1. Introduction
A common European Health Data Space (EHDS) is European Commission’s key initiative that aims to improve interoperability and facilitate the exchange and access of health data within the EU. One of the main goals of the EHDS is to provide a common framework and governance model for the health data use by patients and other purposes such as policymaking, research, and regulation. This includes the development of common standards and protocols for the exchange of electronic health records (EHRs), as well as the creation of a secure infrastructure for the storage and access of different types of health data such as data from EHRs, patient registries and genomics data. [
1,
2,
3]
A key feature of EHDS legislative proposal is ensuring health data interoperability [
4]. The European Interoperability Framework (EIF) is a set of guidelines and recommendations that have been developed by the European Commission to support the development of interoperable digital services across the European Union (EU). The EIF provides a common set of principles and guidance for the design and development of interoperable digital services. [
5] Semantic interoperability describes the unambiguous representation of clinical concepts by use of international standard reference systems and ontologies. Semantic interoperability is mentioned in the legislative proposal, however without providing specifics. [
4,
6]
Interoperable EHRs helps health care practitioners gather, store, and communicate essential health information reliably and securely across care settings to guarantee coordinated and patient-centered care while creating many efficiencies in the delivery of health care [
7]. Utilization possibilities of these data are beyond patient care. EHRs use health-related information pertinent to an individual patient whereas registries are mainly focused on population management and are designed to obtain information on predefined health outcomes data [
8]. Also, public health systems have realized benefits of EHR data. EHR-based surveillance has the potential to provide routine public health surveillance for nonreportable chronic conditions that is timely, actionable, and sustainable and to complement existing disease surveillance systems. [
9]
To achieve a fully integrated EHDS ecosystem leveraging the value of health data, all stakeholders need to overcome many challenges, including implementing common digital health standards and interoperability frameworks. Recent study recommends investing in high standardization of data and interoperability for building EHDS [
3]. Interoperability between EHRs is also a core goal of the standards established by the Office of the National Coordinator for Health Information Technology [
