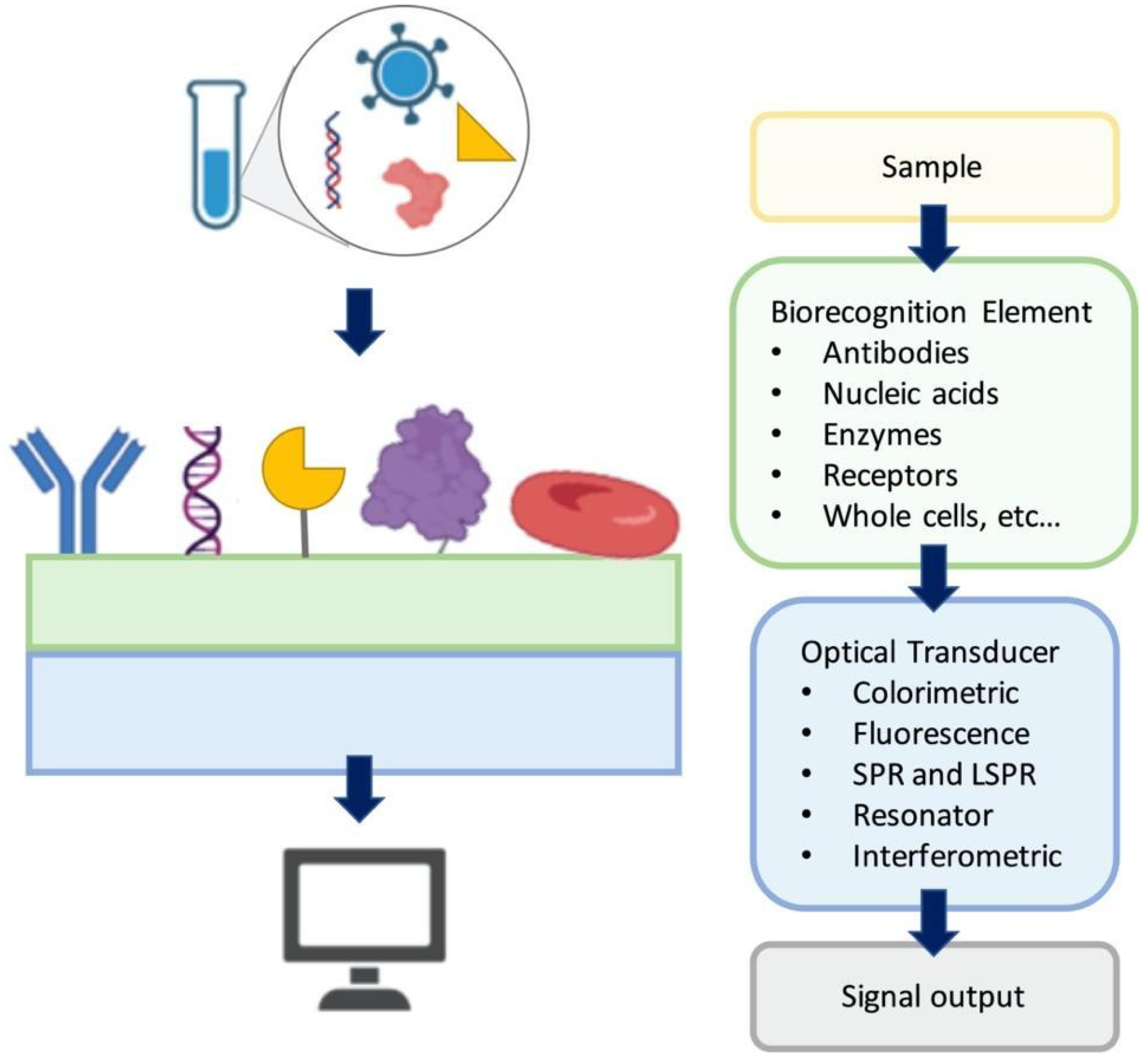
Optical biosensors focus on measurement of the optical signals as the changes in the optical properties and characteristics on the transducer surface in the case of an interaction of the immobilized biorecognition element with the measured substance [
12,
13,
14].
Figure 1 shows a general illustration of an optical biosensor. In general, the optical biosensor typically consists of an optical source, a transduction platform and an optical detector. The optical biosensor can have different types of biological materials such as antibodies, aptamers, peptides, nucleic acids, peptide nucleic acids, proteins, enzymes, whole cells as the biosensing element on the transducer surface which are designed to bind with the target substance specifically [
15,
16,
17,
18,
19]. The optical transducer is integrated with the biosensing element closely, and the optical biosensors are classified based on their transducers. The optical biosensors can also be classified based on the dependence on a label for signal generation as label-based and label-free. In label-free biosensors, the signal produced by the interaction of the target and the biorecognition element is measured directly. In contrast, in the label-based biosensors, the detected signal originates from the complex formed by a label, such as a fluorophore or a chromophore, and the target on the transducer. In this section, we discuss a number of selected surface-based optical biosensor technologies for detection of viruses with an emphasis on their fabrication approaches and application areas.
2.1. Fluorescence-based optical sensors for virus detection
Fluorescence-based optical biosensors employ the fluorescent labels to produce the optical signal which rely on detecting the fluorescence signal as a result of analyte capture on the transducer [
12,
20,
21]. They are widely used in assay development owing to numerous commercially available fluorescent labels, uncomplicated labeling methods, multicolor fluorophores for multiplexed assays, fast response times with localized fluorescence signal, high temporal resolution and sufficient detection sensitivity [
22]. These advantages of fluorescence detection qualify as desirable in the detection of viruses and biological molecules [
23,
24,
25,
26]. However, certain limitations such as fluorophore blinking, photobleaching and insufficient detection limits for some target molecules make fluorescence-based optical biosensors less applicable in certain applications, for instance in detection of low abundance nucleic acids [
27]. Moreover, nonspecific binding of the fluorescent labels to the other components in the sample media remains as an issue in fluorescence detection systems [
28]. Besides, irreversible photobleaching of the fluorescent label restrains observation time, hence, affects the reliability of the test [
29]. Here, we present a number of fluorescence-based biosensor applications for virus detection.
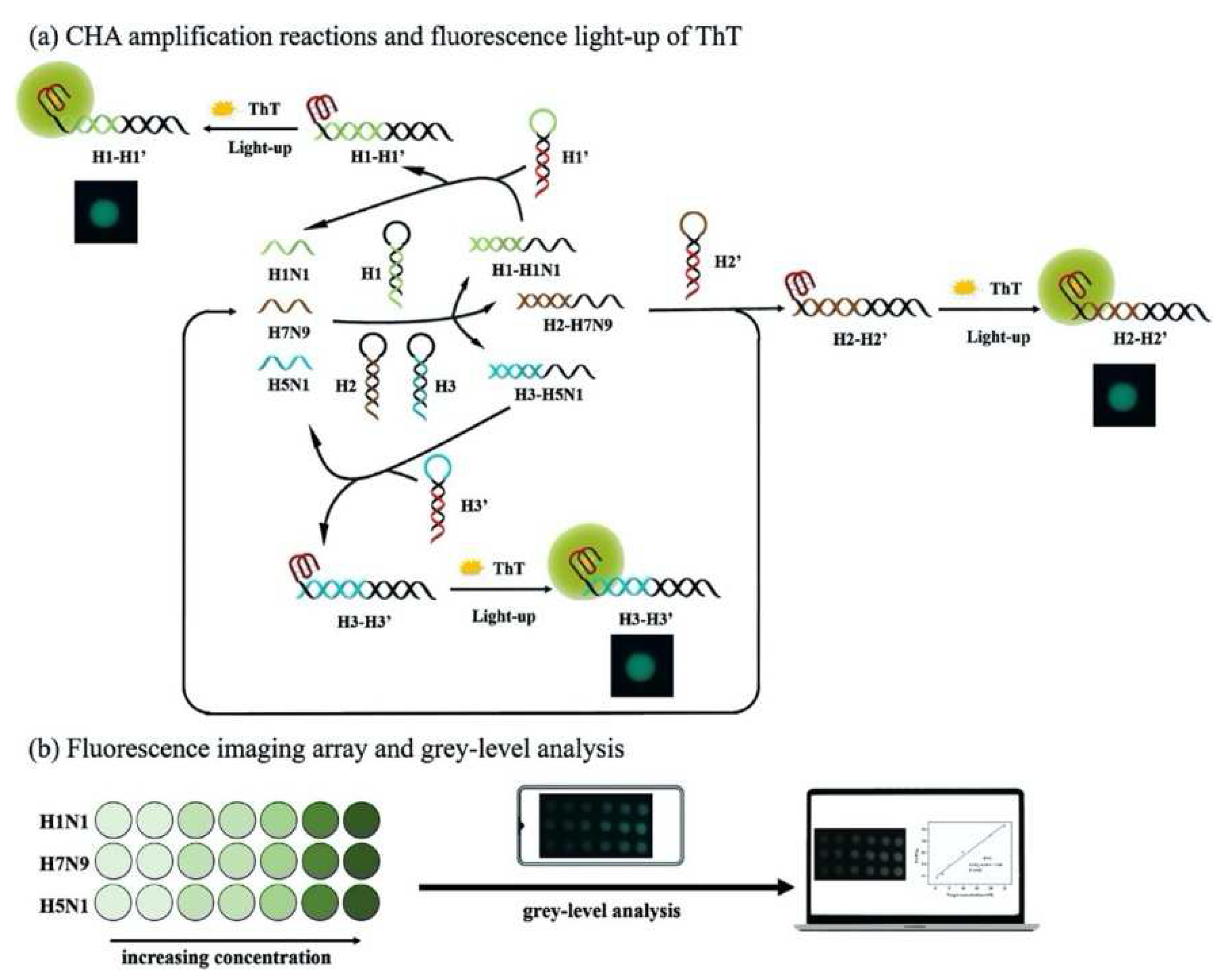
A multiplex imaging array was developed for rapid and low-cost diagnosis of trace avian influenza virus (AIV) using DNA biomarkers by Jiang et al. (
Figure 2) [
30]. They detected three subtypes of AIV DNA biomarkers (H1N1, H7N9 and H5N1) simultaneously using fluorescence imaging and gray-level analysis. They utilized a smartphone for imaging output and completed detection in 20 min. They employed catalytic hairpin assembly (CHA) amplification reactions and utilized thioflavin T, a specific G-quadruplex fluorescence probe for labeling. They reached detection limits of 136 pM, 141 pM and 129 pM for H1N1, H7N9 and H5N1, respectively. Also, the array sensors exhibited excellent anti-interference among the different subtypes and good mismatch discrimination in real sample application. Such a system can easily be applied for early detection of disease diagnostics in low-resource settings.
Another rapid viral detection and identification study was performed by Hepp et al. for detecting influenza virus, avian infectious bronchitis virus and SARS-CoV-2 specifically and quantitatively in approximately 20 min (
Figure 3) [
31]. They used fluorescence in situ hybridization (FISH) protocol, specifically rapid viral FISH protocol (rvFISH), where they used fluorescence microscopy to spatially detect and quantify DNA and RNA inside fixed cells and tissues using complimentary and fluorescently labeled oligos. They were able to detect influenza particles and infectious bronchitis virus (IBV), an avian coronavirus, down to a concentration of 10
