Submitted:
24 April 2023
Posted:
26 April 2023
You are already at the latest version
Abstract
Keywords:
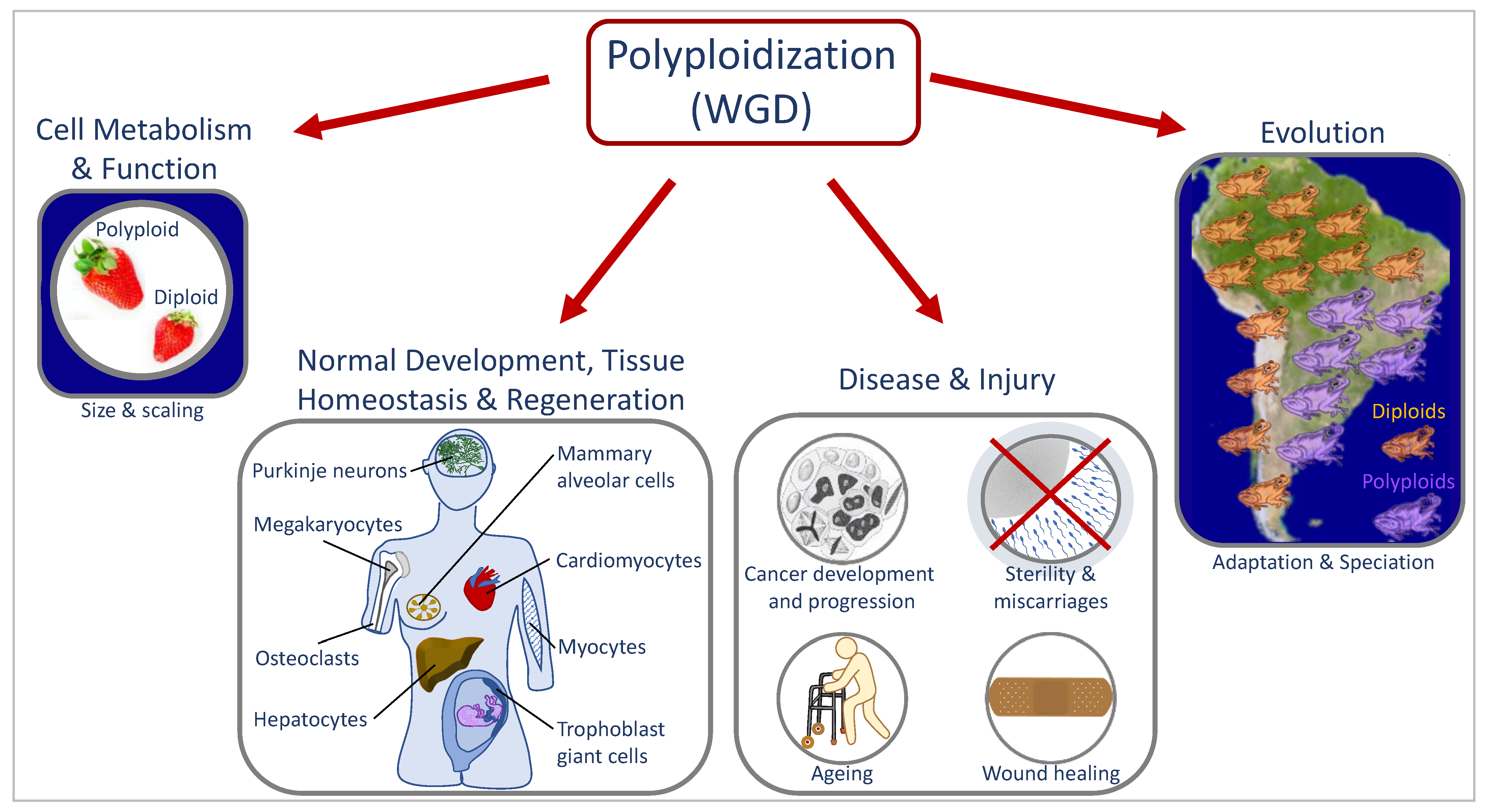
1. Whole genome duplication in development, evolution, and disease
1.1. Causes and downstream effects for WGD
1.2. Laboratory multicellular organism models for WGD
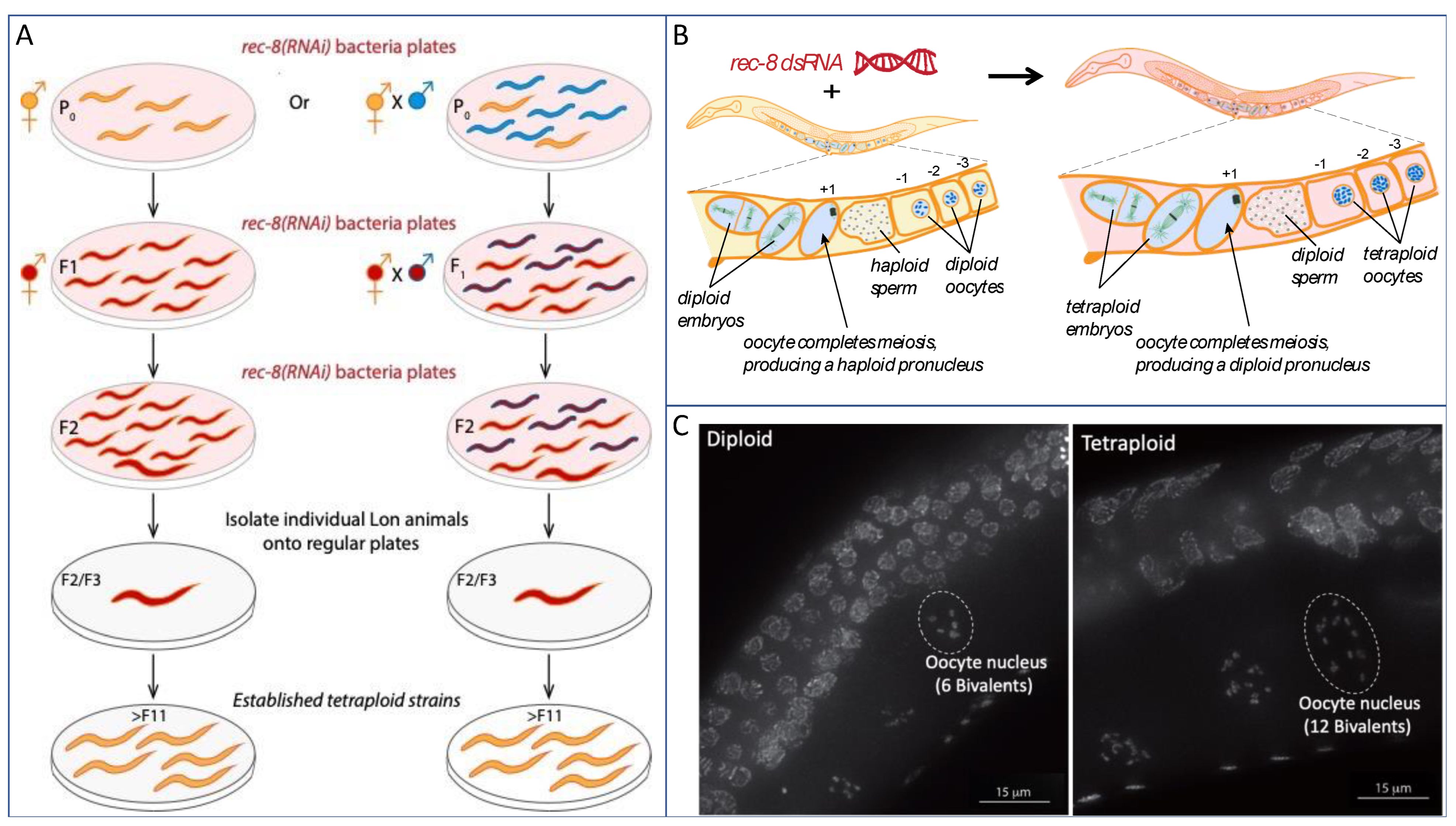
2. Caenorhabditis elegans as an animal laboratory model for understanding WGD
2.1. Polyploidy and aneuploidy in C. elegans

2.2. Polyploid and aneuploid animals uncover C. elegans modes of sex determination and dosage compensation
3. Utilizing polyploid C. elegans as tools to investigate developmental processes
3.1. Understanding early events of meiotic prophase I
3.2. Understanding meiotic and early embryonic cell divisions
4. Utilizing Caenorhabditis to understand the effects of polyploidization
4.1. Polyploid tissues in C. elegans
4.2. Biological size and scaling (allometry)
5. Potential future queries utilizing C. elegans polyploids
Author Contributions
Funding
Conflicts of Interest
References
- Davoli, T.; Lange, T. de The Causes and Consequences of Polyploidy in Normal Development and Cancer. 10.1146/annurev-cellbio-092910-154234 2011. [CrossRef]
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotýnková, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-Genome Doubling Confers Unique Genetic Vulnerabilities on Tumour Cells. Nature 2021, 590, 492–497. [CrossRef]
- Storchova, Z.; Pellman, D. From Polyploidy to Aneuploidy, Genome Instability and Cancer. Nat Rev Mol Cell Bio 2004, 5, 45–54. [CrossRef]
- Storchová, Z.; Breneman, A.; Cande, J.; Dunn, J.; Burbank, K.; O’Toole, E.; Pellman, D. Genome-Wide Genetic Analysis of Polyploidy in Yeast. Nature 2006, 443, 541–547. [CrossRef]
- Nguyen, H.G.; Makitalo, M.; Yang, D.; Chinnappan, D.; St.Hilaire, C.; Ravid, K. Deregulated Aurora-B Induced Tetraploidy Promotes Tumorigenesis. Faseb J 2009, 23, 2741–2748. [CrossRef]
- Bielski, C.M.; Zehir, A.; Penson, A.V.; Donoghue, M.T.A.; Chatila, W.; Armenia, J.; Chang, M.T.; Schram, A.M.; Jonsson, P.; Bandlamudi, C.; et al. Genome Doubling Shapes the Evolution and Prognosis of Advanced Cancers. Nat Genet 2018, 50, 1189–1195. [CrossRef]
- Clarke, E.K.; Gomez, K.A.R.; Mustachi, Z.; Murph, M.C.; Schvarzstein, M. Manipulation of Ploidy in <em>Caenorhabditis Elegans</Em>. J Vis Exp 2018, 57296. [CrossRef]