10]. Without applying appropriate domain-relevant content standards such as applicable terminologies, interoperability is hindered by the need for manual transformation and mapping. These obstacles to interoperability limit the accessibility and reusability of data, thus diminishing its value [
11]. However, the relationship between existing international data interoperability standards and possible new specifications suggested by the Commission may create some uncertainties in development work [
12]. Solutions will have to be found to remedy inconsistencies at different levels of interoperability [
13]. Interoperability is required to enhance the quality, efficiency, and effectiveness of the healthcare system by providing information in the appropriate format whenever and wherever it is needed by eliminating unnecessary replication [14.]. Therefore, our study aims to provide the readers with up-to-date information about the different types of approaches to resolve semantic interoperability and to summarize benefits of these choices.
We aimed to research what evidence literature gives about developing semantic interoperability in EHRs to enhance EHDS goals. Our research questions are as follows: What are the key elements and approaches of building semantic interoperability integrated in EHRs? What kind of goals are driving the development? What kind of clinical benefits are perceived following this development?
2. Materials and Methods
2.1. Methodological Framework
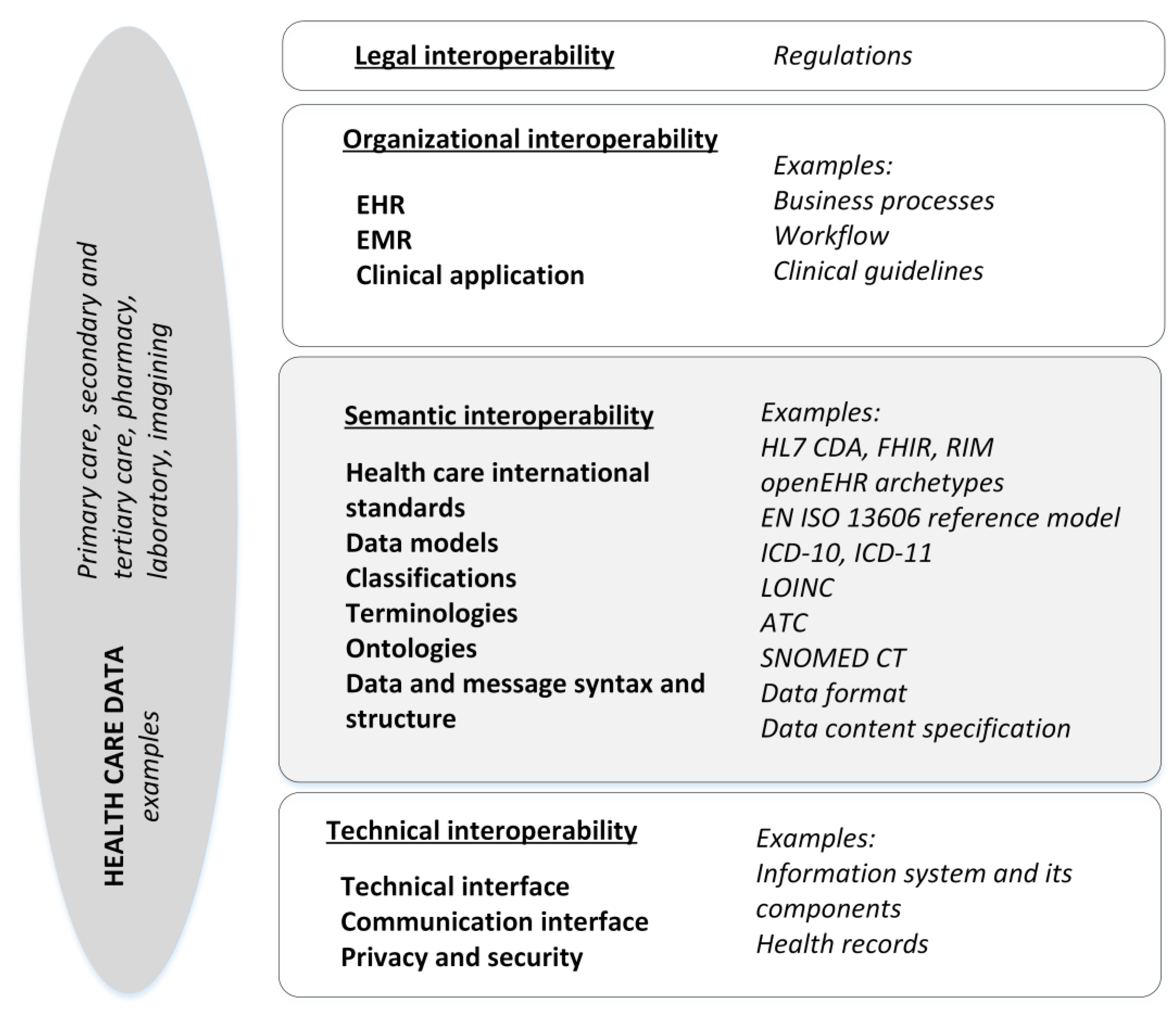
With our research questions as a starting point, we set out to perform a systematic literature review of semantic interoperability. First, we defined our core concepts to refine our literature search strategy. The scope of the review was semantic interoperability, i.e., organizational, legal, and technical interoperability were excluded [
5]. Semantic interoperability was apprehended based on EIF, in which semantic interoperability covers both semantic and syntactic aspects. The semantic aspect refers to the meaning of data elements and their relationships, while the syntactic refers to the format of the information to be exchanged. With semantic interoperability, it is ensured that the data can be shared between organizations and systems in a way that the meaning of data does not change. [
5,
15,
16] However, there are several ways of analyzing interoperability layers [
15]. For example, in comparison to European approach [
5], the Healthcare Information and Management Systems Society HIMSS defines three levels of interoperability for health care technology: foundation, structural and semantic [
16]. Since the EHDS is the point of departure for our review, we chose the EIF definitions that primarily guide our review framework as illustrated in
Figure 1.