5 PFU/ mL and 10
2 PFU/ mL, respectively, in a 20-min assay.
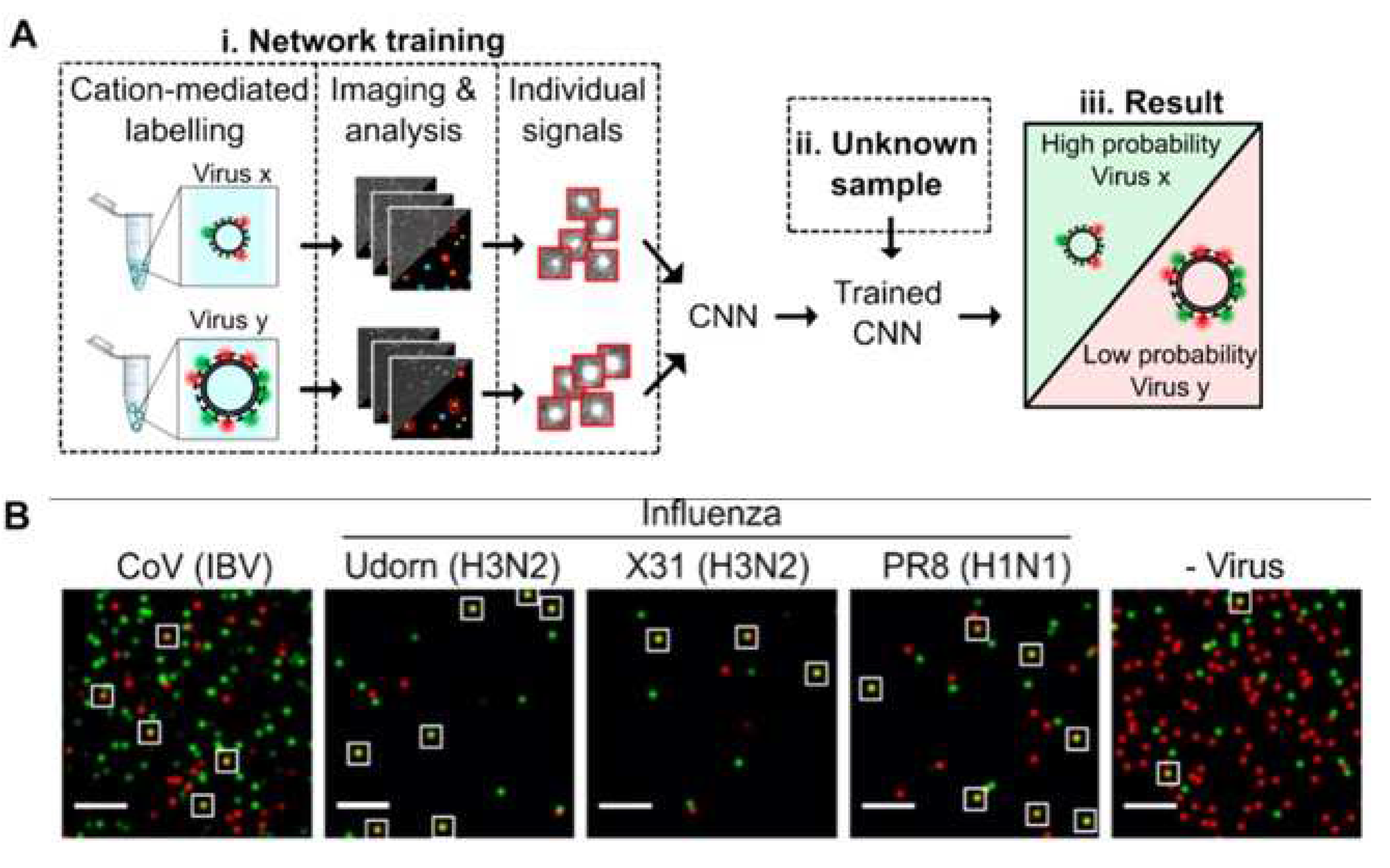
Shiaelis et al. present a methodology for virus detection and identification that uses a convolutional neural network to distinguish between microscopy images of fluorescently labeled intact different viral particles (
Figure 4) [
32]. They used single-particle fluorescence microscopy and deep learning. The assay successfully performed labeling, imaging, and virus identification in less than 5 min and did not require any lysis, purification, or amplification steps. They carried out two clinical tests of the method using 155 patient samples in total, which provided high overall sample accuracies of 98.0% and 97.1%.
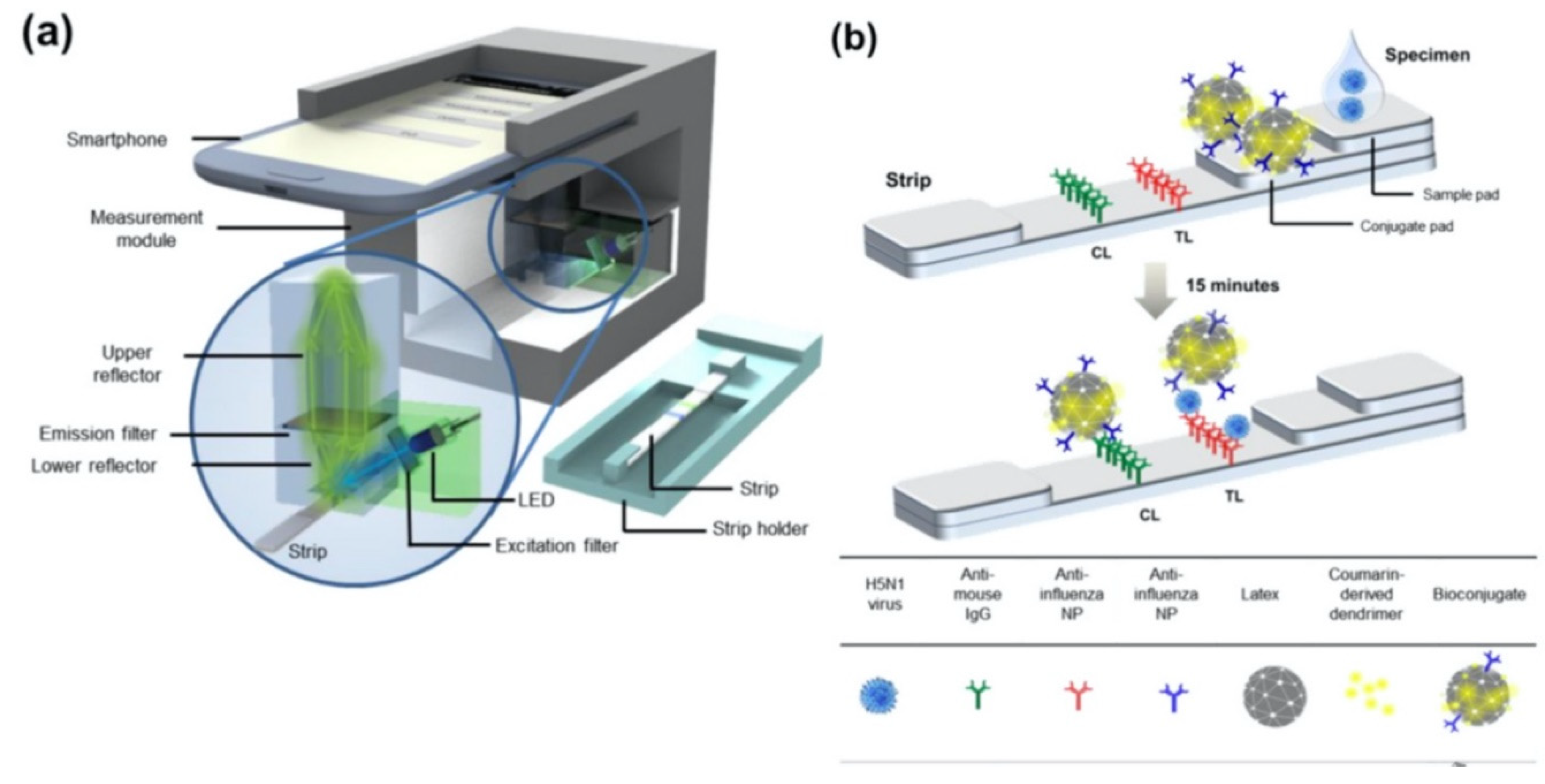
Moreover, Yeo et al. have demonstrated a field-level fluorescent lateral flow immunoassay which was combined with a smartphone-based fluorescent diagnostic device with an efficient reflective light collection module for the detection of avian influenza (AI) (H5N1, H5N3, H7N1, and H9N2) (
Figure 5) [
33]. Fluorescence light efficiency was improved with the use of latex beads along with the coumarin-derived dendrimer-based fluorescent as anti-influenza nanoparticles. lowest detectable virus titers were found as 6.25 × 10
3 PFU/mL for H5N3, 5.34 × 10
2 PFU/mL for H7N1 and 5.23 × 10
1 PFU/mL for H9N2 in throat swab samples. In addition, clinical validation of smartphone-based diagnostic device with H5N1-infected patient samples was completed within 15 min with sensitivity of 96.55% (28/29) [95% confidence interval (CI): 82.24 to 99.91] and a specificity of 98.55% (68/69) (95% CI: 92.19 to 99.96) (P < 0.0001).
2.2. Colorimetric biosensors for virus detection
Colorimetry-based biosensors allows visual detection via a change in color by naked eye or simple, low-cost portable optical detectors. These features make them proper candidates to fabricate point-of-care devices that can be used for rapid and cost-effective virus detection [
34]. They employ a simple platform with quick response and fair sensitivity, selectivity [
35]. In colorimetry-based solid-phase biosensors, on the sensor surface, which is usually a simple test strip, when the sample solution is introduced, a ligand-target complex is formed on the solid support. This complex results in a shift in color which can be easily observed for quantitative measurements. With the recent advancements in nanotechnology, progress has been made in improving the sensitivity of colorimetric detection systems by using various functional nanomaterials such as metal and metal oxide NPs, quantum dots, graphene and derivatives [
36]. In the NP-based approach, colloidal NPs that change color during aggregation or dispersion are conjugated with the biosensing element. Plasmonic-based colorimetric biosensors benefit from the localized surface plasmon resonance (LSPR) extinction coefficient in the visible range of noble metal NPs such as gold NPs (AuNPs). Binding event between the analyte and the AuNPs conjugated bioreceptor cause visible color change to detect viruses [
37]. NP-based colorimetric sensors can be used in a wide range of virus sensing applications. Parker and coworkers designed an immunoassay-based sensing technique which detected hepatitis E virus (HEV) real-time using Ag-decorated AuNPs. While anti-HEV IgG antibodies were used as conjugate to AuNPs in the inner part, in situ silver deposition was achieved on the outer part as a signal amplification strategy. The virus particles were entrapped by the utilized nanocomposites whereas 3,3′,5,5′-tetramethylbenzidine (TMB) and H
2O
2 were added to decompose back Ag-shell to Ag
+. After addition of TMB- H
2O
2, based on the obvious color change, the concentration of HEV was quantified and real-time monitoring of HEV in real sample was realized [
38].
Paper-based lateral flow immunoassays (LFIAs) as point-of-care devices are widely used for early disease diagnostics. Despite its widespread use, it is often limited due to insufficient sensitivity for required sample sizes and short time frames of testing. Loynachan et al., designed a highly sensitive and serum-stable paper-based, nanoparticle catalyst-labeled LFIA for the detection of a viral capsid protein, p24, one of the earliest and most conserved biomarkers of HIV. They used porous platinum core-shell nanocatalysts (PtNCs), and then explored the application of antibody-functionalized PtNCs with high affinity and specificity modified nanobodies toward p24 and established the key larger nanoparticle size regimes needed for efficient amplification and performance in LFIA [
39]. In another study, Wang et al. designed a rapid diagnostic platform integrated with a low-cost reader and a multicolor and a multicolor 4-plex immunoassay to detect and distinguish between dengue virus (DENV) and chikungunya virus (CHIKV) IgM/IgG [
40]. The developed platform employs unique color-mixing encoding and quantitative readout strategy while using an optical reader designed to minimize variations in color detection. This provides a consistent multiplexed detection of dengue and chikungunya IgM/IgG antibodies in human clinical samples within 30 min (
Figure 6). The multiplex assay requires low sample volumes and has ability to test four samples simultaneously, which makes the rapid diagnostic platform a great candidate to be used in resource-limited settings.
2.3. Virus detection with Surface Plasmon Resonance and Localized Surface Plasmon Resonance
Surface plasmon resonance (SPR) is defined as an electromagnetic (EM) phenomenon depending on collective resonant oscillations of free electrons and incoming protons passing through metal-dielectric interface. The working principles of SPR-based optical sensors depend on the detection of changes in refractive index that arise on the dielectric surface near the metal layer [
41,
42]. The properties of this metal layer strongly influence the SPR response that is generated according to refractive index change. The metals which have conduction band electrons, such as gold, silver, aluminum, and copper, show capability for resonating at an appropriate wavelength with the incident light. Gold is the most preferred metal film due to its chemical stability and sensitivity for sensing applications [
43]. A typical SPR experiment consists of three main components (i) immobilized recognition element, (ii) the prism of light, (iii) analyte [
44]. The recognition molecule is immobilized onto the gold surface of the sensor chip. After surface functionalization, a sample solution containing the analyte is passed across the chip surface. Incident light passing through the prism excites the electrons of the metal film to form surface plasmon. As the incident light reaches the medium at various angles, the photons are absorbed by the plasmon wave at a specific angle, as critical angle, which is affected by the refractive index of the medium. When an analyte binds to the immobilized recognition element, due to the mass accumulation on the immobilized layer, the refractive index of the medium near the chip surface changes, which shifts the critical angle for the immobilized molecule [
45,
46,
47]. Thus, any physical changes that cause refractive index change can be monitored without labeling in real time. SPR-based sensing techniques emerge with high potential in diagnostics with their magnificent properties referred above, to accomplish rapid and point of care (POC) detection of viruses. Antibodies against viral antigens and surface proteins are used as bio-receptors to capture viral proteins and intact viruses while preparing sensor chip surfaces. Additionally, artificial recognition sites obtained by molecular imprinting or laboratory made capturing molecules such as DNA and RNA aptamers are used to capture several viruses [
48]. SPR sensing technology, which is highly accurate in detecting biomolecular interactions, also offers various advantages of label-free monitoring, rapid and sensitive outcome, and ability to miniaturize for on-site monitoring [
49]. However, to succeed in early diagnosis of viruses using SPR method, further enhancements in the selectivity and sensitivity are still required [
48]. Here, different SPR-based techniques that can detect viruses are discussed.