- Kawade, K.; Horiguchi, G.; Tsukaya, H. Non-Cell-Autonomously Coordinated Organ Size Regulation in Leaf Development. Development 2010, 137, 4221–4227. [CrossRef]
- Horiguchi, G.; Tsukaya, H. Organ Size Regulation in Plants: Insights from Compensation. Front Plant Sci 2011, 2, 24. [CrossRef]
- Bailey, E.C.; Kobielski, S.; Park, J.; Losick, V.P. Polyploidy in Tissue Repair and Regeneration. Csh Perspect Biol 2021, 13. [CrossRef]
- Donne, R.; Saroul-Aïnama, M.; Cordier, P.; Celton-Morizur, S.; Desdouets, C. Polyploidy in Liver Development, Homeostasis and Disease. Nat Rev Gastroentero 2020, 17, 391–405. [CrossRef]
- Øvrebø, J.I.; Edgar, B.A. Polyploidy in Tissue Homeostasis and Regeneration. Development 2018, 145, dev156034. [CrossRef]
- Besen-McNally, R.; Gjelsvik, K.J.; Losick, V.P. Wound-Induced Polyploidization Is Dependent on Integrin-Yki Signaling. Biol Open 2021, 10, bio055996. [CrossRef]
- Lin, Y.-H.; Zhang, S.; Zhu, M.; Lu, T.; Chen, K.; Wen, Z.; Wang, S.; Xiao, G.; Luo, D.; Jia, Y.; et al. Mice With Increased Numbers of Polyploid Hepatocytes Maintain Regenerative Capacity But Develop Fewer Hepatocellular Carcinomas Following Chronic Liver Injury. Gastroenterology 2020, 158, 1698-1712.e14. [CrossRef]
- Losick, V.P.; Fox, D.T.; Spradling, A.C. Polyploidization and Cell Fusion Contribute to Wound Healing in the Adult Drosophila Epithelium. Curr Biol 2013, 23, 2224–2232. [CrossRef]
- irsten, H.W. Wound Healing Is a First Response in a Cancerous Pathway: Hyperplasia Developments to 4n Cell Cycling in Dysplasia Linked to Rb-Inactivation. J Cancer Ther 2015, 06, 906–916. [CrossRef]
- Losick, V.P. Wound-Induced Polyploidy Is Required for Tissue Repair. Adv Wound Care 2016, 5, 271–278. [CrossRef]
- Berman, J. Ploidy Plasticity: A Rapid and Reversible Strategy for Adaptation to Stress. FEMS Yeast Res 2016, 16. [CrossRef]
- Wood, T.E.; Takebayashi, N.; Barker, M.S.; Mayrose, I.; Greenspoon, P.B.; Rieseberg, L.H. The Frequency of Polyploid Speciation in Vascular Plants. Proc National Acad Sci 2009, 106, 13875–13879. [CrossRef]
- Akagi, T.; Jung, K.; Masuda, K.; Shimizu, K.K. Polyploidy before and after Domestication of Crop Species. Curr Opin Plant Biol 2022, 69, 102255. [CrossRef]
- Hodgkin, J. Exploring the Envelope. Systematic Alteration in the Sex-Determination System of the Nematode Caenorhabditis Elegans. Genetics 2002, 162, 767–780.
- Hodgkin, J. Karyotype, Ploidy, and Gene Dosage. WormBook 2005.
