Submitted:
22 April 2023
Posted:
24 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Results
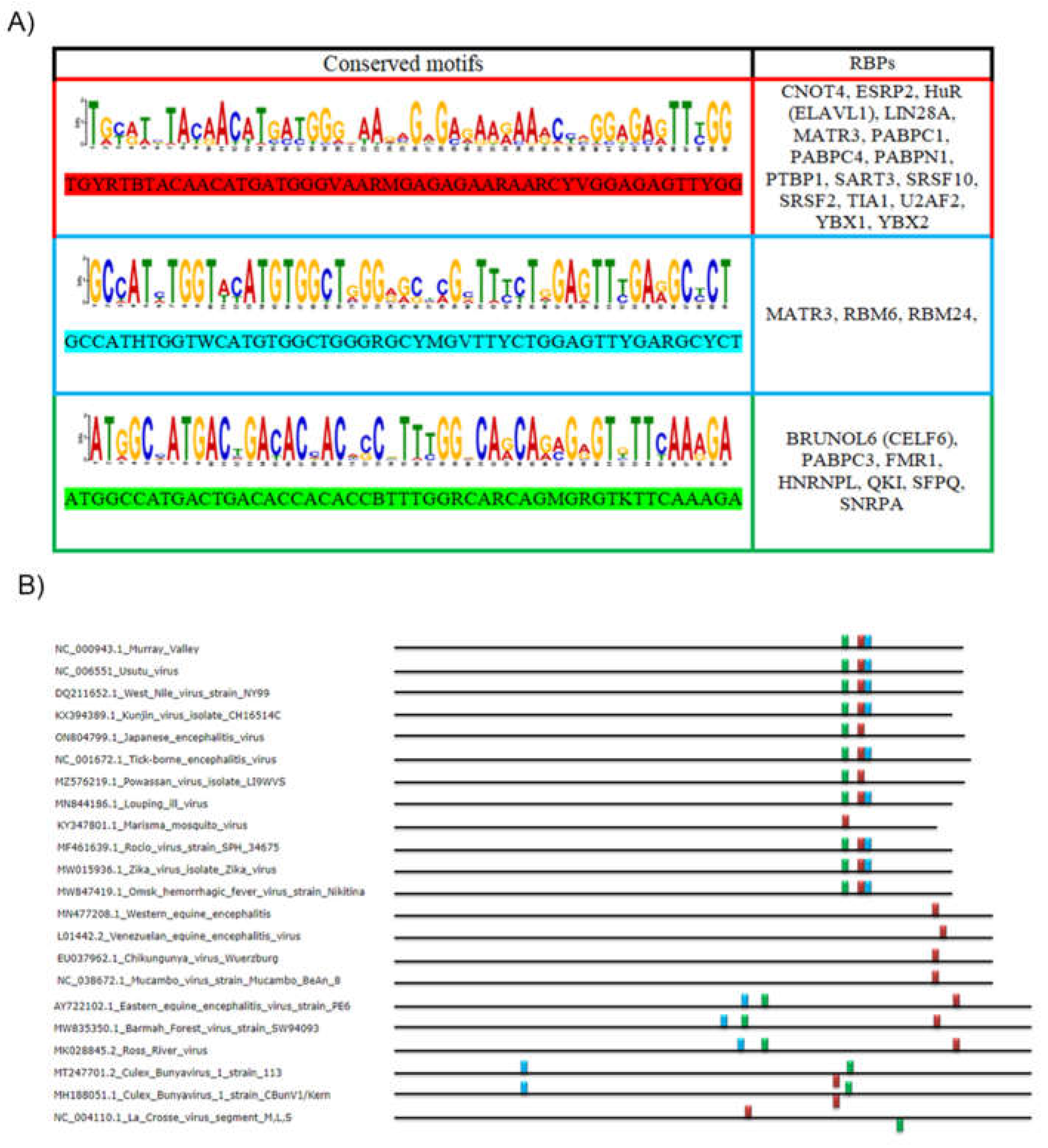
2.1. Analysis of the First Motif (Red Motif)
2.2. Analysis of the Second Motif (Green Motif)
2.3. Analysis of the Third Motif (Light Blue Motif)
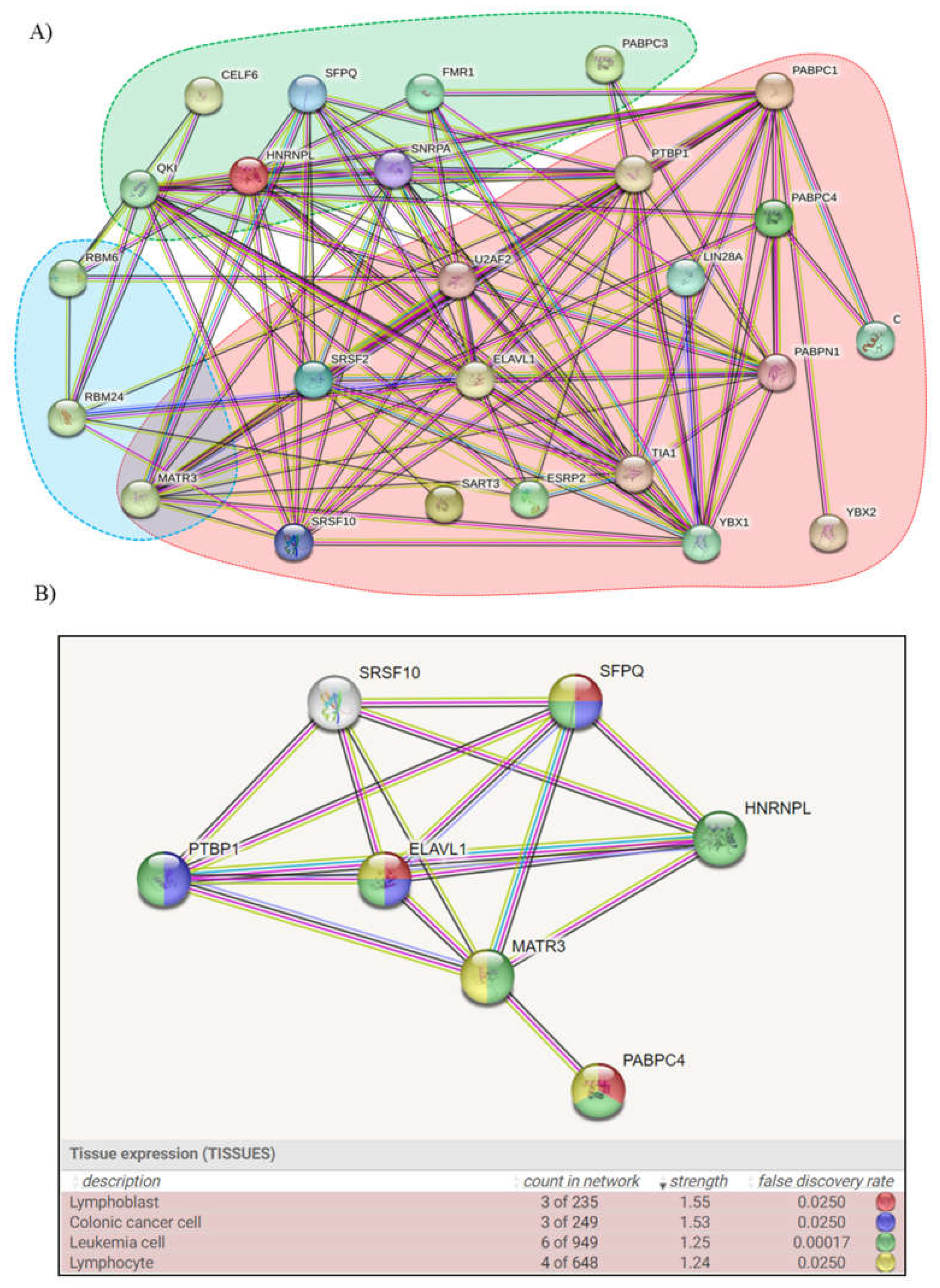
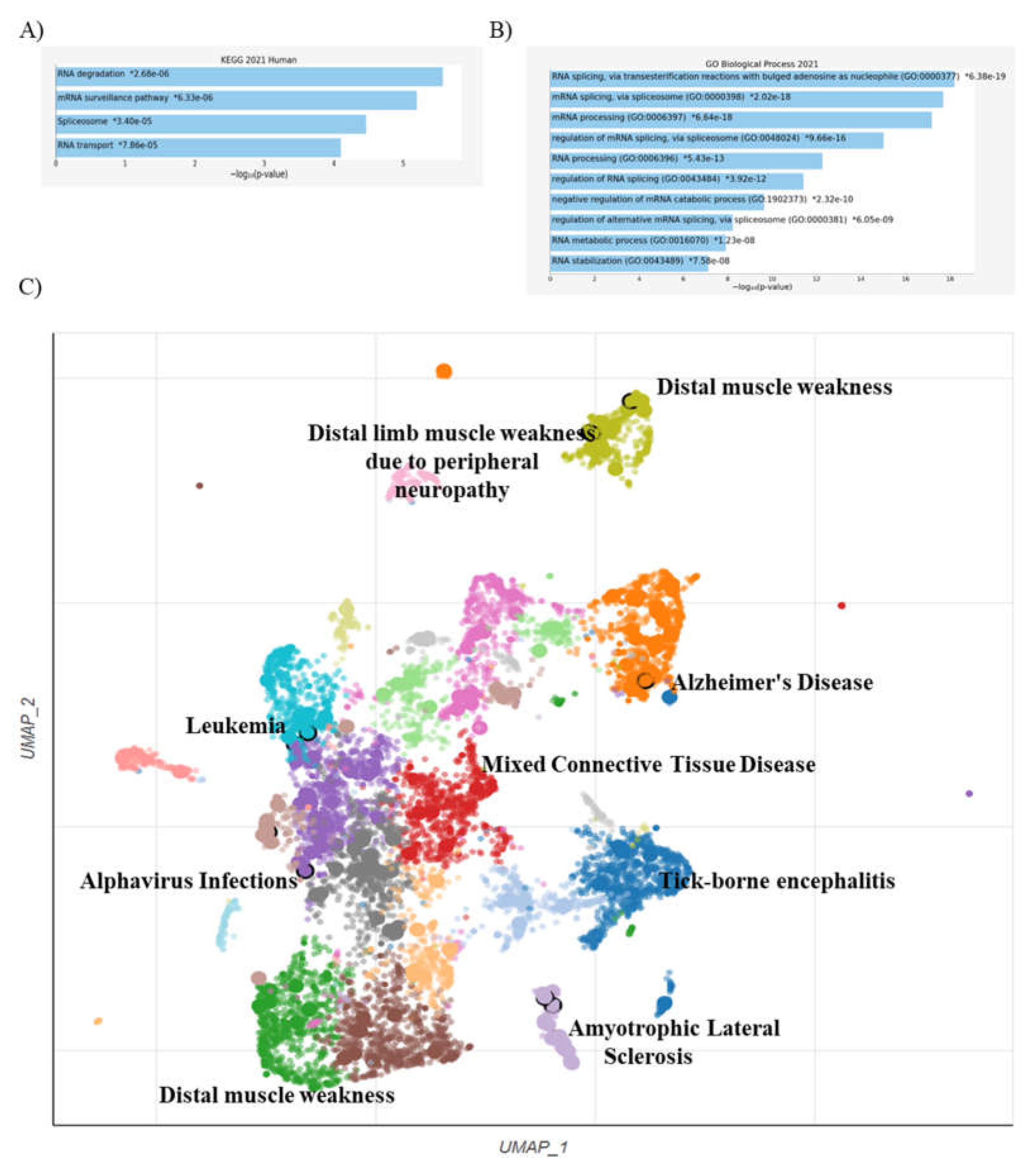
2.4. Enriched Analysis
3. Discussion
4. Materials and Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F.; et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2021, 20, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Sohrabi, F.; Saeidifard, S.; Ghasemi, M.; Asadishad, T.; Hamidi, S.M.; Hosseini, S.M. Role of plasmonics in detection of deadliest viruses: a review. Eur. Phys. J. Plus 2021, 136, 1–71. [Google Scholar] [CrossRef] [PubMed]
- Rossi, B.; Barreca, F.; Benvenuto, D.; Braccialarghe, N.; Campogiani, L.; Lodi, A.; Aguglia, C.; Cavasio, R.A.; Giacalone, M.L.; Kontogiannis, D.; et al. Human Arboviral Infections in Italy: Past, Current, and Future Challenges. Viruses 2023, 15, 368. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, R.; Brown, D.T.; Paredes, A. Structural Differences Observed in Arboviruses of the Alphavirus and Flavivirus Genera. Adv. Virol. 2014, 2014, 1–24. [Google Scholar] [CrossRef]
- Mori, Y.; Otsuki, N.; Sakata, M.; Okamoto, K. Virology of the Family Togaviridae. Uirusu 2011, 61, 211–220. [Google Scholar] [CrossRef]
- Elmasri, Z.; Nasal, B.L.; Jose, J. Alphavirus-Induced Membrane Rearrangements during Replication, Assembly, and Budding. Pathogens 2021, 10, 984. [Google Scholar] [CrossRef]
- Rupp, J.C.; Sokoloski, K.J.; Gebhart, N.N.; Hardy, R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015, 96, 2483–2500. [Google Scholar] [CrossRef]
- Azar, S.R.; Campos, R.K.; Bergren, N.A.; Camargos, V.N.; Rossi, S.L. Epidemic Alphaviruses: Ecology, Emergence and Outbreaks. Microorganisms 2020, 8, 1167. [Google Scholar] [CrossRef]