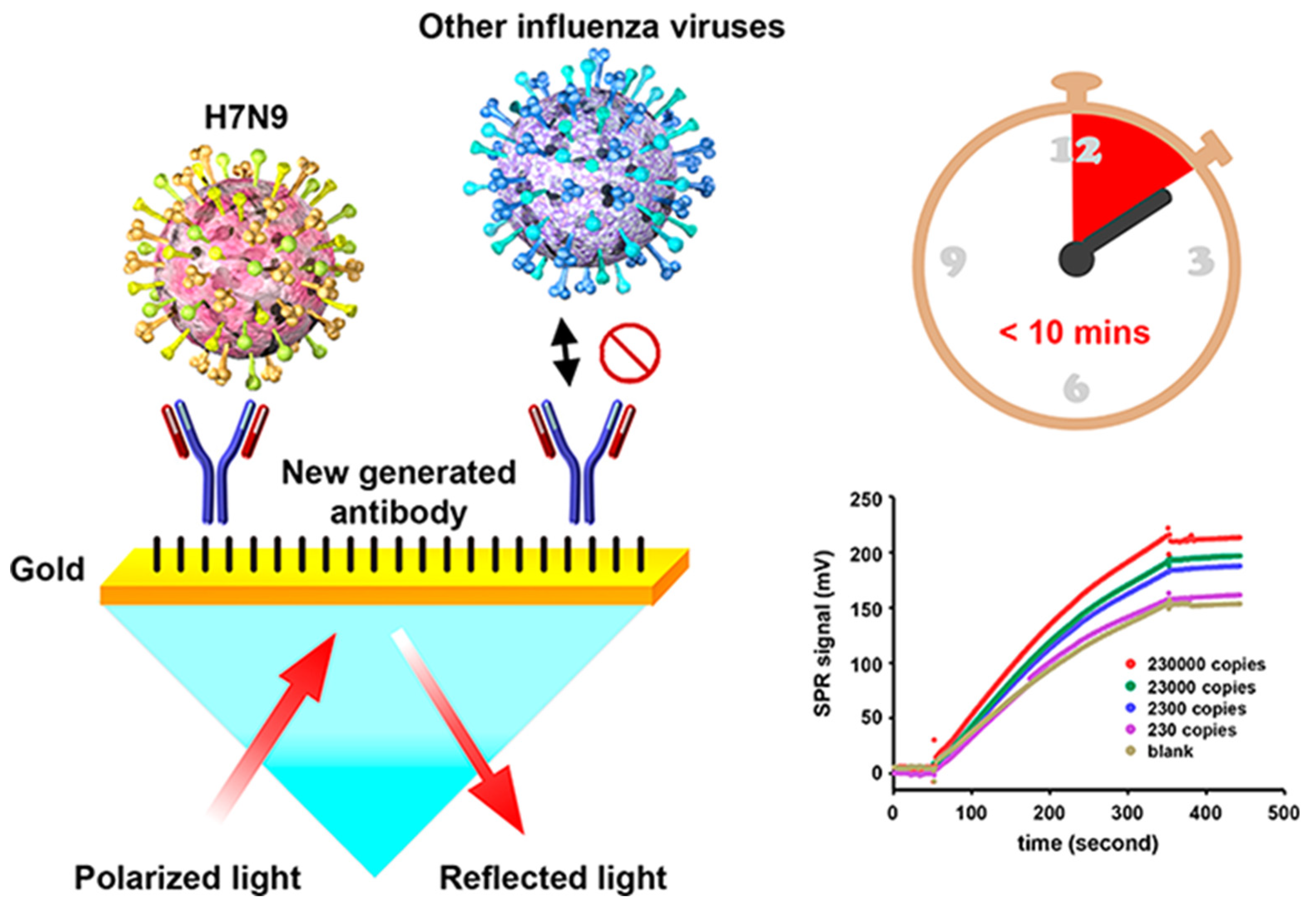
Chang et al. developed an intensity-modulated surface plasmon resonance (IM-SPR) biosensor integrated with a lately generated monoclonal antibody which enables rapid and sensitive detection of H7N9 virus due to the avian influenza A H7N9 virus in China [
50]. The novel antibody displays important specificity in the H7N9 virus detection. They experimentally reported a detection limit of 144 copies/mL using the proposed approach for the H7N9 virus, which is a 20-fold increase in sensitivity compared with homemade target-captured ELISA. They reported less than 10 min assay time. 10 μg/mL of the captured antibody H7-mAb was covalently immobilized to the reaction spot of the SPR chip through mixed self-assembled monolayers of 11-mercaptoundecanoic acid and 6-Mercapto-1-hexanol (1:9) via the amine coupling protocol (
Figure 7). The results demonstrated that the simple SPR-based technology was successfully used in sensitive H7N9 virus detection. They reported that the proposed SPR system can be used in implementations of other emerging virus detection platforms.
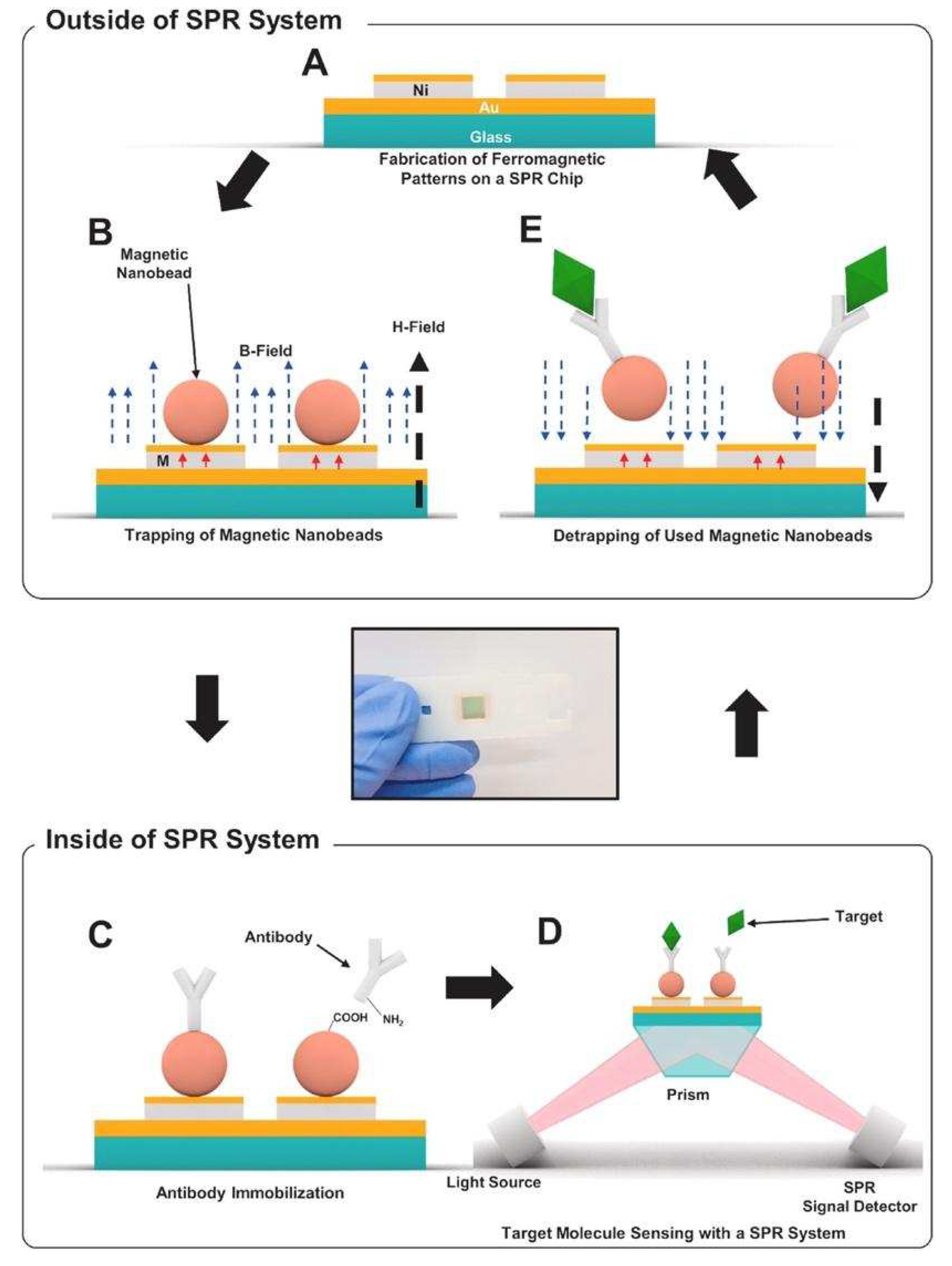
As effective and simple methods are needed for virus detection, Yoo et al. designed a reusable magnetic SPR sensor chip for H1N1 influenza virus detection in a conventional SPR sensor. They used ferromagnetic patterns on an SPR sensor chip to prepare a layer of magnetic particles, and a solid substrate for SPR sensing [
51]. They demonstrated the system platform which enables repetitive use by removing magnetic particles using external magnetic fields at the end of an experiment without the need for antibody-modifying processes which would be a substantial step toward the future application of virus sensor systems.
Figure 8 shows the schematic of the virus detection by antibodies conjugated to magnetic beads on the substrate.
Figure 8A,B depict the ferromagnetic nickel patterns and trapping of magnetic particles on the SPR chips, and
Figure 8C schematizes the antibody immobilization on magnetic particles using EDC-NHS coupling.
Figure 8D,E show the detection of target molecules, and then removal of the magnetic particles by an external magnetic field, respectively. The ferromagnetic patterns on the sensor chip surface are deflected by the strong external fields so that the large aggregation of magnetic particles on the sensor surface was reduced. This study showed the use of a single reusable SPR chip for the detection of nucleoprotein of H1N1 influenza virus without a drastic signal degradation. The cost of the SPR chip was significantly reduced by reusing the SPR repeatedly.
Receptor-analyte interaction occurring on the surface of plasmonic biosensors is also monitored by localized surface plasmon resonance (LSPR). Unlike SPR, LSPR is formed by a light wave absorbed within conductive nano-plasmonic materials which are smaller than the wavelength of incident light [
52]. Owing to the enhanced signal amplification provided with the use of nanomaterials which have important optic, electrical and magnetic features, a low limit of detection can be obtained. While incident light interacts with the metallic nanoparticles (NPs) of the surface, strong localized EM field generated around these nanostructures enables a strong peak in the course of the absorption spectrum collection at resonance [
53]. The height of LSPR peak and the corresponding wavelength are affected by not only the sensing medium but also the material type, size, and the shape of the plasmonic NPs. The utilization of the nanoparticles for the decoration of chip surface also provides large surface area to immobilize a high number of bioreceptor molecules, which increases the sensitivity and specificity of the sensing technique. Several metallic nanostructures such as nanospheres, nanofibers, nanorods, nanoshells and nanowires can be used to fabricate sensing surfaces. The dimensions and the shape of these nanostructures directly affect the plasmonic properties (scattering and absorption ratio, resonance wavelength) of them [
54]. Two main drawbacks of SPR are escaped with LSPR: firstly, temperature sensitivity is not an issue for LSPR since the method depends on a simple absorbance measurement, secondly, less time is required for the whole binding event due to the faster spread of the analyte to the increased surface area of NPs than metallic film [
55,
56]. On the other hand, the response produced for non-specific binding as well as for the refractive index variations is the major drawback limiting the applicability and effectiveness of the sensor to detect analytes in complex media [
57]. In the last decade, a remarkable increase in the number of nanomaterial-based sensing techniques developed for viral diagnosis has been reported.