- Meneely, P.M. Sex Determination in Polyploids of Caenorhabditis Elegans. Genetics 1994, 137, 467–481.
- Madl, J.E.; Herman, R.K. POLYPLOIDS AND SEX DETERMINATION IN CAENORHABDITIS ELEGANS. Genetics 1979, 93, 393–402.
- Lozano, E.; Sáez, A.G.; Flemming, A.J.; Cunha, A.; Leroi, A.M. Regulation of Growth by Ploidy in Caenorhabditis Elegans. Current Biology 2006, 16, 493–498. [CrossRef]
- Hara, Y.; Kimura, A. An Allometric Relationship between Mitotic Spindle Width, Spindle Length, and Ploidy in Caenorhabditis Elegans Embryos. Mol. Biol. Cell 2013, 24, 1411–1419. [CrossRef]
- Woodruff, G.C.; Eke, O.; Baird, S.E.; Félix, M.-A.; Haag, E.S. Insights Into Species Divergence and the Evolution of Hermaphroditism From Fertile Interspecies Hybrids of Caenorhabditis Nematodes. Genetics 2010, 186, 997–1012. [CrossRef]
- Vargas, E.; McNally, K.; Friedman, J.A.; Cortes, D.B.; Wang, D.Y.; Korf, I.F.; McNally, F.J. Autosomal Trisomy and Triploidy Are Corrected During Female Meiosis in Caenorhabditis Elegans. Genetics 2017, genetics.300259.2017. [CrossRef]
- Ozugergin, I.; Mastronardi, K.; Law, C.; Piekny, A. Diverse Mechanisms Regulate Contractile Ring Assembly for Cytokinesis in the Two-Cell Caenorhabditis Elegans Embryo. J Cell Sci 2021, 135. [CrossRef]
- Mlynarczyk-Evans, S.; Roelens, B.; Villeneuve, A.M. Evidence That Masking of Synapsis Imperfections Counterbalances Quality Control to Promote Efficient Meiosis. PLoS Genet 2013, 9, e1003963. [CrossRef]
- Roelens, B.; Schvarzstein, M.; Villeneuve, A.M. Manipulation of Karyotype in Caenorhabditis Elegans Reveals Multiple Inputs Driving Pairwise Chromosome Synapsis During Meiosis. Genetics 2015, 201, 1363–1379. [CrossRef]
- Cortes, D.B.; McNally, K.L.; Mains, P.E.; McNally, F.J. The Asymmetry of Female Meiosis Reduces the Frequency of Inheritance of Unpaired Chromosomes. Elife 2015.
- Orr-Weaver, T.L. When Bigger Is Better: The Role of Polyploidy in Organogenesis. Trends in Genetics 2015, 31, 307–315. [CrossRef]
- Frawley, L.E.; Orr-Weaver, T.L. Polyploidy. Current Biology 2015, 25, R353–R358. [CrossRef]
- Chen, H.-Z.; Ouseph, M.M.; Li, J.; Pécot, T.; Chokshi, V.; Kent, L.; Bae, S.; Byrne, M.; Duran, C.; Comstock, G.; et al. Canonical and Atypical E2Fs Regulate the Mammalian Endocycle. Nat Cell Biol 2012, 14, 1192–1202. [CrossRef]
- Rios, A.C.; Fu, N.Y.; Jamieson, P.R.; Pal, B.; Whitehead, L.; Nicholas, K.R.; Lindeman, G.J.; Visvader, J.E. Essential Role for a Novel Population of Binucleated Mammary Epithelial Cells in Lactation. Nat Commun 2016, 7, 11400. [CrossRef]
- Pandit, S.K.; Westendorp, B.; Nantasanti, S.; Liere, E. van; Tooten, P.C.J.; Cornelissen, P.W.A.; Toussaint, M.J.M.; Lamers, W.H.; Bruin, A. de E2F8 Is Essential for Polyploidization in Mammalian Cells. Nat Cell Biol 2012, 14, 1181–1191. [CrossRef]
- Senyo, S.E.; Steinhauser, M.L.; Pizzimenti, C.L.; Yang, V.K.; Cai, L.; Wang, M.; Wu, T.-D.; Guerquin-Kern, J.-L.; Lechene, C.P.; Lee, R.T. Mammalian Heart Renewal by Pre-Existing Cardiomyocytes. Nature 2013, 493, 433–436. [CrossRef]
- Zanet, J.; Freije, A.; Ruiz, M.; Coulon, V.; Sanz, J.R.; Chiesa, J.; Gandarillas, A. A Mitosis Block Links Active Cell Cycle with Human Epidermal Differentiation and Results in Endoreplication. Plos One 2010, 5, e15701. [CrossRef]
- Mattia, G.; Vulcano, F.; Milazzo, L.; Barca, A.; Macioce, G.; Giampaolo, A.; Hassan, H.J. Different Ploidy Levels of Megakaryocytes Generated from Peripheral or Cord Blood CD34+ Cells Are Correlated with Different Levels of Platelet Release. Blood 2002, 99, 888–897. [CrossRef]
- Zimmet, J.; Ravid, K. Polyploidy Occurrence in Nature, Mechanisms, and Significance for the Megakaryocyte-Platelet System. Exp Hematol 2000, 28, 3–16. [CrossRef]
- Sladky, V.C.; Eichin, F.; Reiberger, T.; Villunger, A. Polyploidy Control in Hepatic Health and Disease. J Hepatol 2021, 75, 1177–1191. [CrossRef]
- Mazzi, S.; Lordier, L.; Debili, N.; Raslova, H.; Vainchenker, W. Megakaryocyte and Polyploidization. Exp Hematol 2018, 57, 1–13. [CrossRef]
- Edgar, B.A.; Zielke, N.; Gutierrez, C. Endocycles: A Recurrent Evolutionary Innovation for Post-Mitotic Cell Growth. Nat Rev Mol Cell Bio 2014, 15, 197–210. [CrossRef]
- Doyle, J.J.; Coate, J.E. Polyploidy, the Nucleotype, and Novelty: The Impact of Genome Doubling on the Biology of the Cell. Int J Plant Sci 2019, 180, 1–52. [CrossRef]
- Ren, R.; Wang, H.; Guo, C.; Zhang, N.; Zeng, L.; Chen, Y.; Ma, H.; Qi, J. Widespread Whole Genome Duplications Contribute to Genome Complexity and Species Diversity in Angiosperms. Mol Plant 2018, 11, 414–428. [CrossRef]
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.-L.; Peer, Y.V. de Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biol 2020, 30, 688–694. [CrossRef]
- Maciver, S.K. Ancestral Eukaryotes Reproduced Asexually, Facilitated by Polyploidy: A Hypothesis. Bioessays 2019, 41, e1900152. [CrossRef]
- Rijnberk, L.M. van; Barrull-Mascaró, R.; Palen, R.L. van der; Schild, E.S.; Korswagen, H.C.; Galli, M. Endomitosis Controls Tissue-Specific Gene Expression during Development. Plos Biol 2022, 20, e3001597. [CrossRef]
- Zhang, J.; Qiao, Q.; Xu, H.; Zhou, R.; Liu, X. Human Cell Polyploidization: The Good and the Evil. Semin Cancer Biol 2022, 81, 54–63. [CrossRef]
- Otto, S.P. The Evolutionary Consequences of Polyploidy. Cell 2007, 131, 452–462. [CrossRef]
- Lazzeri, E.; Angelotti, M.L.; Peired, A.; Conte, C.; Marschner, J.A.; Maggi, L.; Mazzinghi, B.; Lombardi, D.; Melica, M.E.; Nardi, S.; et al. Endocycle-Related Tubular Cell Hypertrophy and Progenitor Proliferation Recover Renal Function after Acute Kidney Injury. Nat Commun 2018, 9, 1344. [CrossRef]
- Li, T.; Wu, Y.; Tsai, H.; Sun, C.; Wu, Y.; Wu, H.; Pei, Y.; Lu, K.; Yen, T.T.; Chang, C.; et al. Intrahepatic Hepatitis B Virus Large Surface Antigen Induces Hepatocyte Hyperploidy via Failure of Cytokinesis. J. Pathol. 2018, 245, 502–513. [CrossRef]