In the
Figure 1, processing, storing, and exchanging health care data in EHRs and between EHRs or other clinical applications is, for example, governed and regulated at the legal layer, arranged as processes and workflows at organizational interoperability layer, resolved at the technical layer while considering the implementation of the principles of data protection and information security. The semantic interoperability layer covers various approaches to resolve interoperability issues, such as more established international or domain specific health care classifications, clinical terminologies, and ontologies and application of international standards for EHRs. Based on EIF, the semantic interoperability covers also syntactic features, such as data format and, for example, structured data content. We identified these key features of semantic interoperability based on previous research [
6,
14,
15,
16,
17]. In our framework, data model is a generic concept that describes various applications of data models from a reference information model to clinical information model. Data models define structure and semantics for storing, exchanging, querying, and processing health care data. Clinical information models can be implemented in an EHR, for example, as archetypes and templates, whereas reference information models refer to standard based approaches to enable health care documentation and messages such as Health Level 7 (HL7) reference information model (RIM) or International Organization for Standards’ ISO 13606 standard for Electronic Health Record Communication. [
16,
18] When designing EHRs, for semantic interoperability, a dual-level method can be applied to represent both information and knowledge levels of interoperability requirements, properties and structures for data. This approach is used, for example, for representing the dual levels of knowledge by an archetype model and information structures by the chosen information reference model. [
14,
17,
18]
2.2. Study Design
In the design of the review, we applied Cochrane review protocol [
19] to add scientific reliability and validity of our review. The search strategy (see
Table 1) was defined based on the framework for semantic interoperability presented in
Figure 1. We made additional test searches with concept “European Health Data Space”, “data space”, “EHDS”, and “European commission” without any results. However, search with “EU” resulted in 2 articles that were already included in the original results. We performed the search in PubMed database in December 2022. To conduct a systematic literature review, PubMed is regarded as a comprehensive database [
20]. Therefore, no further data searches were performed. We documented the search so that it can be reproduced (see
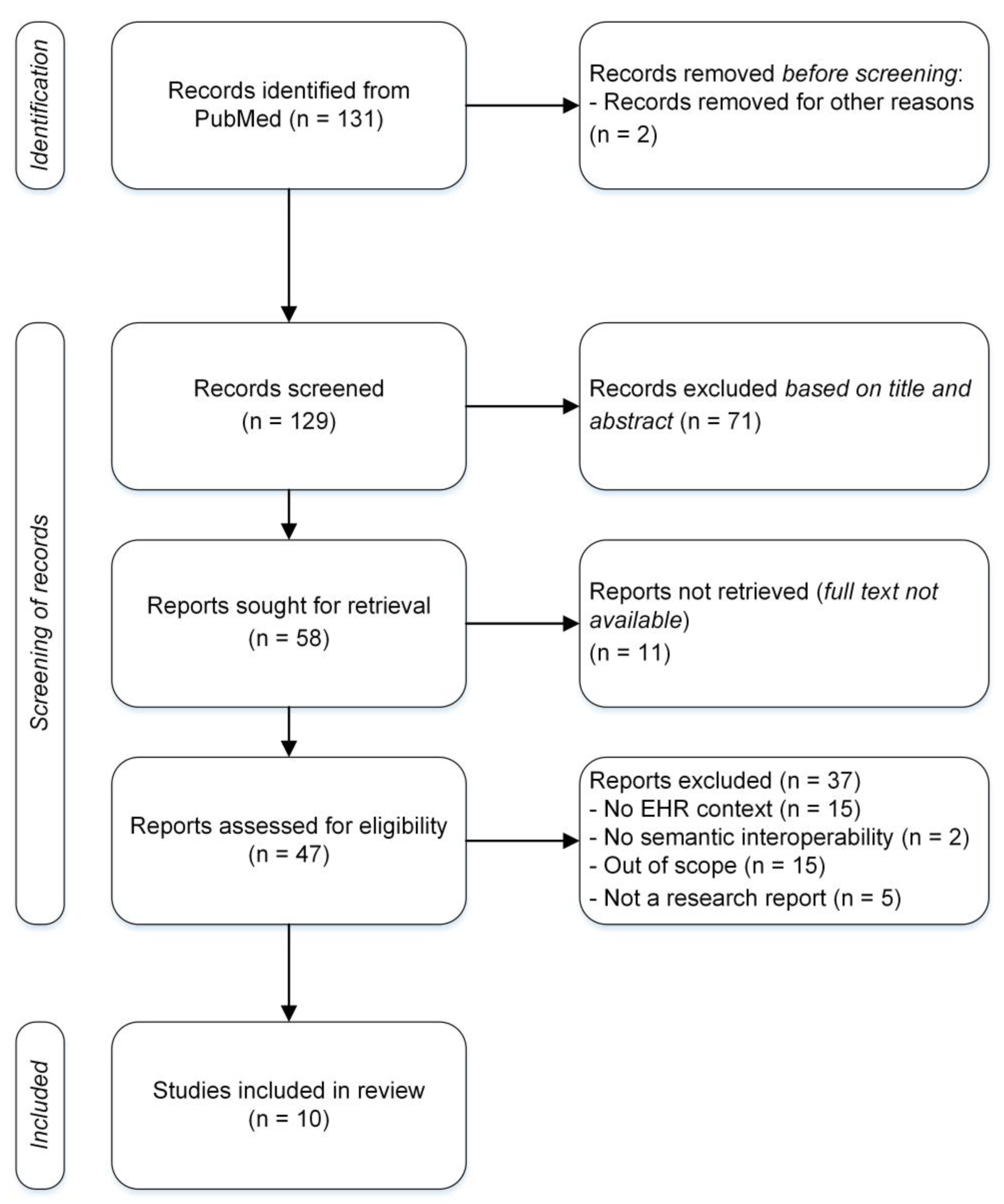
Table 1). The search resulted in 131 unique articles. One article was removed because it did not include abstract, and one was removed because it was not in English. In total, the authors screened 129 articles.