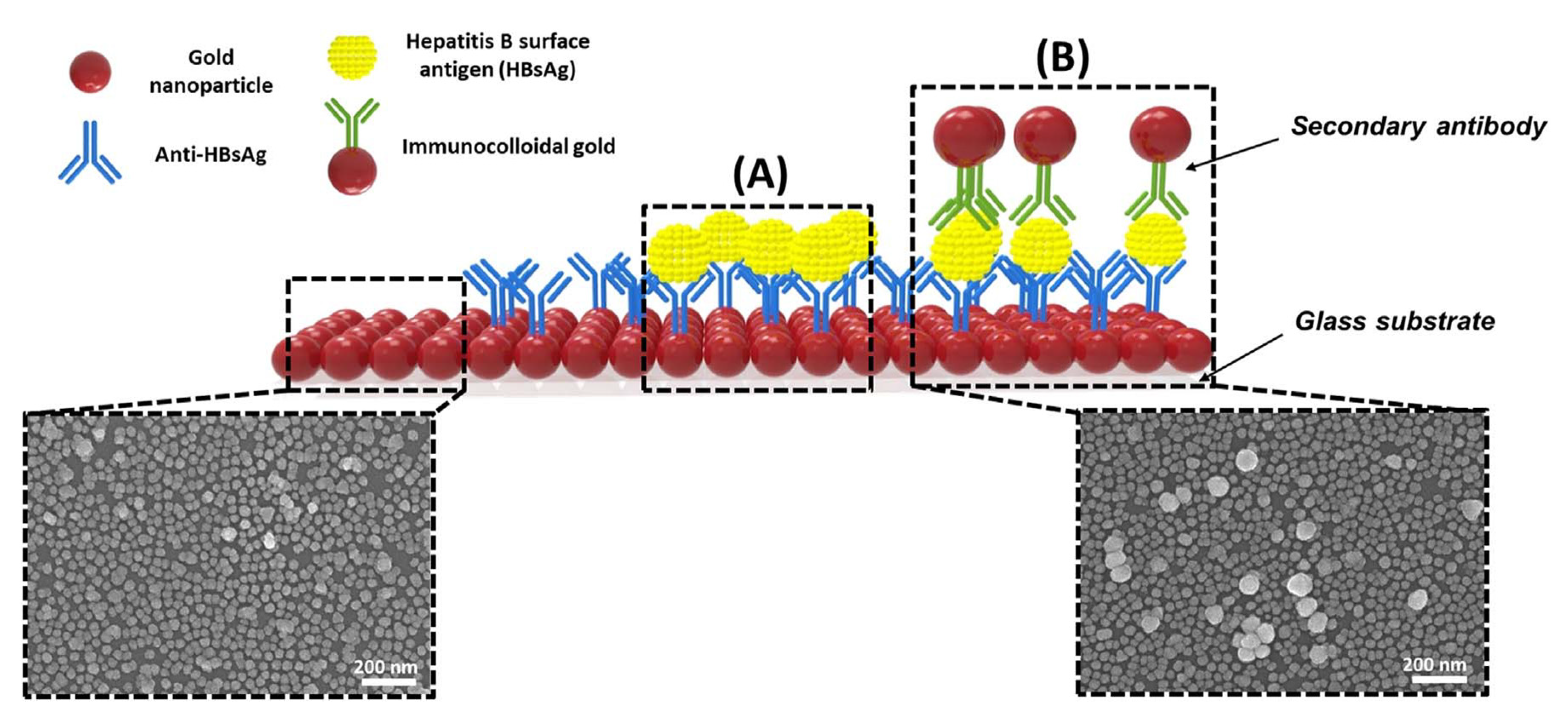
Kim et al. developed a unique structure that used gold nanoparticles (AuNPs) to develop a highly sensitive method for hepatitis B surface antigen (HBsAg) detection [
58]. They designed a single-layered LSPR chip format via antigen-antibody reaction-based detection symmetry using AuNPs. The virus was sandwiched between two different sizes of AuNP on an AuNP-laden substrate. In their study, two AuNPs in close proximity repulse each other in a plasmon resonance state in the presence of the virus, resulting in a stronger peak shift effect than that in the non-sandwich state. The concentration of HBsAg was reported at 10 pg/mL. They fabricated a modified detection format to further improve the detection limit by fixing a secondary antibody to the AuNP monolayer, which detected a 100 times sensitive detection limit. They showed highly sensitive detection of HBsAg, 100 fg/mL within 10–15 min, using a novel-designed sandwich immunoassay LSPR chip. A diagrammatic of the LSPR biosensor chip is shown in
Figure 9. They showed that implementation-based systems have been affected by particle size in using the LSPR.
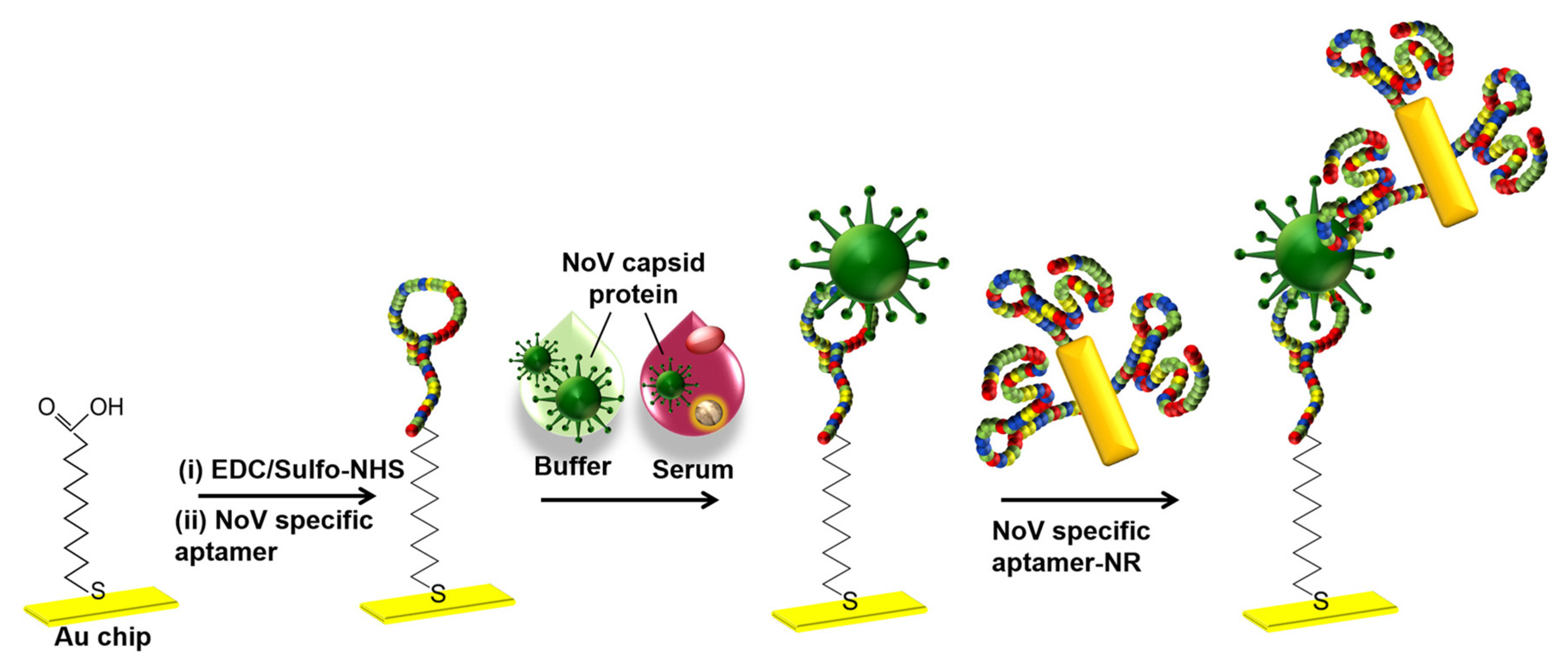
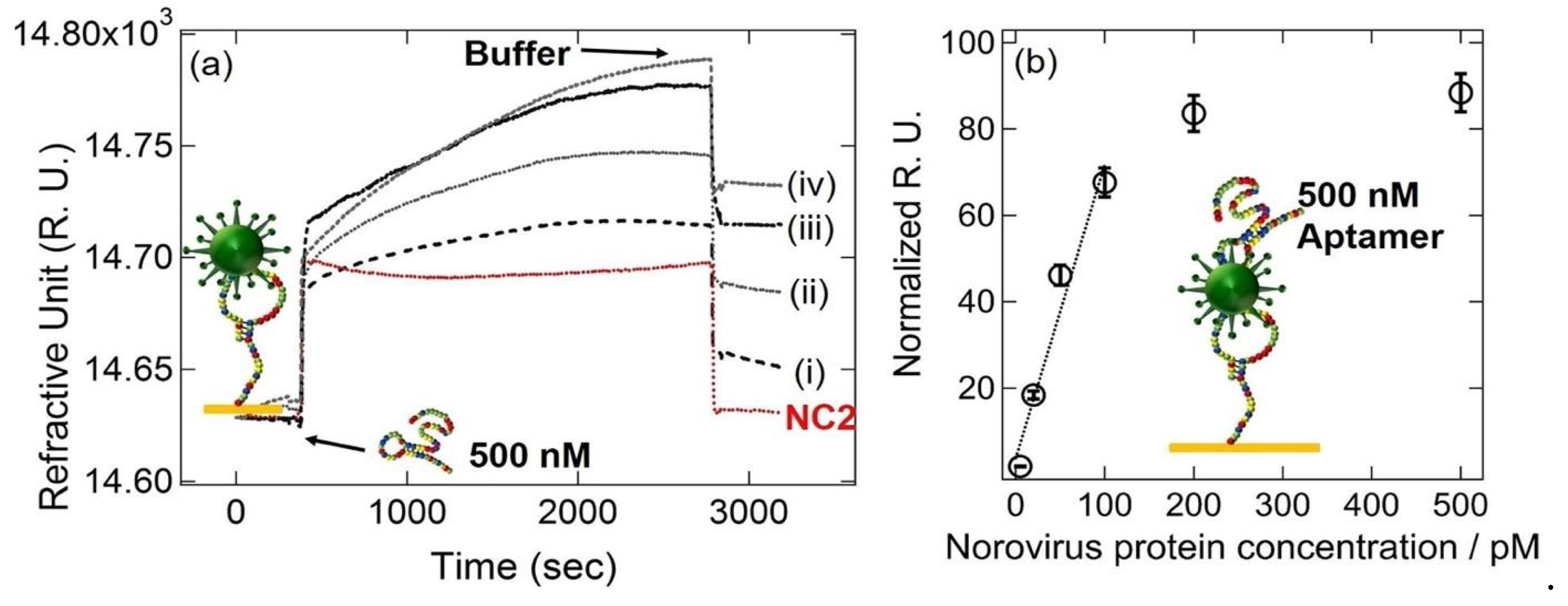
Kim et al. developed a gold nanorod-enhanced surface sandwich assay for norovirus (NoV) capsid protein detection via a novel pair of aptamers, in conjunction with SPR [
59]. They used four different DNA aptamer sequences that were known to be specific for the NoV protein to find the strongest binding constant. The aptamer II sequences were covalently bonded onto a chemically modified thin Au chip surface. For the formation of the surface sandwich complex, the NoV-specific aptamer was attached to the surface of the SPR chip which was modified via 1-Ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride and N-(hydroxy-sulfosuccinimide) solution. Then, NoV capsid protein and gold nanorod- enhanced aptamer were adsorbed. The authors reported a 50 aM limit of detection value for NoV capsid protein after flowing different concentrations of NoV protein solutions over the aptamer-modified chip. The NoV capsid protein concentrations also were analyzed in human serum samples. The schematization of the aptamer-aptamer sandwich assay strategy and representative SPR sensorgrams are shown in
Figure 10.
2.4. Virus detection with Surface-enhanced Raman scattering
Surface-enhanced Raman scattering (SERS)-based sensing platforms have gained attention in the last few decades due to their fascinating advantages: (i) high sensitivity, (ii) capability for multiplex sensing, (iii) applicability as a POC device, (iv) laborless sample preparation [
60]. Although Raman spectroscopy is found to be a beneficial tool for analyte determination by providing fingerprints like spectrum for complex samples, its inherently weak signals limit its use for diagnosis. However, in SERS technology, the limitations of the weak Raman signal of Raman-active material are overcome by enhancing the EM field by using metallic nanostructures. The development of high-sensitivity SERS sensors with advanced EM field is carried out by optimizing the design of plasmonic nanostructures [
53]. The main advantages of SERS technology are the specific analyte determination ability even at very low concentrations without sample pre-treatment and applicability as a POC device. On the other hand, signal-reducing degradation in the substrate over time due to the requirement for close contact between the analyte and the amplification surface is the main challenge that limits the reproducibility of the SERS signal [
61]. SERS-based sensors are dependent on two main methods as direct and indirect. While the direct method relies on the detection of the spectrum of an analyte, in the indirect technique that is constructed as a sandwich-like form SERS signals are getting from the reporter molecule, not the analyte. To differentiate the spectral data of an analyte in the direct technique, main component and linear discriminant analysis need to be performed by comparing the samples of patients and healthy individuals [
62]. In the indirect technique, sensitivity of the method reaches at ultra-low concentrations with combining immunoassay strategy to detect analyte. In order to meet the growing need for accurate and rapid virus detection in recent years, multiplex immunoassay-based SERS has become prominent due to the limitations of PCR-based techniques that only require genetic material for testing [
63].