- Diosa-Toro M, Urcuqui-Inchima S, Smit JM. Arthropod-borne flaviviruses and RNA interference: seeking new approaches for antiviral therapy. Adv Virus Res. 2013, 85, 91–111. [CrossRef]
- Kuno, G.; Chang, G.-J.J.; Tsuchiya, K.R.; Karabatsos, N.; Cropp, C.B. Phylogeny of the Genus Flavivirus. J. Virol. 1998, 72, 73–83. [Google Scholar] [CrossRef]
- Ng, W.C.; Soto-Acosta, R.; Bradrick, S.S.; Garcia-Blanco, M.A.; Ooi, E.E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 2017, 9, 137. [Google Scholar] [CrossRef]
- Perera-Lecoin, M.; Meertens, L.; Carnec, X.; Amara, A. Flavivirus Entry Receptors: An Update. Viruses 2013, 6, 69–88. [Google Scholar] [CrossRef]
- Amor, S. Virus Infections of the Central Nervous System. Manson’s Tropical Diseases. 2009;853-883. [CrossRef]
- de Vries, L.; Harding, A.T. Mechanisms of Neuroinvasion and Neuropathogenesis by Pathologic Flaviviruses. Viruses 2023, 15, 261. [Google Scholar] [CrossRef] [PubMed]
- Braack, L.; De Almeida, A.P.G.; Cornel, A.J.; Swanepoel, R.; De Jager, C. Mosquito-borne arboviruses of African origin: review of key viruses and vectors. Parasites Vectors 2018, 11, 1–26. [Google Scholar] [CrossRef]
- Labuda, M. Arthropod vectors in the evolution of bunyaviruses. . 1991, 35, 98–105. [Google Scholar] [PubMed]
- Liu, J.; Swevers, L.; Kolliopoulou, A.; Smagghe, G. Arboviruses and the Challenge to Establish Systemic and Persistent Infections in Competent Mosquito Vectors: The Interaction With the RNAi Mechanism. Front. Physiol. 2019, 10, 890. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Noble, W.S.; Bailey, T.L. Motif-based analysis of large nucleotide data sets using MEME-ChIP. Nat. Protoc. 2014, 9, 1428–1450. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Stamatoyannopoulos, J.A.; Bailey, T.L.; Noble, W.S. Quantifying similarity between motifs. Genome Biol. 2007, 8, R24. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2022, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Goss, D.J.; Kleiman, F.E. Poly(A) binding proteins: are they all created equal? Wiley Interdiscip. Rev. RNA 2012, 4, 167–179. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Yuan, J.; Zhang, Z.; Chang, X. Cytoplasmic poly(A)-binding protein 1 (PABPC1) interacts with the RNA-binding protein hnRNPLL and thereby regulates immunoglobulin secretion in plasma cells. J. Biol. Chem. 2017, 292, 12285–12295. [Google Scholar] [CrossRef] [PubMed]
- Lemay, J.-F.; Lemieux, C.; St-André, O.; Bachand, F. Crossing the borders: Poly(A)-binding proteins working on both sides of the fence. RNA Biol. 2010, 7, 291–295. [Google Scholar] [CrossRef]
- Kini, H.K.; Kong, J.; Liebhaber, S.A. Cytoplasmic Poly(A) Binding Protein C4 Serves a Critical Role in Erythroid Differentiation. Mol. Cell. Biol. 2014, 34, 1300–1309. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Peng, Q.; Wang, L.; Zhang, X.; Huang, L.; Wang, J.; Qin, Z. Serine/arginine-rich splicing factors: the bridge linking alternative splicing and cancer. Int. J. Biol. Sci. 2020, 16, 2442–2453. [Google Scholar] [CrossRef]
- Elmén, L.; Volpato, C.B.; Kervadec, A.; Pineda, S.; Kalvakuri, S.; Alayari, N.N.; Foco, L.; Pramstaller, P.P.; Ocorr, K.; Rossini, A.; et al. Silencing of CCR4-NOT complex subunits affect heart structure and function. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef]
- Ajam, M.; A Abu-Heija, A.; Shokr, M.; Ajam, F.; Saydain, G. Sinus Bradycardia and QT Interval Prolongation in West Nile Virus Encephalitis: A Case Report. Cureus 2019, 11, e3821. [Google Scholar] [CrossRef]
- David, G.; Reboutier, D.; Deschamps, S.; Méreau, A.; Taylor, W.; Padilla-Parra, S.; Tramier, M.; Audic, Y.; Paillard, L. The RNA-binding proteins CELF1 and ELAVL1 cooperatively control the alternative splicing of CD44. Biochem. Biophys. Res. Commun. 2022, 626, 79–84. [Google Scholar] [CrossRef]