Both authors, i.e., the research team, screened first all the remaining articles by title and abstract. During the first screening 71 articles were excluded from the review after the research team’s discussion due to four reasons; 1) EHR was not a key factor but a contextual factor in the original research setting. 2) The original research did not focus on semantic interoperability but on other level of interoperability. 3) The original study did not entail practical implementation goals, but the focus was theoretical or methodological. 4) The original research was not a research article but, e.g., a poster. The remaining 58 articles were sought for retrieval. For 11 articles, the full text was not available. To evaluate the eligibility, full texts of the 47 articles was read by the research team. At this point, 37 articles were excluded. The previous four reasons for exclusion were complemented by an additional reason: original research was out of scope in a sense that semantic interoperability was not developed with practical goals for EHR implementation but to retrieve and process research data. After agreeing upon the final exclusion within our research team, 10 articles were analyzed for semantic interoperability in EHRs.
Figure 2.
Flowchart of the article identification, screening, and final inclusion.
Figure 2.
Flowchart of the article identification, screening, and final inclusion.
Extracting and documenting the information from the research articles was informed by our research questions, review framework (
Figure 1) and by previous research literature. Based on our framework, the information documentation for the review analysis includes elements of interoperability already identified for the search strategy. As a follow up to this, it is necessary to know, which named elements are typically examined in research and with which methods they are applied in EHRs. [
6,
14,
15,
16] Consequently, we deemed important to document, how semantic interoperability is described in the clinical use context consisting of various EHRs, clinical applications, registers, and other data resources. Lastly, the information documentation should include not only the semantic implementation or use goals or intended benefits, but also practical goals or benefits in the clinical use context. We defined and agreed upon the information documentation categories within our research team to conduct a well-grounded analysis for the review.
3. Results
3.1. Contextual Results
We identified ten (n=10) articles describing semantic interoperability in EHRs, published between 2011-2022 as shown in a
Table 2. Results revealed European advances in the study topic: Most of the research cases (n=8) were affiliated with different types of institutions in the EU member states or EEA countries. One of the publications was co-produced by authors from Columbia and Germany whereas authors of one article represented organizations from the USA, South Korea, and Egypt.
Two of the reported research cases focused on heart failure patients, one on neurosurgical tumor patients, one cancer care patients and one type 1 diabetes patients. Other clinical use domains described were prehospital unit at the site of incident or during transfer to the Emergency Room (ER) and hospital emergency care unit where prehospital patient documentation needs to be re-assessed. Primary care -related case experimented laboratory test results of a population of 230,000 patients. Examples of elderly medication care, and multi-professional health care were part of our sample. One article concerned domain of clinical research utilizing data from different EHR systems.
While all studies concerned data from the EHRs, some studies included more detailed descriptions on data sources. Heart failure summary containing clinical situation data and diagnosis (severity, certainty) as well as heart failure summary covering clinical situation and symptoms data (symptom’s presence, absence, and severity) were represented in the sample. One study focused on diabetes patient history and diabetes care plan (e.g., insulin regimen, diet, and exercise plan) and patient’s self-monitoring of vital signs whereas one was using self-monitoring data on daily activities, side effects and patient reported outcomes (PROMs). One article reported results around diagnosis and laboratory data, one neurosurgical imagining and laboratory data while starting point in paper was diagnosis and medication data. The remaining two studies applied generally prehospital patient case data and emergency care related EHR data, and laboratory data.
A summary of study and sample characteristics is provided in
Table 2.
3.2. Interoperability Results
In our sample, data is transferred and shared between different EHRs and clinical applications with no loss of data or changes in its meaning. Most studies (n=4) were aimed at developing semantic interoperability between different EHRs or within different EHR modules such as medication module in one EHR system. One (n=1) case concentrated specifically on an EHR and a clinical application. Two (n=2) articles reported results around interoperability between EHR and Personal Health Record (PHR). Interoperability on laboratory system and EHR was focus of the study in two cases (n=2). One study (n=1) reported advances in interoperability development between EHR and clinical research resources.