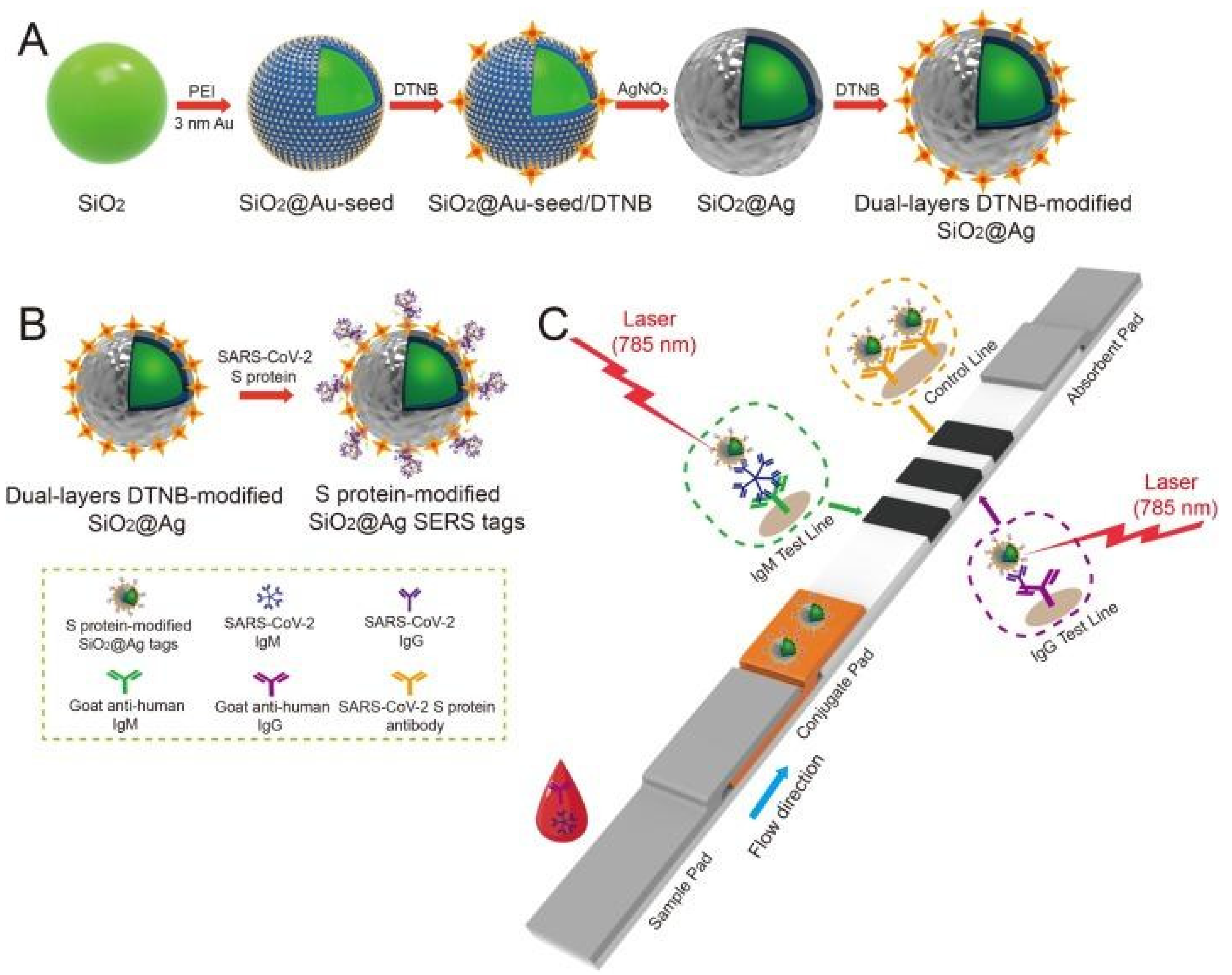
Liu et al. used SERS-based lateral flow immunoassay (LFIA) to determine COVID-19 at the point of care [
64]. They used Raman molecules to functionalize dual layers of silver shell on SiO
2 core NPs as SERS tags. Anti-human IgM and IgG were immobilized onto the two test lines of the strip to capture the formed SiO
2-Ag-spike (S) protein-anti-SARS-CoV-2 IgM/IgG immunocomplexes. The author used a 785 nm excitation with 10.0 mW laser power. The detection value was 1 ng/mL of the S-protein antibody which was 800 times higher than that of standard gold NPs-based LFIA for template IgM and IgG. A schematic of the dual-layers of Raman reporter molecule 5,5′-dithiobis-2-nitrobenzoic acid (DTNB) modified silica-Ag NPs (SiO
2-Ag) via LFIA is shown in
Figure 11. They revealed that the results showed high accuracy and specificity for patients with SARS-CoV-2 infection using the designed method.
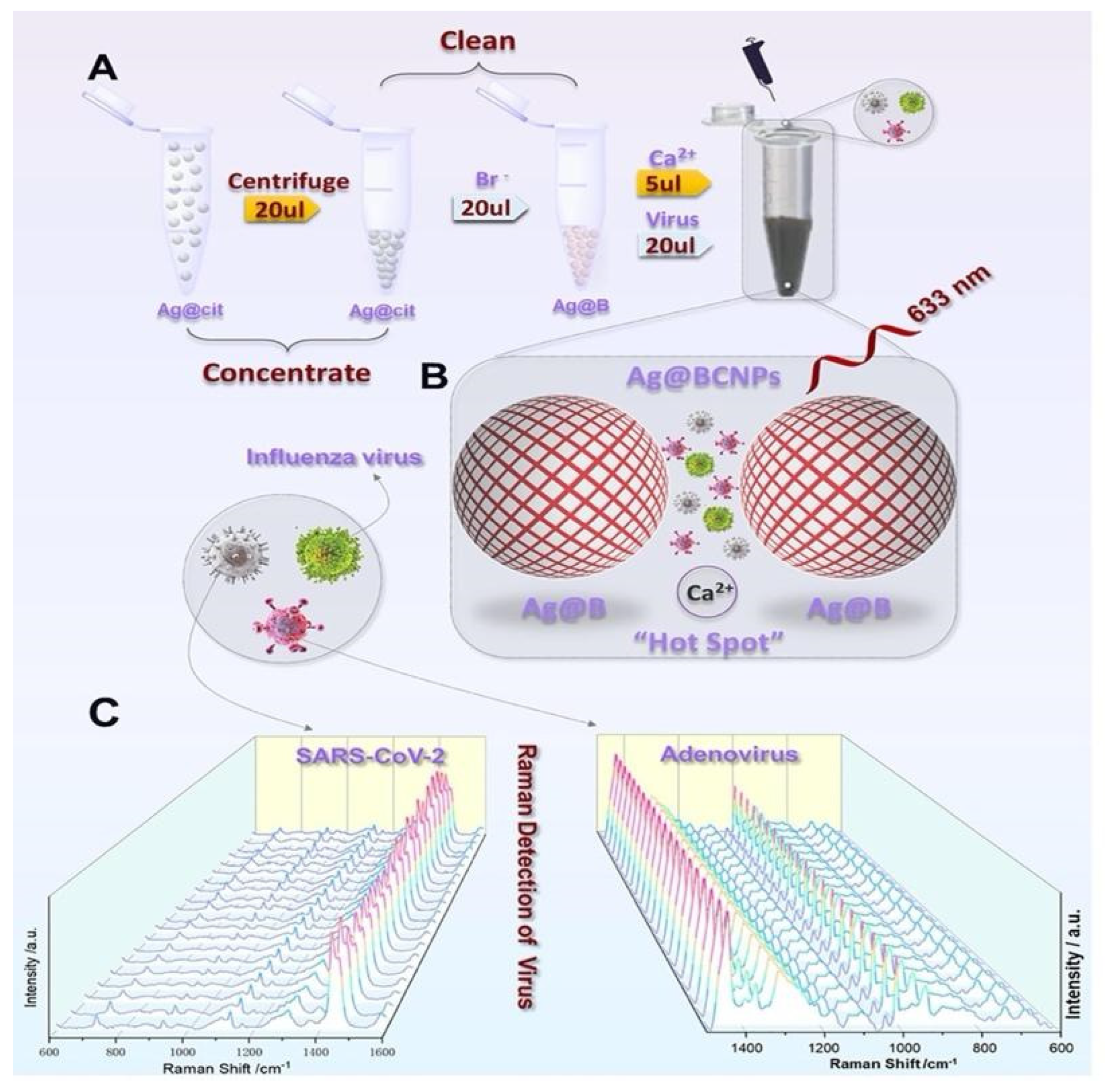
A new SERS detection system was developed by Zhang et al. They used citrate to reduce Ag NPs and added acetonitrile solvent to form a superb hot spot proper for viruses by aggregating Ca
2+, and exploring the human adenovirus, SARS-CoV-2 without difference and marker [
65]. Then, they showed 100 PFU/test detection limits of viruses within 1 to 2 min when combined with machine learning.
Figure 12 shows the experimental process where Ag NPs were modified by bromide ions modified and acetonitrile and Ca ions.
2.5. Optical resonators for virus detection
Optical resonator sensor systems have attracted significant attention in recent years as a powerful tool for detecting a range of biological and chemical analytes with high sensitivity and specificity [
66]. These sensors operate by measuring the spectral changes in the resonant frequency of an optical cavity when the analyte is introduced into the cavity. The main principle of an optical resonator sensor is based on the detection of light intensity changes induced by changes in the refractive index of the medium surrounding the resonator. The resonator consists of a thin film layer or a ring resonator that supports resonant modes, which are excited by a laser beam. The resonant modes of the resonator are highly sensitive to changes in the refractive index of the surrounding medium. When a target molecule or virus binds to the resonator surface, it causes a change in the refractive index, which is detected as a shift in the resonant frequency of the resonator [
67].
The basic design of an optical resonator sensor consists of a high-quality factor (Q-factor) resonator, such as a microdisk or microring, and a waveguide coupled to the resonator. The resonator acts as a sensitive transducer that is capable of detecting changes in the refractive index of the surrounding environment caused by the analyte binding to the surface of the resonator. The change in the resonant frequency is then measured by monitoring the light transmitted through the waveguide. Fabry-Perot cavities, whispering gallery mode (WGM) cavities, photonic crystal cavities (PC) and plasmonic resonators are some of the most common types of optical resonators [
68,
69].
For biological particle detection, optical resonators such as microspheres and microtoroids have been used to detect individual virus particles in the size about 100 nm in a label-free format [
70,
71]. WGM cavities are highly efficient optical resonators, with high Q-factors which allows for the detection of very small changes in the refractive index of the cavity, making them useful for a variety of sensing applications. He et al. developed a WGM microresonator using frequency splitting in a microlaser and showed detection of Influenza A virus on this sensor [
72]. Their method relies on measuring the changes in the beat frequency as an ultra-narrow emission line from a WGM microlaser is split into two modes as a result of nanoparticle binding. Before nanoparticles arrive, there is a single laser mode and the laser intensity is constant. The lasing mode splits into two modes when the first nanoparticle binds, leading to a beat note with a frequency that is equal to the difference in frequency between the two modes. Using this approach, they were able to detect sizes down to 15 nm for polystyrene nanoparticles and 10 nm gold nanoparticles as well as Influenza A virus. However, this system was tested with purified nanoparticle and virus solutions and has yet to show multiplexed virus detection from complex biological systems.
As another type of resonator-based sensors, ring resonators have been preferred owing to their unique potential for the ability to be coupled in high-throughput arrays efficiently for multiplexed analysis [
73,
74]. A rapid detection of an Ebola biomarker with optical microring resonators was performed by Qavi et al. [
75]. Soluble glycoprotein (sGP) is the primary product of the glycoprotein (GP) gene of Ebola virus (EBOV), which is a nonstructural secreted GP that is expressed from the unedited RNA transcript. There are several roles sGP appears to play in EBOV pathogenesis, therefore, it is very useful to be utilized as a biomarker in virus detection. In this study, the authors developed a sensor by adapting a silicon photon microring resonator platform to detect EBOV sGP (
Figure 13A). The microring resonator sensor detected sGP in under 40 min with a LOD as 1.00 ng/mL in serum which was much lower analytical sensitivity compared to the ELISA tests.