- Wu, K.; Ahmad, T.; Eri, R. LIN28A: A multifunctional versatile molecule with future therapeutic potential. World J. Biol. Chem. 2022, 13, 35–46. [Google Scholar] [CrossRef]
- Bantle, C.M.; Phillips, A.T.; Smeyne, R.J.; Rocha, S.M.; Olson, K.E.; Tjalkens, R.B. Infection with mosquito-borne alphavirus induces selective loss of dopaminergic neurons, neuroinflammation and widespread protein aggregation. npj Park. Dis. 2019, 5, 1–15. [Google Scholar] [CrossRef]
- Malik, A.M.; Barmada, S.J. Matrin 3 in neuromuscular disease: physiology and pathophysiology. J. Clin. Investig. 2021, 6. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, Z.; Ma, L.; Huang, X.; Zhan, Q.; Song, Y. PTBP1 promotes IRES-mediated translation of cyclin B1in cancer. Acta Biochim. et Biophys. Sin. 2022, 54, 696–707. [Google Scholar] [CrossRef]
- Vavougios, G.D. SARS-CoV-2 dysregulation of PTBP1 and YWHAE/Z gene expression: A primer of neurodegeneration. Med Hypotheses 2020, 144, 110212–110212. [Google Scholar] [CrossRef] [PubMed]
- Long, L.; Thelen, J.P.; Furgason, M.; Haj-Yahya, M.; Brik, A.; Cheng, D.; Peng, J.; Yao, T. The U4/U6 Recycling Factor SART3 Has Histone Chaperone Activity and Associates with USP15 to Regulate H2B Deubiquitination. J. Biol. Chem. 2014, 289, 8916–8930. [Google Scholar] [CrossRef] [PubMed]
- Sherman, E.J.; Mitchell, D.C.; Garner, A.L. The RNA-binding protein SART3 promotes miR-34a biogenesis and G1 cell cycle arrest in lung cancer cells. J. Biol. Chem. 2019, 294, 17188–17196. [Google Scholar] [CrossRef] [PubMed]
- Velasco, B.R.; Izquierdo, J.M. T-Cell Intracellular Antigen 1-Like Protein in Physiology and Pathology. Int. J. Mol. Sci. 2022, 23, 7836. [Google Scholar] [CrossRef]
- Schott, G.; Galarza-Muñoz, G.; Trevino, N.; Chen, X.; Weirauch, M.T.; Gregory, S.G.; Bradrick, S.S.; Garcia-Blanco, M.A. U2AF2 binds IL7R exon 6 ectopically and represses its inclusion. RNA 2021, 27, 571–583. [Google Scholar] [CrossRef]