Of the state of development most of studies (n=4) were categorized ‘in development’. The next largest category was ‘in use’ (n=3). Two articles (n=2) reported results regarding testing phase and the remaining one study (n=1) was in an implementation stage.
All articles reported clinical benefits of the selected approach to enhance semantic interoperability. One case described that patient care would benefit from better availability of data e.g., to enhance quality and effectiveness of care. Among the advantages of semantic interoperability, it was described that data transfer into EHR and thus, availability of data improves quality of treatment. Resource effectiveness and optimization of patient care planning and monitoring and better patient management as well as continuity of care because of reuse of data were documented benefits. Interoperable and accessible data facilitate making informed decisions. Two of the examples described benefits on a general level as availability of data, provision of homogenous access to heterogeneous data sets and ensuring information flow across EHRs and clinical applications. It was also documented that semantic interoperability enables data availability from various EHRs and clinical applications specifically for post-marketing research.
In our analysis, semantic development goals were closely coupled with the need to build towards usable and computable data out of heterogeneous medical information that is accessible through various EHRs and databases, e.g., registers. Semantic development goals were described in 4 research (n = 4) as harmonizing data or otherwise processing semantically equivalent data across different medical domains, among different clinical data sources and applications, and, thus, facilitating clinician’s data usage. In 3 research cases (n = 3), semantic development goals emphasized better access to data regardless of underlying standards and data structures or EHRs in use. Underlying assumption with better access to data is that it facilitates better communication between professionals and continuity of care. Research (n = 1) reported increasing data quality as the semantic interoperability development goals to decrease information overload of clinicians by standardized data content. In 2 research case (n = 2), main semantic development goals concentrated in advancing interoperability of EHR data and patient generated data or sensor data for monitoring the situation of chronic patients. One of these research cases also identified data reuse needs for semantic development.
Features of semantic interoperability were described in all 10 articles (n=10). In more detail, the aspects of semantic interoperability were described as follows: ontologies were the chosen aspect in 3 research cases (n = 3), terminologies in 5 (n = 5) and classifications in 3 (n = 3), various clinical documentation standards in 7 (n = 7), and different data models in 8 research cases (n = 8). In this categorization, data model refers to various semantic model layers, namely to utilization of various types of data models that include, for example, data content specifications, reference information models and clinical information models depending on the development context. A dual model is discussed in 2 of the cases for application of data models.
Closely related to the aspects of interoperability, several interoperability standard solutions were named. Named ontology solutions were A Top-Domain Ontology for the Life Sciences BioTopLite in 2 cases, a HL7 FHIR (Fast Health Interoperability Resources) and semantic sensor network (SSN) based T1D Ontology (FASTO) for type 1 diabetes data and a system of several ontologies to be utilized for building EHR interoperability. SNOMED CT clinical terminology was the common terminology application in 5 cases, whereas classification systems were applied in more heterogeneous ways. Of international classifications, following were named: International Classification of Diseases with versions ICD-10 and ICD-9-CM, The Anatomical Therapeutic Chemical (ATC) Classification System and Logical Observation Identifiers Names and Codes LOINC. Applied health care specific standards were the open standard specification in health informatics, openEHR (n = 5), Digital Imaging and Communications in Medicine DICOM (n = 1), HL7 FHIR (n = 5) and the HL7 Clinical Document Architecture, CDA (n = 2). Regarding data model or reference information models, several types were applied for different use environments. These included the Observational Medical Outcomes Partnership OMOP common data model, an EHR specific data component model, i2b2 common data model for data warehouse development, HL7 FHIR reference information model and ISO EN 13606 standard based model. Moreover, one case mentioned using openEHR for data model reference.
Method for applying an interoperability framework or approach is related to the overall design of the data use purposes and needs driving the semantic development. A chosen methodology for semantic development was based on ontology development or application of an ontology framework in 4 research cases (n = 4), on data model-based development in 3 research cases (n = 3), in archetype development in 1 case (n = 1) and in clinical data warehouse development to enhance access and processing of data in 1 case (n = 1). In data model-based approaches, use cases documented method’s capability in separating different semantic level development, i.e., system level or application level and clinical user interface level or patient information level development. Reusability of data-model based semantic approaches and related methods was assessed to result in resource savings in time and cost in development projects, and thus, justifying choice of the approach. For example, clinical knowledge model-based development may allow recycling archetypes that further promote semantic interoperability.