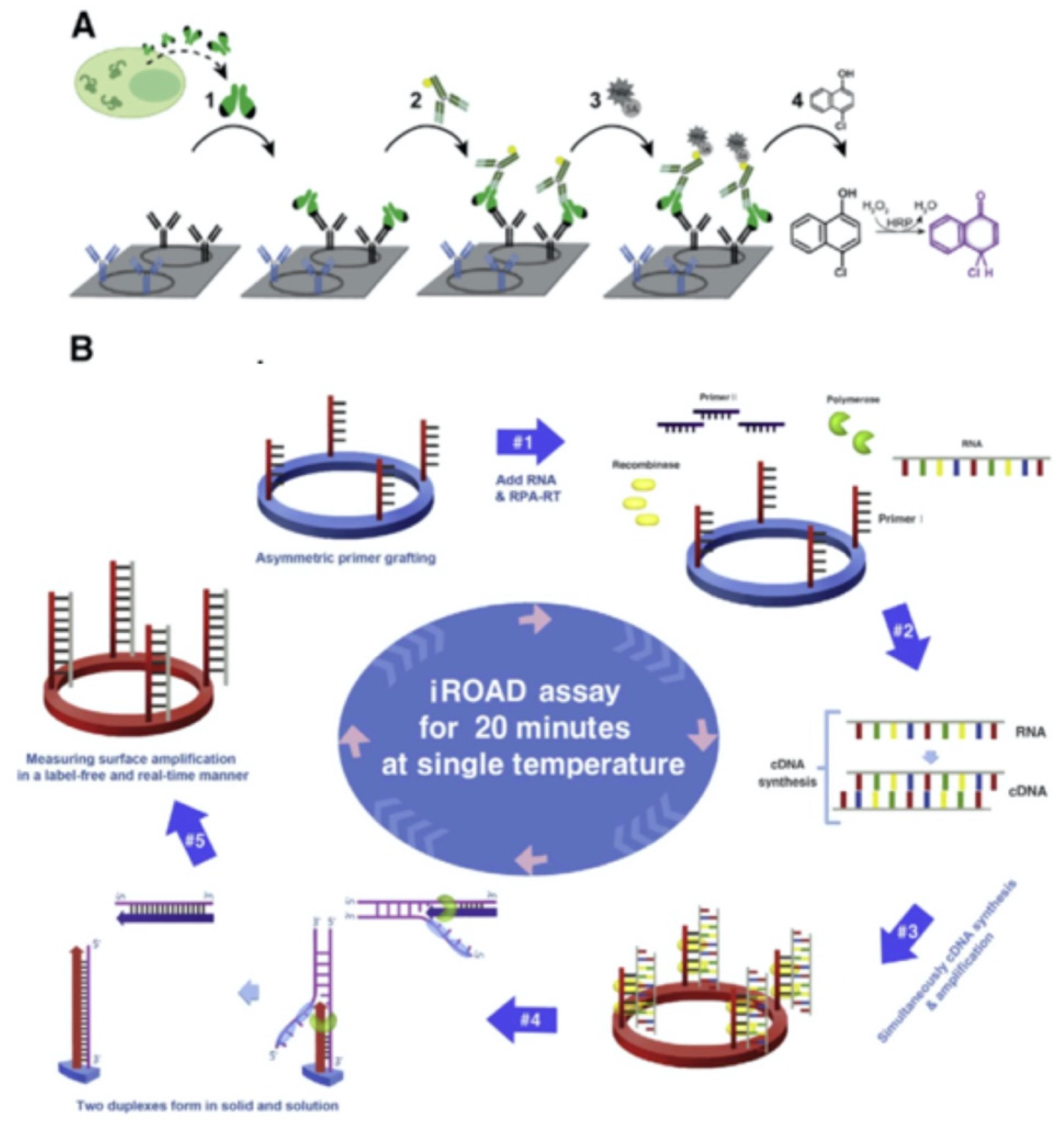
Koo et al reported an isothermal, label-free, one-step RNA amplification and detection system, termed as iROAD, for the diagnosis of respiratory diseases based on silicon microring resonators [
76].
Figure 13B shows the chip fabrication and functionalization for the assay. The iROAD assay achieved a one-step viral RNA amplification/detection example to rapid analysis (< 20 min). They have obtained a LOD for the iROAD assay to be 10-times more sensitive than that of real-time reverse transcription-PCR method. The authors tested the iROAD system on 63 human respiratory samples and confirmed its utilization as a more robust operation by using an array of microrings for multiple detection in clinical use.
Optical resonators offer several advantages over traditional optical devices, including high sensitivity, selectivity, and miniaturization. Optical resonators can be designed to have a very high Q-factor, which allows for the efficient coupling between the optical signal and the surrounding environment. However, optical resonators also have some limitations, including sensitivity to temperature and fabrication complexity.
2.6. Interferometry-based sensor platforms
Viruses are difficult to detect using conventional light microscopy which, for the most part, relies on measuring the scattered light by the imaged objects. This is due to their small size (typically 20 – 300 nm in diameter) and low contrast. Light-particle interaction for small-sized particles can be represented by an induced dipole. The strength of an induced dipole is directly proportional to the polarizability of the particle which can be given as
where
R is the radius of the particle, and
εp and
εm are the permittivity of the particle and medium, respectively. The optical techniques that detect scattered light intensity generate a signal proportional to the |
Es|
2 which scales with |
α|
2 thus
R6. Therefore, the scattering signal recorded at the detector drops below the shot-noise limit for small particles. On the contrary, interferometric imaging utilizes a strong reference beam (
Er) which interacts with the weak scattered fields (
Es) from the particle and modifies the intensity obtained at the detector as
where
is the phase angle difference between the reference and scattered fields. As the particles size gets smaller, the scattered field, the second term in the Equation 2, becomes very small compared to the other two terms representing the reference field and the interference signal. Once the reference field is subtracted, the signal recorded at the detector is proportional to the multiplication of the reference and scattered fields, thus proportional to
R3 as opposed to
R6. As a result, interferometric imaging makes it possible to detect smaller particles as well as higher dynamic range of particles sizes. Due to these advantages, interferometry has been utilized for both ensemble-based measurements and single nanoparticle detection in previous studies [
77]. One example of ensemble-based measurement technique is biolayer interferometry (BLI), which is a label-free, real-time characterization technique for biomolecular interactions [
78]. In this technique, a fiber optic biosensor is used to illuminate the sensor area with white light and the resulting shift in the wavelength of the reflected light is recorded. Although this technique was shown to perform antibody detection with similar sensitivity to ELISA, there are some disadvantages associated with it such as lack of single nanoparticle detection ability and signal jumps as the new solutions are introduced to the well [
79]. Interferometry-based nanoparticle imaging techniques have also been developed for single virus detection [
80,
81]. However, these techniques use cost-inefficient lasers as the light source and can be time consuming due to small measurement area that is on the order of nanometers.
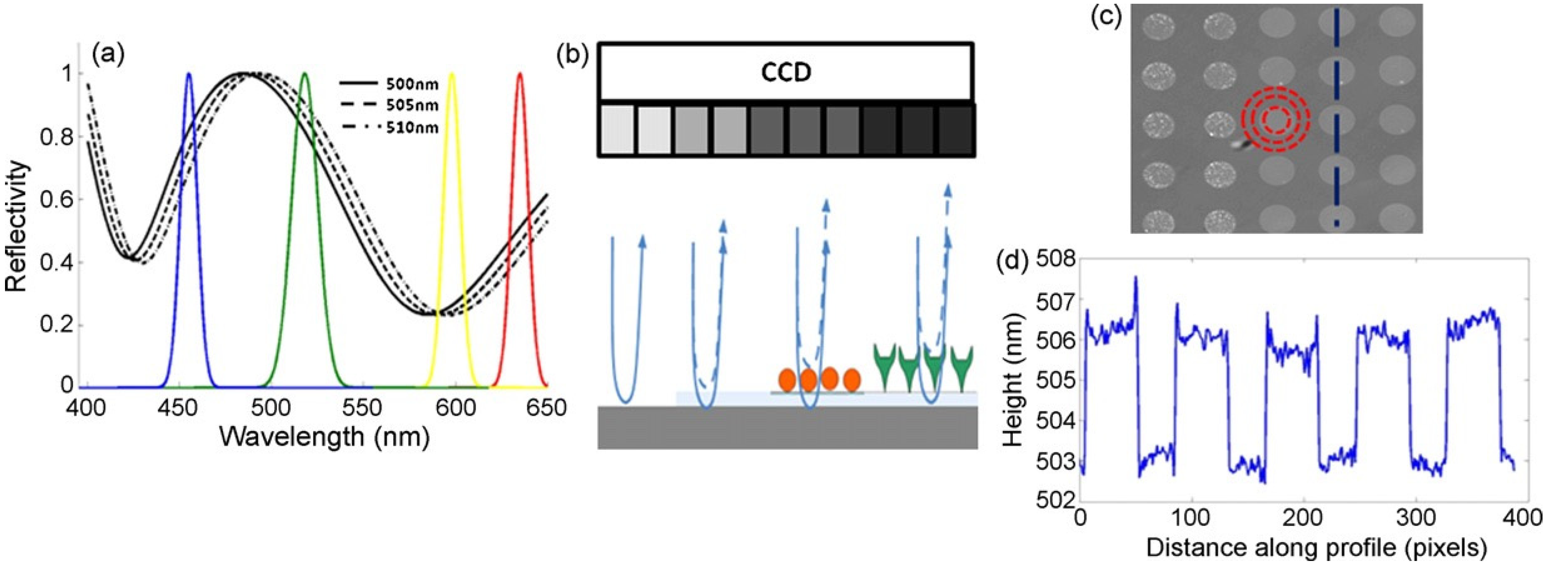
Interferometric reflectance imaging sensor (IRIS), developed by Ozkumur et al., is a label-free biosensor that can probe bimolecular interactions on a silicon/silicon dioxide (Si/SiO
2) substrate in a multiplexed microarray format [
82,
83]. Detection mechanism of IRIS depends on obtaining the interference signature of the reflected light from Si/SiO
2 substrate and measuring the optical thickness of the top layer (
Figure 14). The substrate is a silicon chip with a thermally grown oxide layer which is spotted with capture probes that are specific for the target molecule. Binding of target molecules in the solution to the surface causes height increase in the transparent oxide film which in turn changes the optical path length difference (OPD). The intensity of the reflected light at a given wavelength is determined by the OPD between the top of the biomass layer and the Si/SiO
2 interface. The thicker the biomass layer gets, the higher the OPD becomes, and this increase in the OPD leads to a shift in the spectral reflectivity curve (
Figure 14a) as well as change in the reflected light intensity at a specific wavelength. In IRIS optical setup, four LEDs with different wavelengths (455, 518, 598, and 635 nm) illuminate the substrate sequentially and the intensity of the reflected light is recorded by a CCD to generate an intensity image of the chip surface. Then, each pixel in this intensity image is fitted to the reflection function from which the thickness of the transparent film (oxide layer plus any biomass layer) is obtained. The calculated thickness difference for a given spot between two time points (before and after analyte incubation) is converted to biomass density by using simple conversion factors and an average mass density is calculated from replicate spots [
84].
