





