- Ishii, H.; Saitoh, M.; Sakamoto, K.; Kondo, T.; Katoh, R.; Tanaka, S.; Motizuki, M.; Masuyama, K.; Miyazawa, K. Epithelial Splicing Regulatory Proteins 1 (ESRP1) and 2 (ESRP2) Suppress Cancer Cell Motility via Different Mechanisms. J. Biol. Chem. 2014, 289, 27386–27399. [Google Scholar] [CrossRef]
- Gupta, K.; Metgud, R. Evidences Suggesting Involvement of Viruses in Oral Squamous Cell Carcinoma. Pathol. Res. Int. 2013, 2013, 1–17. [Google Scholar] [CrossRef]
- Diosa-Toro, M.; Kennedy, D.R.; Chuo, V.; Popov, V.L.; Pompon, J.; Garcia-Blanco, M.A. Y-Box Binding Protein 1 Interacts with Dengue Virus Nucleocapsid and Mediates Viral Assembly. Mbio 2022, 13, e0019622. [Google Scholar] [CrossRef]
- Aliakbari, F.; Eshghifar, N.; Mirfakhraie, R.; Pourghorban, P.; Azizi, F. Coding and Non-Coding RNAs, as Male Fertility and Infertility Biomarkers. 15. [CrossRef]
- Kuassivi, O.N.; Abiven, H.; Satie, A.-P.; Cartron, M.; Mahé, D.; Aubry, F.; Mathieu, R.; Rebours, V.; Le Tortorec, A.; Dejucq-Rainsford, N. Human Testicular Germ Cells, a Reservoir for Zika Virus, Lack Antiviral Response Upon Zika or Poly(I:C) Exposure. Front. Immunol. 2022, 13, 909341. [Google Scholar] [CrossRef] [PubMed]
- Adinolfi, S.; Bagni, C.; Musco, G.; Gibson, T.; Mazzarella, L.; Pastore, A. Dissecting FMR1, the protein responsible for fragile X syndrome, in its structural and functional domains. RNA 1999, 5, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Katoh, H.; Mori, Y.; Kambara, H.; Abe, T.; Fukuhara, T.; Morita, E.; Moriishi, K.; Kamitani, W.; Matsuura, Y. Heterogeneous Nuclear Ribonucleoprotein A2 Participates in the Replication of Japanese Encephalitis Virus through an Interaction with Viral Proteins and RNA. J. Virol. 2011, 85, 10976–10988. [Google Scholar] [CrossRef]