IRIS has been shown to be a versatile platform for monitoring protein-protein [
85,
86], DNA-DNA [
87], and DNA-protein interactions [
88], with applications into cytokine detection, identification of single nucleotide polymorphisms (SNPs), study of DNA binding proteins such as transcription factors, and antibody affinity measurements. IRIS was also used to detect intact vesicular stomatitis virus (VSV) particles with a detection limit close to 10
5 PFU/mL as well as internal viral proteins, such as nucleocapsid and matrix proteins by lysing the viruses with detergent [
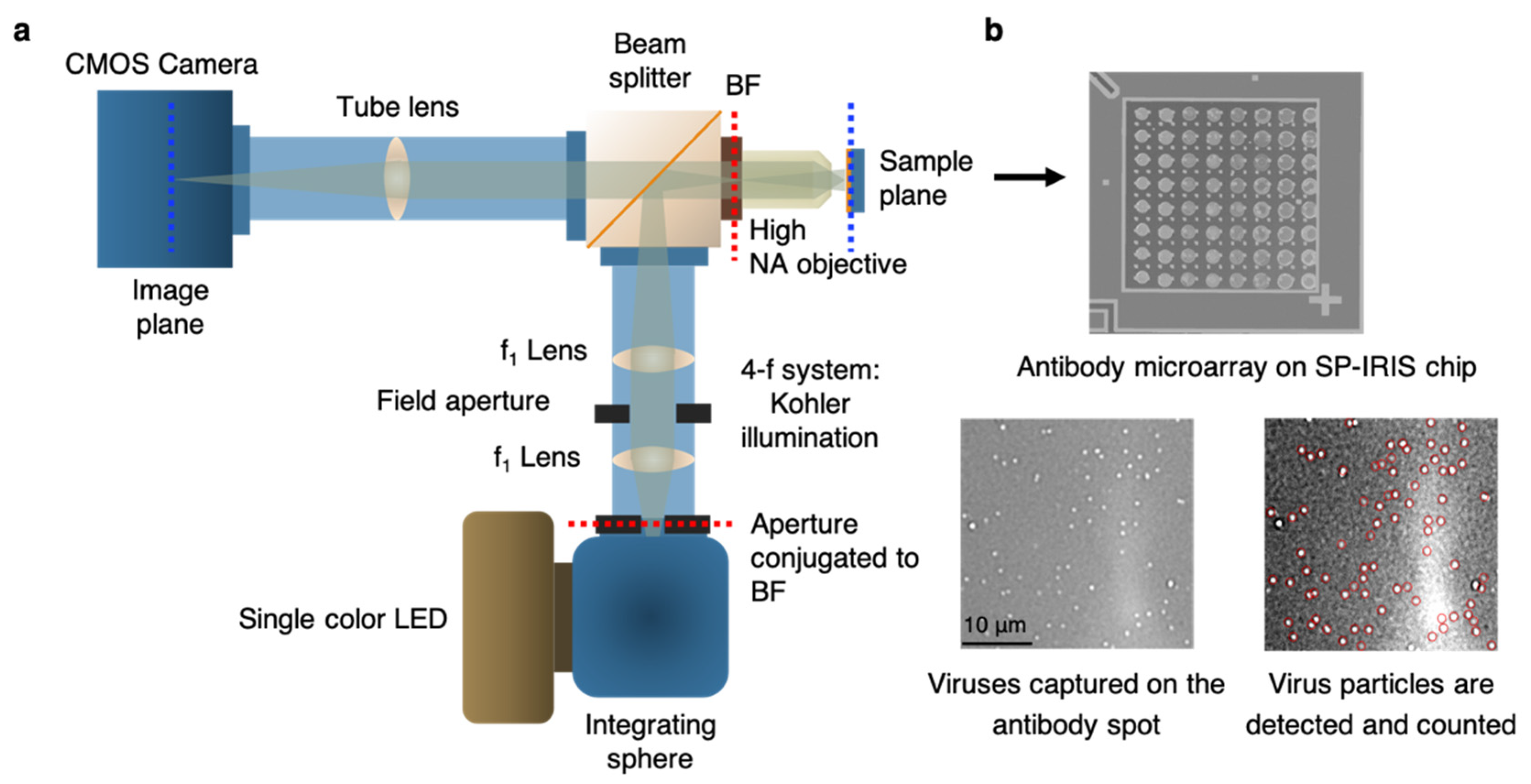
89]. IRIS platform was later modified to generate a digital detection modality to allow for the visualization and counting of single nanoparticles. This platform, referred to as single-particle IRIS (SP-IRIS), is composed of a single wavelength LED (525 nm) for illumination of the substrate, a high numerical aperture (NA = 0.8) objective to obtain a high spatial resolution image, and a CCD camera [
90]. The thickness of the oxide layer was adjusted to optimize the interference of the particle scattered field with the reference field. The schematic of the optical setup for SP-IRIS is shown in
Figure 15a.
For detecting biological nanoparticles such as viruses, an array of high affinity capture probes is generated on the surface that can selectively bind to the target virus. Capture probe immobilization is achieved by using a 3-D copolymer coating that provides NHS groups for the covalent attachment of amine-containing biomolecules [
91]. When virus particles bind to the capture antibodies, scattered light from the particles interferes with the reference field reflecting from the Si/SiO
2 interface, which enhances the signal detected on the CCD camera. Particles captured on the chip surface show as bright dots in the recorded image (
Figure 15b). SP-IRIS takes images of the spots in a microarray, and then these spot images are analyzed using custom software that finds particle-associated intensity peaks that correlate with a Gaussian profile. A forward model is applied to associate background normalized intensities (contrast) of the particles to particle size [
90]. Therefore, for a given spot, the diffraction-limited dots with expected size range are selected using a Gaussian filter (red circles in Fig. 15b) and counted to obtain the number of virus particles bound to the spot. Pre-incubation particle count is subtracted from post-incubation particle count and the net virus count is divided by the spot area to obtain the bound virus density (number of particles per mm
2). Single particle detection ability of SP-IRIS offers a great advantage over ensemble-based methods, such as BLI, where many binding events need to occur to record a signal above the background noise. The high sensitivity achieved by single virus counting, when combined with the on-chip multiplexing ability, renders SP-IRIS an attractive platform for virus diagnostics applications.
2.7. Virus diagnostics applications of SP-IRIS
SP-IRIS can individually count and size the nanoparticles bound to capture probes on the sensor surface over a large sensor area, orders of magnitude larger than other virus imaging techniques such as electron microscopy. It allows for a large range of nanoparticle detection including both natural nanoparticles (e.g. viruses) and synthetic nanoparticles (e.g. gold nanospheres, gold nanorods) in a highly-multiplexed microarray format. So far, SP-IRIS has been shown to detect many different biological targets such as viruses [
93], allergen-specific antibodies [
94], extracellular vesicles [
95], bacteria [
96], and microRNA [
97]. When the target is a nanoparticle itself, such as viruses, the detection can be done directly without using any secondary labels. If the biomolecule being searched for is below the size limit of the SP-IRIS (~ 30 nm), the target binding can be monitored by using specific detection probes attached to nanoparticle barcodes such as gold nanoparticles. Since this review’s main topic is optical virus detection techniques, in this section, we will review the SP-IRIS studies demonstrating this application.
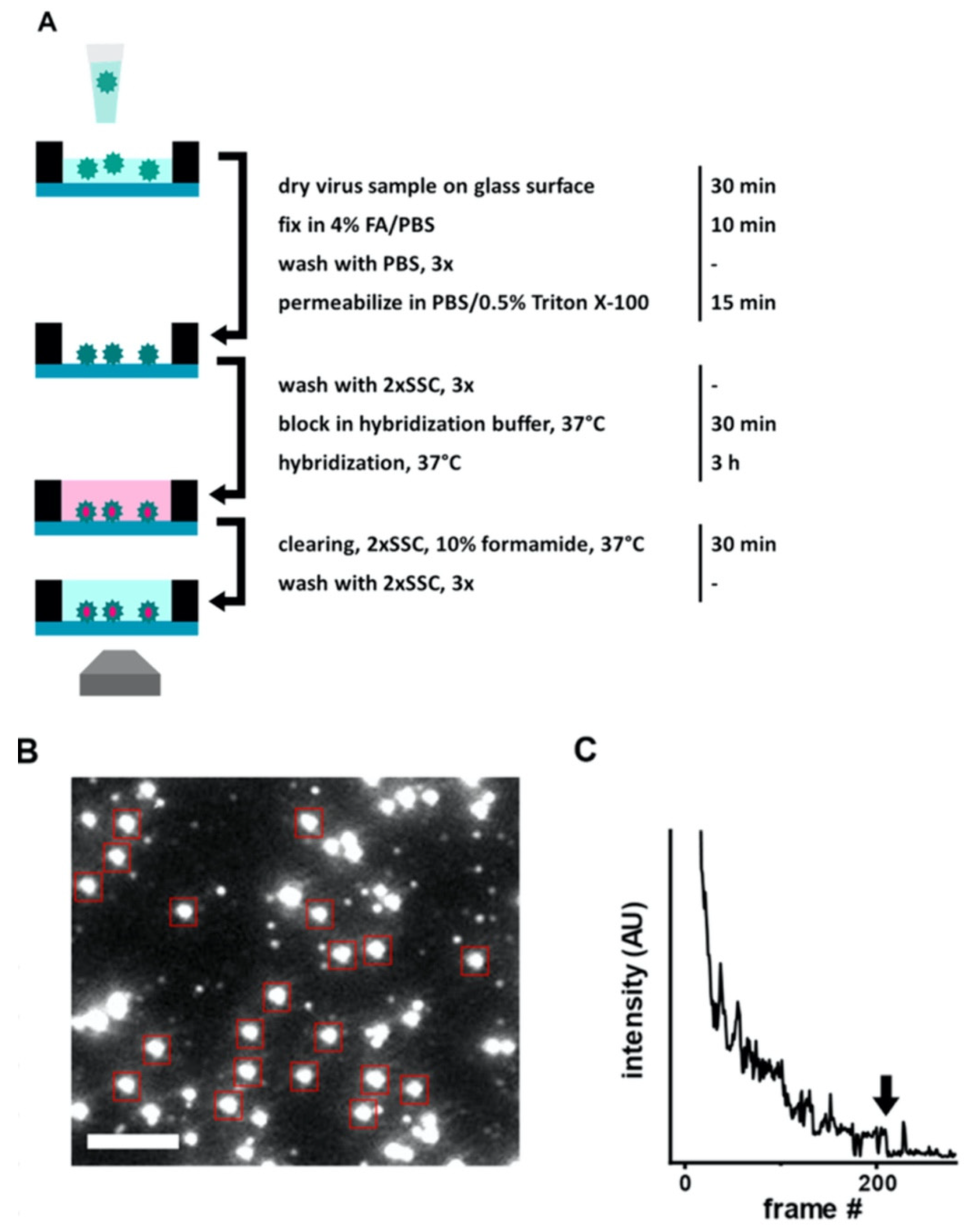
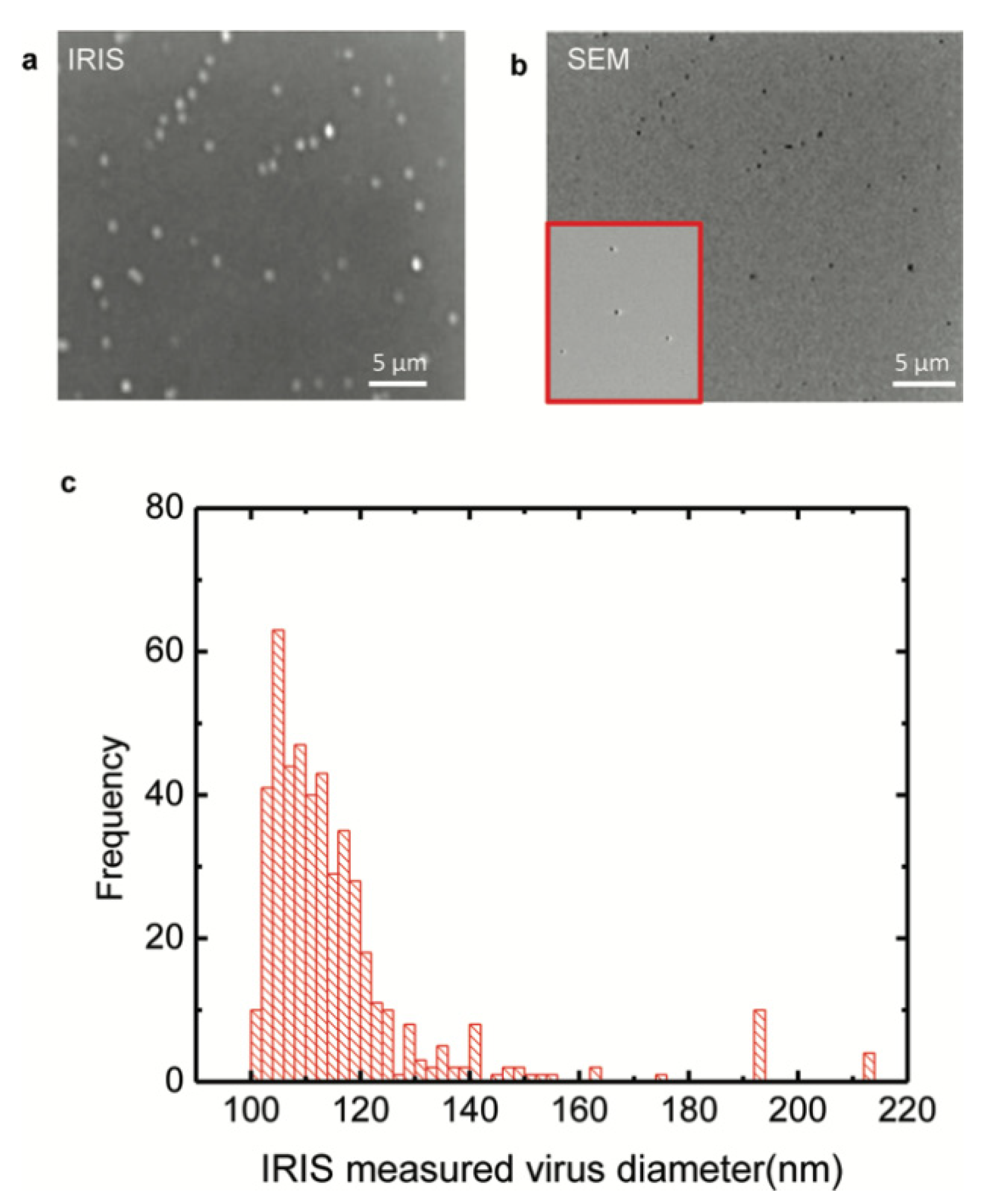
The work by Daaboul et al. was the first report to show SP-IRIS as a virus detection platform by demonstrating detection and sizing of individual H1N1 viruses [
90]. In this work, H1N1 virus was immobilized on the sensor surface and imaged using both SP-IRIS and scanning electron microscopy (SEM) for the exact same field of view. Their results showed a one-to-one correspondence between SP-IRIS and SEM images confirming the particles observed in the SP-IRIS system are indeed virus particles, proving the sensor’s ability to detect individual viruses (
Figure 16). In the same work, by using the forward model mentioned before, they measured the mean size of the H1N1 particles as 116 nm with a size distribution of 17 nm, which is in good agreement with the reported H1N1virus size in the literature.