- David, K.T.; Halanych, K.M. Spatial Proximity between Polyploids across South American Frog Genera. J Biogeogr 2021, 48, 991–1000. [CrossRef]
- Glennon, K.L.; Ritchie, M.E.; Segraves, K.A. Evidence for Shared Broad-scale Climatic Niches of Diploid and Polyploid Plants. Ecol Lett 2014, 17, 574–582. [CrossRef]
- Arnold, B.; Kim, S.-T.; Bomblies, K. Single Geographic Origin of a Widespread Autotetraploid Arabidopsis Arenosa Lineage Followed by Interploidy Admixture. Mol. Biol. Evol. 2015, 32, 1382–1395. [CrossRef]
- Molina-Henao, Y.F.; Hopkins, R. Autopolyploid Lineage Shows Climatic Niche Expansion but Not Divergence in Arabidopsis Arenosa. Am J Bot 2019, 106, 61–70. [CrossRef]
- Ramsey, J. Polyploidy and Ecological Adaptation in Wild Yarrow. Proc National Acad Sci 2011, 108, 7096–7101. [CrossRef]
- Martin, S.L.; Husband, B.C. ADAPTATION OF DIPLOID AND TETRAPLOID CHAMERION ANGUSTIFOLIUM TO ELEVATION BUT NOT LOCAL ENVIRONMENT: ADAPTATION OF CHAMERION ANGUSTIFOLIUM PLOIDIES. Evolution 2013, 67, 1780–1791. [CrossRef]
- Schoenfelder, K.P.; Fox, D.T. The Expanding Implications of Polyploidy. J Cell Biol 2015, 209, 485–491. [CrossRef]
- Bomblies, K. When Everything Changes at Once: Finding a New Normal after Genome Duplication. Proc Royal Soc B Biological Sci 2020, 287, 20202154. [CrossRef]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R.; et al. Polyploidy Can Drive Rapid Adaptation in Yeast. Nature 2015, 519, 349–352. [CrossRef]
- Peer, Y.V. de; Mizrachi, E.; Marchal, K. The Evolutionary Significance of Polyploidy. Nat Rev Genet 2017, 18, 411–424. [CrossRef]
- Zack, T.I.; Schumacher, S.E.; Carter, S.L.; Cherniack, A.D.; Saksena, G.; Tabak, B.; Lawrence, M.S.; Zhang, C.-Z.; Wala, J.; Mermel, C.H.; et al. Pan-Cancer Patterns of Somatic Copy Number Alteration. Nat Genet 2013, 45, 1134–1140. [CrossRef]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and Clinical Implications of Chromosomal Instability in Cancer. Nat Rev Clin Oncol 2018, 15, 139–150. [CrossRef]
- Fujiwara, T.; Bandi, M.; Nitta, M.; Ivanova, E.V.; Bronson, R.T.; Pellman, D. Cytokinesis Failure Generating Tetraploids Promotes Tumorigenesis in P53-Null Cells. Nature 2005, 437, 1043–1047. [CrossRef]
- Matsumoto, T.; Wakefield, L.; Peters, A.; Peto, M.; Spellman, P.; Grompe, M. Proliferative Polyploid Cells Give Rise to Tumors via Ploidy Reduction. Nat Commun 2021, 12, 646. [CrossRef]
- Lambuta, R.A.; Nanni, L.; Liu, Y.; Diaz-Miyar, J.; Iyer, A.; Tavernari, D.; Katanayeva, N.; Ciriello, G.; Oricchio, E. Whole-Genome Doubling Drives Oncogenic Loss of Chromatin Segregation. Nature 2023, 1–9. [CrossRef]
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Grönroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of Whole-Genome Doubling Propagates Chromosomal Instability and Accelerates Cancer Genome Evolution. Cancer Discov 2014, 4, 175–185. [CrossRef]
- Adams, K.L.; Wendel, J.F. Polyploidy and Genome Evolution in Plants. Current Opinion in Plant Biology 2005, 8, 135–141. [CrossRef]
- Cheng, A.; Hanafiah, N.M.; Harikrishna, J.A.; Eem, L.P.; Baisakh, N.; Mispan, M.S. A Reappraisal of Polyploidy Events in Grasses (Poaceae) in a Rapidly Changing World. Biology 2022, 11, 636. [CrossRef]
- Park, C.-W.; Bhandari, G.S.; Won, H.; Park, J.H.; Park, D.S. Polyploidy and Introgression in Invasive Giant Knotweed (Fallopia Sachalinensis) during the Colonization of Remote Volcanic Islands. Sci Rep-uk 2018, 8, 16021. [CrossRef]
- Demin, S.Yu.; Berdieva, M.A.; Goodkov, A.V. Cyclic Polyploidy in Obligate Agamic Amoebae. Cell Tissue Biology 2019, 13, 242–246. [CrossRef]
- Albertin, W.; Marullo, P. Polyploidy in Fungi: Evolution after Whole-Genome Duplication. Proc Royal Soc B Biological Sci 2012, 279, 2497–2509. [CrossRef]
- Soppa, J. Polyploidy in Archaea and Bacteria: About Desiccation Resistance, Giant Cell Size, Long-Term Survival, Enforcement by a Eukaryotic Host and Additional Aspects. J Mol Microb Biotech 2015, 24, 409–419. [CrossRef]
- Markov, A.V.; Kaznacheev, I.S. Evolutionary Consequences of Polyploidy in Prokaryotes and the Origin of Mitosis and Meiosis. Biol Direct 2016, 11, 28. [CrossRef]
- Wertheim, B.; Beukeboom, L.W.; Zande, L. van de Polyploidy in Animals: Effects of Gene Expression on Sex Determination, Evolution and Ecology. Cytogenet Genome Res 2013, 140, 256–269. [CrossRef]
- Mable, B.K.; Alexandrou, M.A.; Taylor, M.I. Genome Duplication in Amphibians and Fish: An Extended Synthesis. J Zool 2011, 284, 151–182. [CrossRef]
- Sacerdot, C.; Louis, A.; Bon, C.; Berthelot, C.; Crollius, H.R. Chromosome Evolution at the Origin of the Ancestral Vertebrate Genome. Genome Biol 2018, 19, 166. [CrossRef]
- Comai, L. The Advantages and Disadvantages of Being Polyploid. Nat Rev Genet 2005, 6, 836–846. [CrossRef]
- Monnahan, P.; Kolář, F.; Baduel, P.; Sailer, C.; Koch, J.; Horvath, R.; Laenen, B.; Schmickl, R.; Paajanen, P.; Šrámková, G.; et al. Pervasive Population Genomic Consequences of Genome Duplication in Arabidopsis Arenosa. Nat Ecol Evol 2019, 3, 457–468. [CrossRef]
- Yamazaki, W.; Takahashi, M.; Kawahara, M. Restricted Development of Mouse Triploid Fetuses with Disorganized Expression of Imprinted Genes. Zygote 2015, 23, 874–884. [CrossRef]
- Menon, T.; Nair, S. Experimental Manipulation of Ploidy in Zebrafish Embryos and Its Application in Genetic Screens. Methods Mol Biology Clifton N J 2019, 1920, 111–128. [CrossRef]
- Eakin, G.S.; Hadjantonakis, A.-K.; Papaioannou, V.E.; Behringer, R.R. Developmental Potential and Behavior of Tetraploid Cells in the Mouse Embryo. Dev Biol 2005, 288, 150–159. [CrossRef]
- Nigon; V. Les Modalites de La Reproduction et Le Determinisme Du Sexe Chez Quelques Nematodes Libres. Annales de Sciences Naturelles - Zool. Biol. Anim. 1949, 11, 1–132.