Table 3 summarizes results on semantic interoperability in EHRs with details about each case.
4. Discussion
One of the main goals of proposed EHDS regulation is to support the use of health data for e.g., better healthcare delivery and better research. Comprehensive and timely availability of EHR data is known to improve quality of care and patient safety. Concurrently the lack of not only technical or organizational but also semantic interoperability has been recognized as one of the barriers for the cross-border exchange of health data in Europe.[
1,
2,
3,
4,
5,
6] Therefore, commonly agreed interoperability standards and methods for the harmonization of the semantic interoperability are needed.
The EHDS regulation proposal [
4] highlights the need to follow-up the implementation of interoperable applications that enhance continuity of care and ensure access to safe and high-quality healthcare. In our review, reported clinical benefits of developing semantic interoperability reflect well the major goals of EHDS regulation. Results in our sample show that, an evident goal driving the development in these studies is the following assumption: through increased access to patient information a better quality and outcomes in care can be achieved [
21,
23,
25,
27]. Better communication based on easily accessible data across EHRs is facilitated not only between clinicians and but also between professionals and the patients [
26,
28,
30]. Further advances are related to efficiency and subsequent economic factors, for example, lessening the clinicians’ workload related to documenting and evaluating extensive patient data to avoid information overload and to support multi-professional care processes [
23,
24,
25,
28,
29]. In addition, interoperable patient data provides opportunities for a wide range of EHR-related clinical development, for example, regarding decision-making support, clinical research, or other type of secondary use [
22,
25,
29,
30]. Essentially, the interoperability cases in our review demonstrated a well-documented selection of development goals in EHRs including considerations of patient generated, self-monitoring data interoperability features.
A main goal of semantic interoperability development is to enable patient data usage no matter in which EHRs the data is originated and by what terminologies, classifications, or other semantic features it is supported [
14,
15,
16,
17,
18]. Of the documented terminologies, SNOMED CT seemed to prevail as the dominant choice for a clinical terminology [
21,
22,
23,
27,
30]. For international classifications that are typically integrated into EHRs, a selection of well-established classifications was documented [
22,
29,
30]. Likewise, several health care specific standards [
21,
22,
23,
25,
29,
30], ontologies [
21,
24,
27,
28]and data models [
22,
24,
25,
26,
27,
28,
29,
30] were presented even though here in a relatively small sample. One possible factor affecting the selection of interoperability features such as international standards may be open availability and level of cost in the standard specific resources and their deployment. Consequently, shared implementation experiences and recommendations from previous projects or from international communities may promote and facilitate decision-making concerning future implementations.
For ontologies and data models, several layers may be deployed to address semantic interoperability development needs. For ontologies, deploying a system of ontologies seeks to bridge, for example, between domain specific ontologies and application specific ontologies. In our sample, a case with data model-based development approach enhanced communicating clinical information between the application. The application was used by the patients in self-monitoring and the EHR served as a clinical data repository to avoid loss of meaningful information. In general, when applying data model-based approach, a dual or multi-model approach may be needed to address different semantic interoperability issues during the development – from the real-world clinician as an EHR user to system transactions level.
Finally, when reflecting the semantic interoperability results in our review, there is evidently not one universal approach available to tackle all interoperability related needs and challenges. One reason for this is that interoperability is to be achieved in diffuse and disconnected clinical care environment and in research organizations across borders. However, regulations and related recommendations can support choosing common tools and standards for building interoperability for patient data generated in various EHRs and clinical applications. That may be the strongest selling point for EHDS regulation, supposed it has entered into force: it can achieve a unified toolkit of the most crucial means for building European eHealth interoperability. By these means, common solutions and standards can be agreed upon to remedy existing inconsistencies and to avoid possible future imparities that hinder the goals of EHDS to be realized. It would require a joint in-depth discussion on which level of interoperability should be reached, when the operating environment consists of diverse set of clinical practices and related data needs, i.e., between public and private care or between primary and specialized care. Additionally, it may be worthwhile to consider whether instead of promoting a single approach, semantic interoperability requirements should be assessed through several levels of semantic requirements, i.e., standards, data-models, classifications, terminologies etc. Moreover, developing necessary skills and increasing capabilities is an essential part of the European development package.