- Liao, K.-C.; Chuo, V.; Ng, W.C.; Neo, S.P.; Pompon, J.; Gunaratne, J.; Ooi, E.E.; Garcia-Blanco, M.A. Identification and characterization of host proteins bound to dengue virus 3′ UTR reveal an antiviral role for quaking proteins. RNA 2018, 24, 803–814. [Google Scholar] [CrossRef] [PubMed]
- Lim, Y.W.; James, D.; Huang, J.; Lee, M. The Emerging Role of the RNA-Binding Protein SFPQ in Neuronal Function and Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 7151. [Google Scholar] [CrossRef]
- Yuan, M.; Yu, C.; Chen, X.; Wu, Y. Investigation on Potential Correlation Between Small Nuclear Ribonucleoprotein Polypeptide A and Lung Cancer. Front. Genet. 2021, 11. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, F.; Zhong, W.; Ling, H.; Wang, J.; Cui, J.; Xie, T.; Wen, S.; Chen, J. RNA-binding protein RBM6 as a tumor suppressor gene represses the growth and progression in laryngocarcinoma. Gene 2019, 697, 26–34. [Google Scholar] [CrossRef]
- Grifone, R.; Saquet, A.; Desgres, M.; Sangiorgi, C.; Gargano, C.; Li, Z.; Coletti, D.; Shi, D.-L. Rbm24 displays dynamic functions required for myogenic differentiation during muscle regeneration. Sci. Rep. 2021, 11, 1–15. [Google Scholar] [CrossRef]
- Baker, R.E.; Mahmud, A.S.; Miller, I.F.; Rajeev, M.; Rasambainarivo, F.; Rice, B.L.; Takahashi, S.; Tatem, A.J.; Wagner, C.E.; Wang, L.-F.; et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 2021, 20, 193–205. [Google Scholar] [CrossRef]
- Hassell, J.M.; Begon, M.; Ward, M.J.; Fèvre, E.M. Urbanization and Disease Emergence: Dynamics at the Wildlife–Livestock–Human Interface. Trends Ecol. Evol. 2016, 32, 55–67. [Google Scholar] [CrossRef]
- Ellwanger, J.H.; Chies, J.A.B. Zoonotic spillover: Understanding basic aspects for better prevention. Genet. Mol. Biol. 2021, 44, e20200355. [Google Scholar] [CrossRef]
- Dafale, N.A.; Srivastava, S.; Purohit, H.J. Zoonosis: An Emerging Link to Antibiotic Resistance Under “One Health Approach”. Indian J. Microbiol. 2020, 60, 139–152. [Google Scholar] [CrossRef]