- Hodgkin, J. MALE PHENOTYPES AND MATING EFFICIENCY IN CAENORHABDITIS ELEGANS. Genetics 1983, 103, 43–64. [CrossRef]
- Hodgkin, J. Primary Sex Determination in the Nematode C. Elegans. Development 1987, 101, 5–16.
- Nigon, V.M.; Félix, M.-A. History of Research on C. Elegans and Other Free-Living Nematodes as Model Organisms. Wormbook Online Rev C Elegans Biology 2017, 2017, 1–84. [CrossRef]
- Brenner, S. The Genetics of Caenorhabditis Elegans. Genetics 1974, 77, 71–94.
- Hedgecock, E.M.; White, J.G. Polyploid Tissues in the Nematode Caenorhabditis Elegans. Developmental Biology 1985, 107, 128–133. [CrossRef]
- Flemming, A.J.; Shen, Z.-Z.; Cunha, A.; Emmons, S.W.; Leroi, A.M. Somatic Polyploidization and Cellular Proliferation Drive Body Size Evolution in Nematodes. Proceedings of the National Academy of Sciences 2000, 97, 5285–5290. [CrossRef]
- Hodgkin, J.; Horvitz, H.R.; Brenner, S. NONDISJUNCTION MUTANTS OF THE NEMATODE CAENORHABDITIS ELEGANS. Genetics 1979, 91, 67–94. [CrossRef]
- Sigurdson, D.C.; Spanier, G.J.; Herman, R.K. CAENORHABDITIS ELEGANS DEFICIENCY MAPPING. Genetics 1984, 108, 331–345. [CrossRef]
- Haack, H.; Hodgkin, J. Tests for Parental Imprinting in the Nematode Caenorhabditis Elegans. Molec. Gen. Genet. 1991, 228, 482–485. [CrossRef]
- Larkin, K.; Tucci, C.; Neiman, M. Effects of Polyploidy and Reproductive Mode on Life History Trait Expression. Ecol Evol 2016, 6, 765–778. [CrossRef]
- Schoonmaker, A.; Hao, Y.; Bird, D.McK.; Conant, G.C. A Single, Shared Triploidy in Three Species of Parasitic Nematodes. G3 Genes Genomes Genetics 2019, 10, 225–233. [CrossRef]
- Nigon; V. Effets de La Polyploïdie Chez Un Nématode Libre. C. R. Acad. Sc. Paris 1949, D 228, 1161-1162.
- Nigon, V. Polyploidie Experimentale Chez Un Nematode Libre, Rhabditis Elegans. Maupas. Bull. Biol. Fr. Belg. 1951, 85, 187–225. [CrossRef]
- V, N. Effets de La Polyploïdie Chez Un Nématode Libre. C. R. Acad. Sc. 1949, 228, 1161-1162.
- Meyer, B.J. X-Chromosome Dosage Compensation. Wormbook 2005, 1–14. [CrossRef]
- Meyer, B.J. The X Chromosome in C. Elegans Sex Determination and Dosage Compensation. Curr Opin Genet Dev 2022, 74, 101912. [CrossRef]
- Schvarzstein, M.; Wignall, S.M.; Villeneuve, A.M. Coordinating Cohesion, Co-Orientation, and Congression during Meiosis: Lessons from Holocentric Chromosomes. Genes & Development 2010, 24, 219–228. [CrossRef]
- MacQueen, A.J.; Phillips, C.M.; Bhalla, N.; Weiser, P.; Villeneuve, A.M.; Dernburg, A.F. Chromosome Sites Play Dual Roles to Establish Homologous Synapsis during Meiosis in C. Elegans. Cell 2005, 123, 1037–1050. [CrossRef]
- Meyerzon, M.; Gao, Z.; Liu, J.; Wu, J.-C.; Malone, C.J.; Starr, D.A. Centrosome Attachment to the C. Elegans Male Pronucleus Is Dependent on the Surface Area of the Nuclear Envelope. Developmental Biology 2009, 327, 433–446. [CrossRef]
- Schierenberg, E.; Wood, W.B. Control of Cell-Cycle Timing in Early Embryos of Caenorhabditis Elegans. Dev Biol 1985, 107, 337–354. [CrossRef]
- Sadler, P.L.; Shakes, D.C. Anucleate Caenorhabditis Elegans Sperm Can Crawl, Fertilize Oocytes and Direct Anterior-Posterior Polarization of the 1-Cell Embryo. Development 2000, 127, 355–366. [CrossRef]
- Schierenberg, E. Cell Fusion. 1987, 409–418. [CrossRef]
- Oegema, K.; Hyman, A.A. Cell Division. Wormbook 2006, 1–40. [CrossRef]
- Malone, C.J.; Misner, L.; Bot, N.L.; Tsai, M.-C.; Campbell, J.M.; Ahringer, J.; White, J.G. The C. Elegans Hook Protein, ZYG-12, Mediates the Essential Attachment between the Centrosome and Nucleus. Cell 2003, 115, 825–836. [CrossRef]
- Gönczy, P.; Pichler, S.; Kirkham, M.; Hyman, A.A. Cytoplasmic Dynein Is Required for Distinct Aspects of Mtoc Positioning, Including Centrosome Separation, in the One Cell Stage Caenorhabditis Elegans Embryo. J Cell Biology 1999, 147, 135–150. [CrossRef]
- Newport, J.; Kirschner, M. A Major Developmental Transition in Early Xenopus Embryos: I. Characterization and Timing of Cellular Changes at the Midblastula Stage. Cell 1982, 30, 675–686. [CrossRef]