Our data set was limited by a small sample size of ten (n=10) articles. Therefore, there is potential for bias, and findings can only be regarded as descriptive in nature. Relatively large heterogeneity in study environments and selected research approaches limit us from drawing strong conclusions. Despite this, within limitations this review demonstrates potentially feasible approaches in promoting semantic interoperability towards the EHDS. Additional real-world studies accounting semantic interoperability are needed to reinforce understanding on most promising, scalable examples. Moreover, one sample category, namely “development status” was for some of the studies difficult to define. This was due to varying levels of details in study reports where some of the studies provided wealth of details while some were more restricted in their scope.
Future research directions are twofold from the EHDS perspective. Firstly, evidence-based recommendations on semantic interoperability features, such as data models and terminologies, are needed. Secondly, more experiences of interoperability development should be reported in peer-reviewed research literature to add on evidence on successful and not so successful experiences instead of leaning on solely individual expert opinions. Assumably, due to the early implementation status of semantic interoperability cases illustrated in research literature, systematic research-based evaluation of benefits and outcomes is still scarce. Despite of well-established standard-based development, implementation of semantic interoperability tends to require a lot of time and resources. Therefore, choices should be well prepared, thought out and justified to meet sustainable and long-term outcomes.
5. Conclusions
We conclude that, based on our review, research literature highlights valuable aspects in promoting semantic interoperability in terms of efficiency and feasibility of solutions integrated in EHRs and possibly for enhancing care. However, when heading towards semantic harmonization as outlined in the EHDS proposal, more data, experiences, and analyses are needed to assess how applicable the chosen specific solutions are for standardization and semantic interoperability of patient data. From that point of view, it is possible to outline the future directions in selecting semantic interoperability standards for realization of the EHDS goals.
Author Contributions
All authors have contributed equally in conceptualization, methodology, formal analysis, investigation, writing—original draft preparation, writing—review and editing, and visualization. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by The Finnish Governmental Research Funding granted for Helsinki University Hospital District, HUS Group/ Diagnostic Centre, grant number TYH2021319.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kohl, S. Public consultation for European health data space is open. Eur J Hosp Pharm. 2021, 28, 239–240. [Google Scholar] [CrossRef] [PubMed]
- Das, M. Stakeholders welcome proposal on the European Health Data Space. Lancet Oncol. 2022, 23, 1492. [Google Scholar] [CrossRef] [PubMed]
- Hussein R, Scherdel L, Nicolet F, Martin-Sanchez F. Towards the European Health Data Space (EHDS) ecosystem: A survey research on future health data scenarios. Int J Med Inform. 2023, 170, 104949. [Google Scholar] [CrossRef] [PubMed]
- Proposal for a REGULATION OF THE EUROPEAN PARLIAMENT AND OF THE COUNCIL on the European Health Data Space. COM/2022/197 final. https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A52022PC0197. 5202.
- Kouroubali A, Katehakis DG. The new European interoperability framework as a facilitator of digital transformation for citizen empowerment. J Biomed Inform. 2019, 94, 103166. [Google Scholar] [CrossRef]
- Stellmach C, Muzoora MR, Thun S. Digitalization of Health Data: Interoperability of the Proposed European Health Data Space. Stud Health Technol Inform. 2022, 298, 132–136. [Google Scholar] [CrossRef]
- Gottumukkala, M. Design, Development, and Evaluation of an Automated Solution for Electronic Information Exchange Between Acute and Long-term Postacute Care Facilities: Design Science Research. JMIR Form Res. 2023, 7, e43758. [Google Scholar] [CrossRef] [PubMed]
- Carlson J, Laryea J. Electronic Health Record-Based Registries: Clinical Research Using Registries in Colon and Rectal Surgery. Clin Colon Rectal Surg. 2019, 32, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Hohman KH, Martinez AK, Klompas M, et al. Leveraging Electronic Health Record Data for Timely Chronic Disease Surveillance: The Multi-State EHR-Based Network for Disease Surveillance. J Public Health Manag Pract. 2023, 29, 162–173. [Google Scholar] [CrossRef]
- 2015 Edition Health Information Technology (Health IT) Certification Criteria, 2015 Edition Base Electronic Health Record (EHR) Definition, and ONC Health IT Certification Program Modifications. Available online: https://www.federalregister.gov/documents/2015/10/16/2015-25597/2015-edition-health-information-technology-health-it-certification-criteria-2015-edition-base (accessed on 21 February 2023).