- Singh, K.; Mehta, D.; Dumka, S.; Chauhan, A.S.; Kumar, S. Quasispecies Nature of RNA Viruses: Lessons from the Past. Vaccines 2023, 11, 308. [Google Scholar] [CrossRef]
- Domingo, E.; Sheldon, J.; Perales, C. Viral Quasispecies Evolution. Microbiol. Mol. Biol. Rev. 2012, 76, 159–216. [Google Scholar] [CrossRef]
- Domingo E, Escarmís C, Menéndez-Arias L, et al. Viral Quasispecies: Dynamics, Interactions, and Pathogenesis. Origin and Evolution of Viruses. 2008;87-118. [CrossRef]
- Peck, K.M.; Lauring, A.S. Complexities of Viral Mutation Rates. J. Virol. 2018, 92. [Google Scholar] [CrossRef]
- Almehdi, A.M.; Khoder, G.; Alchakee, A.S.; Alsayyid, A.T.; Sarg, N.H.; Soliman, S.S.M. SARS-CoV-2 spike protein: pathogenesis, vaccines, and potential therapies. Infection 2021, 49, 855–876. [Google Scholar] [CrossRef] [PubMed]
- Pesti, R.; Kontra, L.; Paul, K.; Vass, I.; Csorba, T.; Havelda, Z.; Várallyay. Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLOS ONE 2019, 14, e0216618. [Google Scholar] [CrossRef] [PubMed]
- E Randall, R.; E Griffin, D. Within host RNA virus persistence: mechanisms and consequences. Curr. Opin. Virol. 2017, 23, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, S.; Federspiel, J.D.; Hartenian, E.; Cristea, I.M.; Glaunsinger, B. Changes in mRNA abundance drive shuttling of RNA binding proteins, linking cytoplasmic RNA degradation to transcription. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Chetta, M.; Tarsitano, M.; Oro, M.; Rivieccio, M.; Bukvic, N. An in silico pipeline approach uncovers a potentially intricate network involving spike SARS-CoV-2 RNA, RNA vaccines, host RNA-binding proteins (RBPs), and host miRNAs at the cellular level. J. Genet. Eng. Biotechnol. 2022, 20, 1–11. [Google Scholar] [CrossRef]
- Lundstrom, K. Alphaviruses in Immunotherapy and Anticancer Therapy. Biomedicines 2022, 10, 2263. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