- Sachsenmaier, W.; Remy, U.; Plattner-Schobel, R. Initiation of Synchronous Mitosis in Physarum Polycephalum A Model of the Control of Cell Division in Eukariots. Exp Cell Res 1972, 73, 41–48. [CrossRef]
- Fantes, P.A. Isolation of Cell Size Mutants of a Fission Yeast by a New Selective Method: Characterization of Mutants and Implications for Division Control Mechanisms. J Bacteriol 1981, 146, 746–754. [CrossRef]
- Matson, J.P.; Cook, J.G. Cell Cycle Proliferation Decisions: The Impact of Single Cell Analyses. Febs J 2017, 284, 362–375. [CrossRef]
- Arata, Y.; Takagi, H. Quantitative Studies for Cell-Division Cycle Control. Front Physiol 2019, 10, 1022. [CrossRef]
- Koreth, J.; Heuvel, S. van den Cell-Cycle Control in Caenorhabditis Elegans: How the Worm Moves from G1 to S. Oncogene 2005, 24, 2756–2764. [CrossRef]
- Wang, Z. Regulation of Cell Cycle Progression by Growth Factor-Induced Cell Signaling. Cells 2021, 10, 3327. [CrossRef]
- Dumont, S.; Mitchison, T.J. Compression Regulates Mitotic Spindle Length by a Mechanochemical Switch at the Poles. Curr Biol 2009, 19, 1086–1095. [CrossRef]
- Itabashi, T.; Takagi, J.; Shimamoto, Y.; Onoe, H.; Kuwana, K.; Shimoyama, I.; Gaetz, J.; Kapoor, T.M.; Ishiwata, S. Probing the Mechanical Architecture of the Vertebrate Meiotic Spindle. Nat Methods 2009, 6, 167–172. [CrossRef]
- Hara, Y.; Iwabuchi, M.; Ohsumi, K.; Kimura, A. Intranuclear DNA Density Affects Chromosome Condensation in Metazoans. Mol Biol Cell 2013, 24, 2442–2453. [CrossRef]
- Mains, P.E.; Kemphues, K.J.; Sprunger, S.A.; Sulston, I.A.; Wood, W.B. Mutations Affecting the Meiotic and Mitotic Divisions of the Early Caenorhabditis Elegans Embryo. Genetics 1990, 126, 593–605. [CrossRef]
- Quintin, S.; Mains, P.E.; Zinke, A.; Hyman, A.A. The Mbk-2 Kinase Is Required for Inactivation of MEI-1/Katanin in the One-cell Caenorhabditis Elegans Embryo. Embo Rep 2003, 4, 1175–1181. [CrossRef]
- Segbert, C.; Barkus, R.; Powers, J.; Strome, S.; Saxton, W.M.; Bossinger, O. KLP-18, a Klp2 Kinesin, Is Required for Assembly of Acentrosomal Meiotic Spindles in Caenorhabditis Elegans. Mol Biol Cell 2003, 14, 4458–4469. [CrossRef]
- Maddox, A.S.; Habermann, B.; Desai, A.; Oegema, K. Distinct Roles for Two C. Elegans Anillins in the Gonad and Early Embryo. Development 2005, 132, 2837–2848. [CrossRef]
- Dorn, J.F.; Zhang, L.; Paradis, V.; Edoh-Bedi, D.; Jusu, S.; Maddox, P.S.; Maddox, A.S. Actomyosin Tube Formation in Polar Body Cytokinesis Requires Anillin in C. Elegans. Curr Biol 2010, 20, 2046–2051. [CrossRef]
- Fankhauser, G. Cytological Studies on Egg Fragments of the Salamander Triton. V. Chromosome Number and Chromosome Individuality in the Cleavage Mitoses of Merogonic Fragments. J. Exp. Zool. 1934, 68, 1–57. [CrossRef]
- Conklin, E.G. Cell Size and Nuclear Size. J. Exp. Zool. 1912, 12, 1–98. [CrossRef]
- Berardino, M.A.D. The Karyotype of Rana Pipiens and Investigation of Its Stability during Embryonic Differentiation. Dev Biol 1962, 5, 101–126. [CrossRef]
- Belmont, A.S.; Sedat, J.W.; Agard, D.A. A Three-Dimensional Approach to Mitotic Chromosome Structure: Evidence for a Complex Hierarchical Organization. J Cell Biology 1987, 105, 77–92. [CrossRef]
- Sönnichsen, B.; Koski, L.B.; Walsh, A.; Marschall, P.; Neumann, B.; Brehm, M.; Alleaume, A.-M.; Artelt, J.; Bettencourt, P.; Cassin, E.; et al. Full-Genome RNAi Profiling of Early Embryogenesis in Caenorhabditis Elegans. Nature 2005, 434, 462–469. [CrossRef]
- Deppe, U.; Schierenberg, E.; Cole, T.; Krieg, C.; Schmitt, D.; Yoder, B.; Ehrenstein, G. von Cell Lineages of the Embryo of the Nematode Caenorhabditis Elegans. Proc National Acad Sci 1978, 75, 376–380. [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The Embryonic Cell Lineage of the Nematode Caenorhabditis Elegans. Dev Biol 1983, 100, 64–119. [CrossRef]
- I., G. Development of the Vulva. In C. elegans II; R., R.L., T., B., J., M.B., R., P.J., Eds.; Cold Spring Harbor Laboratory Press, Cold Spring Harbor: New York, 1997; Vol. Chapter 19, pp. 519–541.