- Kush RD, Warzel D, Kush MA, et al. FAIR data sharing: The roles of common data elements and harmonization. J Biomed Inform. 2020, 107, 103421. [Google Scholar] [CrossRef]
- Horgan D, Hajduch M, Vrana M, et al. European Health Data Space-An Opportunity Now to Grasp the Future of Data-Driven Healthcare. Healthcare 2022, 10, 1629. [Google Scholar] [CrossRef]
- Palojoki S, Vakkuri A, Vuokko R. The European Cross-Border Health Data Exchange: Focus on Clinically Relevant Data. Stud Health Technol Inform. 2021, 281, 442–446. [Google Scholar] [CrossRef]
- Gamal A, Barakat S, Rezk A. Standardized electronic health record data modeling and persistence: A comparative review. J Biomed Inform. 2021, 114, 103670. [Google Scholar] [CrossRef]
- Lee, A. R. , Kim, I. K., & Lee, E. Developing a Transnational Health Record Framework with Level-Specific Interoperability Guidelines Based on a Related Literature Review. Healthcare 2021, 9, 67. [Google Scholar] [CrossRef] [PubMed]
- de Mello, B. H. , Rigo, S. J., da Costa, C. A., da Rosa Righi, R., Donida, B., Bez, M. R., & Schunke, L. C. Semantic interoperability in health records standards: a systematic literature review. Health and technology 2022, 12, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K. H. , Chung, K. I., Chung, M. A., & Choi, D. Review of semantically interoperable electronic health records for ubiquitous healthcare. Healthcare informatics research 2010, 16, 1–5. [Google Scholar] [CrossRef]
- Moreno-Conde A, Moner D, Cruz WD, et al. Clinical information modeling processes for semantic interoperability of electronic health records: systematic review and inductive analysis. J Am Med Inform Assoc. 2015, 22, 925–934. [Google Scholar] [CrossRef]
- Higgins J, Thomas J, Chandler J, Cumpston M, Li T, Page M. Cochrane Handbook for Systematic Reviews of Interventions version 6.2. Cochrane Training. Available online: http://www.training.cochrane.org/handbook (accessed on 27 January 2023).
- Gusenbauer M, Haddaway NR. Which academic search systems are suitable for systematic reviews or meta-analyses? Evaluating retrieval qualities of Google Scholar, PubMed, and 26 other resources. Res Synth Methods. 2020, 11, 181–217. [Google Scholar] [CrossRef]
- Martínez-Costa C, Schulz S. Ontology content patterns as bridge for the semantic representation of clinical information. Appl Clin Inform. 2014, 5, 660–669. [Google Scholar] [CrossRef]
- Sun H, Depraetere K, De Roo J, et al. Semantic processing of EHR data for clinical research. J Biomed Inform. 2015, 58, 247–259. [Google Scholar] [CrossRef]
- Andersen SNL, Brandsborg CM, Pape-Haugaard L. Use of Semantic Interoperability to Improve the Urgent Continuity of Care in Danish ERs. Stud Health Technol Inform. 2021, 281, 203–207. [Google Scholar] [CrossRef]
- González C, Blobel BG, López DM. Ontology-based framework for electronic health records interoperability. Stud Health Technol Inform. 2011, 169, 694–698. [Google Scholar]
- Kropf S, Chalopin C, Lindner D, Denecke K. Domain Modeling and Application Development of an Archetype- and XML-based EHRS. Practical Experiences and Lessons Learnt. Appl Clin Inform. 2017, 8, 660–679. [Google Scholar] [CrossRef]
- Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG. Archetype-based data warehouse environment to enable the reuse of electronic health record data. Int J Med Inform. 2015, 84, 702–714. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Costa C, Cornet R, Karlsson D, Schulz S, Kalra D. Semantic enrichment of clinical models towards semantic interoperability. The heart failure summary use case. J Am Med Inform Assoc. 2015, 22, 565–576. [Google Scholar] [CrossRef]
- El-Sappagh S, Ali F, Hendawi A, Jang JH, Kwak KS. A mobile health monitoring-and-treatment system based on integration of the SSN sensor ontology and the HL7 FHIR standard. BMC Med Inform Decis Mak. 2019, 19, 97. [Google Scholar] [CrossRef]
- Boussadi A, Zapletal E. A Fast Healthcare Interoperability Resources (FHIR) layer implemented over i2b2. BMC Med Inform Decis Mak. 2017, 17, 120. [Google Scholar] [CrossRef]
- Frid S, Fuentes Expósito MA, Grau-Corral I, et al. Successful Integration of EN/ISO 13606-Standardized Extracts From a Patient Mobile App Into an Electronic Health Record: Description of a Methodology. JMIR Med Inform. 2022, 10, e40344. [Google Scholar] [CrossRef]
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).