- Simske, J.S.; Hardin, J. Getting into Shape: Epidermal Morphogenesis in Caenorhabditis Elegans Embryos. Bioessays 2001, 23, 12–23. [CrossRef]
- Wong, M.; Schwarzbauer, J.E. Gonad Morphogenesis and Distal Tip Cell Migration in the Caenorhabditis Elegans Hermaphrodite. Wiley Interdiscip Rev Dev Biology 2012, 1, 519–531. [CrossRef]
- Shemer, G.; Podbilewicz, B. Fusomorphogenesis: Cell Fusion in Organ Formation. Dev. Dyn. 2000, 218, 30–51. [CrossRef]
- Tuck, S. The Control of Cell Growth and Body Size in Caenorhabditis Elegans. Exp Cell Res 2014, 321, 71–76. [CrossRef]
- Tsou, M.-F.B.; Stearns, T. Mechanism Limiting Centrosome Duplication to Once per Cell Cycle. Nature 2006, 442, 947–951. [CrossRef]
- Tsou, M.-F.B.; Stearns, T. Controlling Centrosome Number: Licenses and Blocks. Current Opinion in Cell Biology 2006, 18, 74–78. [CrossRef]
- Duensing, A.; Duensing, S. Polyploidization and Cancer. Adv Exp Med Biol 2010, 676, 93–103. [CrossRef]
- Lu, Y.; Roy, R. Centrosome/Cell Cycle Uncoupling and Elimination in the Endoreduplicating Intestinal Cells of C. Elegans. PLoS ONE 2014, 9, e110958-17. [CrossRef]
- Lee, H.O.; Davidson, J.M.; Duronio, R.J. Endoreplication: Polyploidy with Purpose. Gene Dev 2009, 23, 2461–2477. [CrossRef]
- Zhang, L.; Ward, J.D.; Cheng, Z.; Dernburg, A.F. The Auxin-Inducible Degradation (AID) System Enables Versatile Conditional Protein Depletion in C. Elegans. Development 2015, 142, 4374–4384. [CrossRef]
- Jorgensen, P.; Edgington, N.P.; Schneider, B.L.; Rupeš, I.; Tyers, M.; Futcher, B. The Size of the Nucleus Increases as Yeast Cells Grow. Mol Biol Cell 2007, 18, 3523–3532. [CrossRef]
- Yahya, G.; Menges, P.; Amponsah, P.S.; Ngandiri, D.A.; Schulz, D.; Wallek, A.; Kulak, N.; Mann, M.; Cramer, P.; Savage, V.; et al. Sublinear Scaling of the Cellular Proteome with Ploidy. Nat Commun 2022, 13, 6182. [CrossRef]
- Neumann, F.R.; Nurse, P. Nuclear Size Control in Fission Yeast. J Cell Biology 2007, 179, 593–600. [CrossRef]
- Gillooly, J.F.; Hein, A.; Damiani, R. Nuclear DNA Content Varies with Cell Size across Human Cell Types. Csh Perspect Biol 2015, 7, a019091. [CrossRef]
- Katagiri, Y.; Hasegawa, J.; Fujikura, U.; Hoshino, R.; Matsunaga, S.; Tsukaya, H. The Coordination of Ploidy and Cell Size Differs between Cell Layers in Leaves. Development 2016, 143, 1120–1125. [CrossRef]
- Cadart, C.; Heald, R. Scaling of Biosynthesis and Metabolism with Cell Size. Mol Biol Cell 2022, 33, pe5. [CrossRef]
- Robinson, D.O.; Coate, J.E.; Singh, A.; Hong, L.; Bush, M.; Doyle, J.J.; Roeder, A.H.K. Ploidy and Size at Multiple Scales in the Arabidopsis Sepal. Plant Cell 2018, 30, 2308–2329. [CrossRef]
- Cantwell, H.; Dey, G. Nuclear Size and Shape Control. Semin Cell Dev Biol 2022, 130, 90–97. [CrossRef]
- Weber, G.M.; Wiens, G.D.; Welch, T.J.; Hostuttler, M.A.; Leeds, T.D. Comparison of Disease Resistance between Diploid, Induced-Triploid, and Intercross-Triploid Rainbow Trout Including Trout Selected for Resistance to Flavobacterium Psychrophilum. Aquaculture 2013, 410–411, 66–71. [CrossRef]
- Horiguchi, G.; Tsukaya, H. Organ Size Regulation in Plants: Insights from Compensation. Front Plant Sci 2011, 2, 24. [CrossRef]
- Kawade, K.; Horiguchi, G.; Tsukaya, H. Non-Cell-Autonomously Coordinated Organ Size Regulation in Leaf Development. Development 2010, 137, 4221–4227. [CrossRef]
- Tsukaya, H. Re-Examination of the Role of Endoreduplication on Cell-Size Control in Leaves. J Plant Res 2019, 132, 571–580. [CrossRef]
- Nyström, J.; Shen, Z.-Z.; Aili, M.; Flemming, A.J.; Leroi, A.; Tuck, S. Increased or Decreased Levels of Caenorhabditis Elegans Lon-3, a Gene Encoding a Collagen, Cause Reciprocal Changes in Body Length. Genetics 2002, 161, 83–97.
- Morita, K.; Flemming, A.J.; Sugihara, Y.; Mochii, M.; Suzuki, Y.; Yoshida, S.; Wood, W.B.; Kohara, Y.; Leroi, A.M.; Ueno, N. A Caenorhabditis Elegans TGF-β, DBL-1, Controls the Expression of LON-1, a PR-related Protein, That Regulates Polyploidization and Body Length. The EMBO Journal 2002, 21, 1063–1073. [CrossRef]
- Morita, K.; Chow, K.L.; Ueno, N. Regulation of Body Length and Male Tail Ray Pattern Formation of Caenorhabditis Elegans by a Member of TGF-Beta Family. Dev Camb Engl 1999, 126, 1337–1347. [CrossRef]
- Hirose, T.; Nakano, Y.; Nagamatsu, Y.; Misumi, T.; Ohta, H.; Ohshima, Y. Cyclic GMP-Dependent Protein Kinase EGL-4 Controls Body Size and Lifespan in C. Elegans. Development 2003, 130, 1089–1099. [CrossRef]
- Nagamatsu, Y.; Ohshima, Y. Mechanisms for the Control of Body Size by a G-kinase and a Downstream TGFβ Signal Pathway in Caenorhabditis Elegans. Genes Cells 2004, 9, 39–47. [CrossRef]
- Wang, J.; Tokarz, R.; Savage-Dunn, C. The Expression of TGFβ Signal Transducers in the Hypodermis Regulates Body Size in C. Elegans. Development 2002, 129, 4989–4998. [CrossRef]
- Parisi, T.; Beck, A.R.; Rougier, N.; McNeil, T.; Lucian, L.; Werb, Z.; Amati, B. Cyclins E1 and E2 Are Required for Endoreplication in Placental Trophoblast Giant Cells. Embo J 2003, 22, 4794–4803. [CrossRef]
- Geng, Y.; Yu, Q.; Sicinska, E.; Das, M.; Schneider, J.E.; Bhattacharya, S.; Rideout, W.M.; Bronson, R.T.; Gardner, H.; Sicinski, P. Cyclin E Ablation in the Mouse. Cell 2003, 114, 431–443. [CrossRef]
- Suzuki, Y.; Yandell, M.D.; Roy, P.J.; Krishna, S.; Savage-Dunn, C.; Ross, R.M.; Padgett, R.W.; Wood, W.B. A BMP Homolog Acts as a Dose-Dependent Regulator of Body Size and Male Tail Patterning in Caenorhabditis Elegans. Development 1999, 126, 241–250. [CrossRef]
- SAVAGE-DUNN, C.; YU, L.; GILL, K.; AWAN, M.; FERNANDO, T. Non-Stringent Tissue-Source Requirements for BMP Ligand Expression in Regulation of Body Size in Caenorhabditis Elegans. Genet Res 2011, 93, 427–432. [CrossRef]
- Savage-Dunn, C. Targets of TGFβ-Related Signaling in Caenorhabditis Elegans. Cytokine Growth F R 2001, 12, 305–312. [CrossRef]
- Savage-Dunn, C.; Tokarz, R.; Wang, H.; Cohen, S.; Giannikas, C.; Padgett, R.W. SMA-3 Smad Has Specific and Critical Functions in DBL-1/SMA-6 TGFβ-Related Signaling. Dev Biol 2000, 223, 70–76. [CrossRef]
- Szyperski, T.; Fernández, C.; Mumenthaler, C.; Wüthrich, K. Structure Comparison of Human Glioma Pathogenesis-Related Protein GliPR and the Plant Pathogenesis-Related Protein P14a Indicates a Functional Link between the Human Immune System and a Plant Defense System. Proc National Acad Sci 1998, 95, 2262–2266. [CrossRef]
- Cai, Q.; Wang, W.; Gao, Y.; Yang, Y.; Zhu, Z.; Fan, Q. Ce-wts-1 Plays Important Roles in Caenorhabditis Elegans Development. Febs Lett 2009, 583, 3158–3164. [CrossRef]
- Jones, K.T.; Greer, E.R.; Pearce, D.; Ashrafi, K. Rictor/TORC2 Regulates Caenorhabditis Elegans Fat Storage, Body Size, and Development through Sgk-1. Plos Biol 2009, 7, e1000060. [CrossRef]
- Oldham, S.; Böhni, R.; Stocker, H.; Brogiolo, W.; Hafen, E. Genetic Control of Size in Drosophila. Philosophical Transactions Royal Soc Lond Ser B Biological Sci 2000, 355, 945–952. [CrossRef]
- Schmelzle, T.; Hall, M.N. TOR, a Central Controller of Cell Growth. Cell 2000, 103, 253–262. [CrossRef]
- ZHANG, W.; LIU, H.T. MAPK Signal Pathways in the Regulation of Cell Proliferation in Mammalian Cells. Cell Res 2002, 12, 9–18. [CrossRef]
- Watanabe, N.; Nagamatsu, Y.; Gengyo-Ando, K.; Mitani, S.; Ohshima, Y. Control of Body Size by SMA-5, a Homolog of MAP Kinase BMK1/ERK5, in C. Elegans. Development 2005, 132, 3175–3184. [CrossRef]
- Hyun, S. Body Size Regulation and Insulin-like Growth Factor Signaling. Cell Mol Life Sci 2013, 70, 2351–2365. [CrossRef]
- McKeown, C.; Praitis, V.; Austin, J. Sma-1 Encodes a ΒH-Spectrin Homolog Required for Caenorhabditis Elegans Morphogenesis. Development 1998, 125, 2087–2098. [CrossRef]
- Hayashi, S.; Yokoyama, H.; Tamura, K. Roles of Hippo Signaling Pathway in Size Control of Organ Regeneration. Dev Growth Differ 2015, 57, 341–351. [CrossRef]
- Goodman, M.B.; Savage-Dunn, C. Reciprocal Interactions between Transforming Growth Factor Beta Signaling and Collagens: Insights from Caenorhabditis Elegans. Dev. Dyn. 2022, 251, 47–60. [CrossRef]
- Song, Q.; Ando, A.; Jiang, N.; Ikeda, Y.; Chen, Z.J. Single-Cell RNA-Seq Analysis Reveals Ploidy-Dependent and Cell-Specific Transcriptome Changes in Arabidopsis Female Gametophytes. Genome Biol 2020, 21, 178. [CrossRef]
- Wu, C.-Y.; Rolfe, P.A.; Gifford, D.K.; Fink, G.R. Control of Transcription by Cell Size. Plos Biol 2010, 8, e1000523. [CrossRef]
- Marguerat, S.; Bähler, J. Coordinating Genome Expression with Cell Size. Trends Genet 2012, 28, 560–565. [CrossRef]
- Coate, J.E.; Doyle, J.J. Variation in Transcriptome Size: Are We Getting the Message? Chromosoma 2015, 124, 27–43. [CrossRef]
- LEITCH, I.J.; BENNETT, M.D. Genome Downsizing in Polyploid Plants. Biol J Linn Soc 2004, 82, 651–663. [CrossRef]
- Zenil-Ferguson, R.; Ponciano, J.M.; Burleigh, J.G. Evaluating the Role of Genome Downsizing and Size Thresholds from Genome Size Distributions in Angiosperms. Am J Bot 2016, 103, 1175–1186. [CrossRef]
- Pellicer, J.; Hidalgo, O.; Dodsworth, S.; Leitch, I.J. Genome Size Diversity and Its Impact on the Evolution of Land Plants. Genes-basel 2018, 9, 88. [CrossRef]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene Retention, Fractionation and Subgenome Differences in Polyploid Plants. Nat Plants 2018, 4, 258–268. [CrossRef]
- Baduel, P.; Quadrana, L.; Hunter, B.; Bomblies, K.; Colot, V. Relaxed Purifying Selection in Autopolyploids Drives Transposable Element Over-Accumulation Which Provides Variants for Local Adaptation. Nat Commun 2019, 10, 5818. [CrossRef]
- Raina, S.N.; Parida, A.; Koul, K.K.; Salimath, S.S.; Bisht, M.S.; Raja, V.; Khoshoo, T.N. Associated Chromosomal DNA Changes in Polyploids. Genome 1994, 37, 560–564. [CrossRef]
- Lovén, J.; Orlando, D.A.; Sigova, A.A.; Lin, C.Y.; Rahl, P.B.; Burge, C.B.; Levens, D.L.; Lee, T.I.; Young, R.A. Revisiting Global Gene Expression Analysis. Cell 2012, 151, 476–482. [CrossRef]
- Bourdon, M.; Pirrello, J.; Cheniclet, C.; Coriton, O.; Bourge, M.; Brown, S.; Moïse, A.; Peypelut, M.; Rouyère, V.; Renaudin, J.-P.; et al. Evidence for Karyoplasmic Homeostasis during Endoreduplication and a Ploidy-Dependent Increase in Gene Transcription during Tomato Fruit Growth. Development 2012, 139, 3817–3826. [CrossRef]
- Voichek, Y.; Bar-Ziv, R.; Barkai, N. Expression Homeostasis during DNA Replication. Science 2016, 351, 1087–1090. [CrossRef]
| Karyotype | X/A ratio | Sexual Fate |
|---|---|---|
| 2A;2X | 1 | female |
| 2A;1X | 0.5 | male |
| 3A;3X | 1 | female |
| 3A;2X | 0.67* | male |
| 4A;4X | 1 | female |
| 4A;3X | 0.75* | female |
| 4A;2X | 0.5 | male |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).