Following the first virus detection demonstration with immobilized H1N1 virus, SP-IRIS was shown to perform sensitive and multiplexed detection of whole viruses from serum and blood samples [
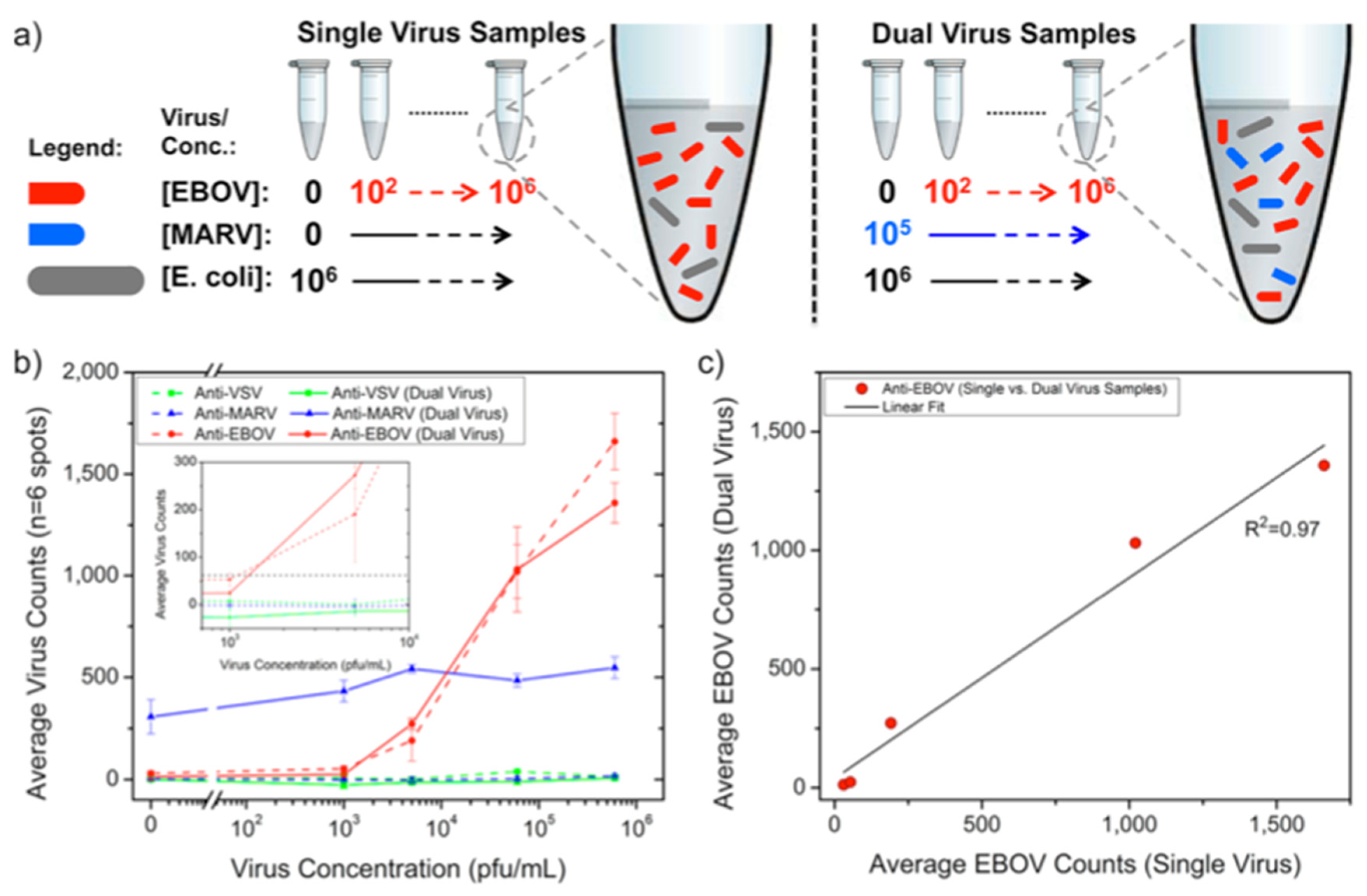
93]. For this work, Daaboul et al. used genetically engineered vesicular stomatitis virus (VSV) pseudotypes that express surface glycoproteins of Ebola and Marburg viruses (rVSV-EBOV and rVSV-MARV) (
Figure 17). They first arrayed EBOV and MARV specific antibodies on SP-IRIS chips and incubated the chips with either increasing concentrations of rVSV-EBOV only or the same increasing rVSV-EBOV concentrations in the presence of a constant concentration of rVSV-MARV. The virus solutions were prepared in serum containing 10
6 CFU/mL
E.coli K12 to mimic a complex solution environment. Their results showed specific detection of rVSV-EBOV with increasing virus particles on anti-EBOV spots whereas anti-MARV spots had a constant signal in dual virus samples. The limit of detection (LOD) reported for rVSV-EBOV detection from serum and blood was 5 × 10
3 PFU/mL. A similar level of sensitivity was also reached for rVSV-MARV detection in the same study. This work demonstrated that SP-IRIS has a great potential to be used as a virus diagnostic technique with its ability of direct detection of the target viruses from complex samples without labeling and complicated sample preparation.
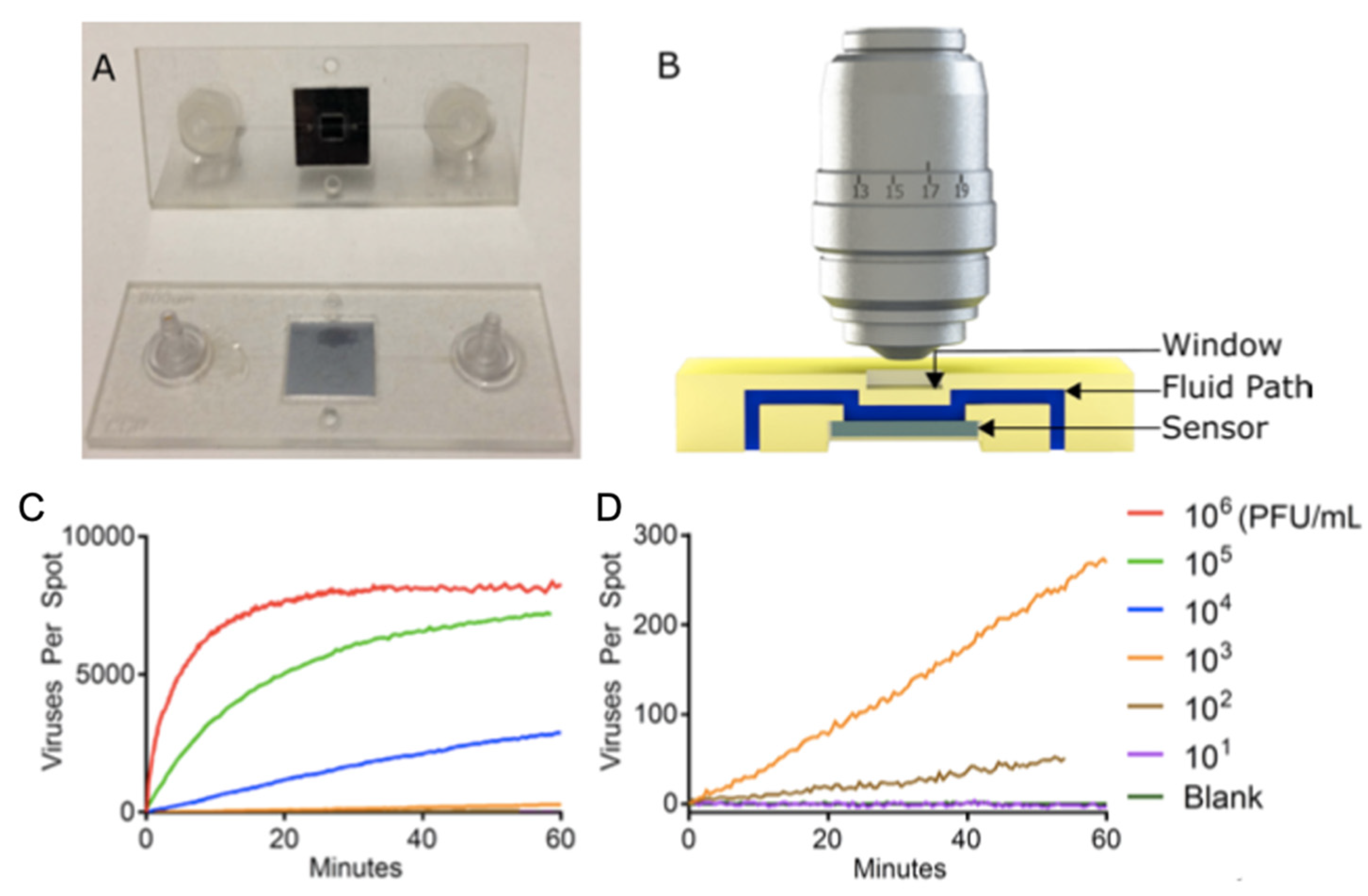
SP-IRIS was further advanced to implement the ability of visualizing virus particles in-liquid environment, rendering the system a real-time imaging platform and eliminating washing and drying steps (
Figure 18). In order to increase the contrast of the virus particles in liquid, some changes to the optical setup have been made such as use of a 40×, 0.9 NA objective and adjustment of the oxide thickness of the sensor chip. In this setup, the SP-IRIS chip is mounted in a disposable active microfluidic cartridge via a pressure-sensitive adhesive and the cartridge is fixed on the SP-IRIS stage. Scherr et al. reported a 50-fold increase in sensitivity compared to in-air measurements, leading to an LOD of 100 PFU/mL, for the detection of rVSV-EBOV from serum samples [
98].
To demonstrate the applicability of SP-IRIS to point-of-care diagnostics as a rapid detection method, a disposable passive microfluidic cartridge was designed with a multilayer polymer laminate structure and an integrated absorbent paper to establish capillary flow of the sample in the cartridge. This passive-flow integrated SP-IRIS achieved a better sensitivity than ELISA and a commercial rapid antigen test by detecting 10
4 PFU/mL rVSV-EBOV in less than 20 min [
99,
100]. A different study by Daaboul et al, demonstrated the usability of SP-IRIS for detection and characterization of a variety of virus sizes ranging from 40 nm for Zika virus to 360 nm for Vaccinia virus as well as filamentous virus particles like Ebola virus [
101]. In addition, recently, Yurdakul et al. showed a different modality of SP-IRIS, referred to as single-particle interferometric microscopy, for obtaining shape and size information which will enable in depth morphological studies of viruses [
102]. Collectively, these studies demonstrated the potential of SP-IRIS as a sensitive, fast, and multiplexed virus detection platform in a label-free and sample-to-answer format.
Besides the microfluidics integration and improvements in the optical setup of SP-IRIS, sensor chip surface chemistry has also been studied in an effort to increase the sensitivity of detection. By using a technique called DNA-directed antibody immobilization (DDI), Seymour et al. showed that capture antibodies can be elevated over the surface (~ 14 nm) through the use of DNA linkers attached to the antibodies [
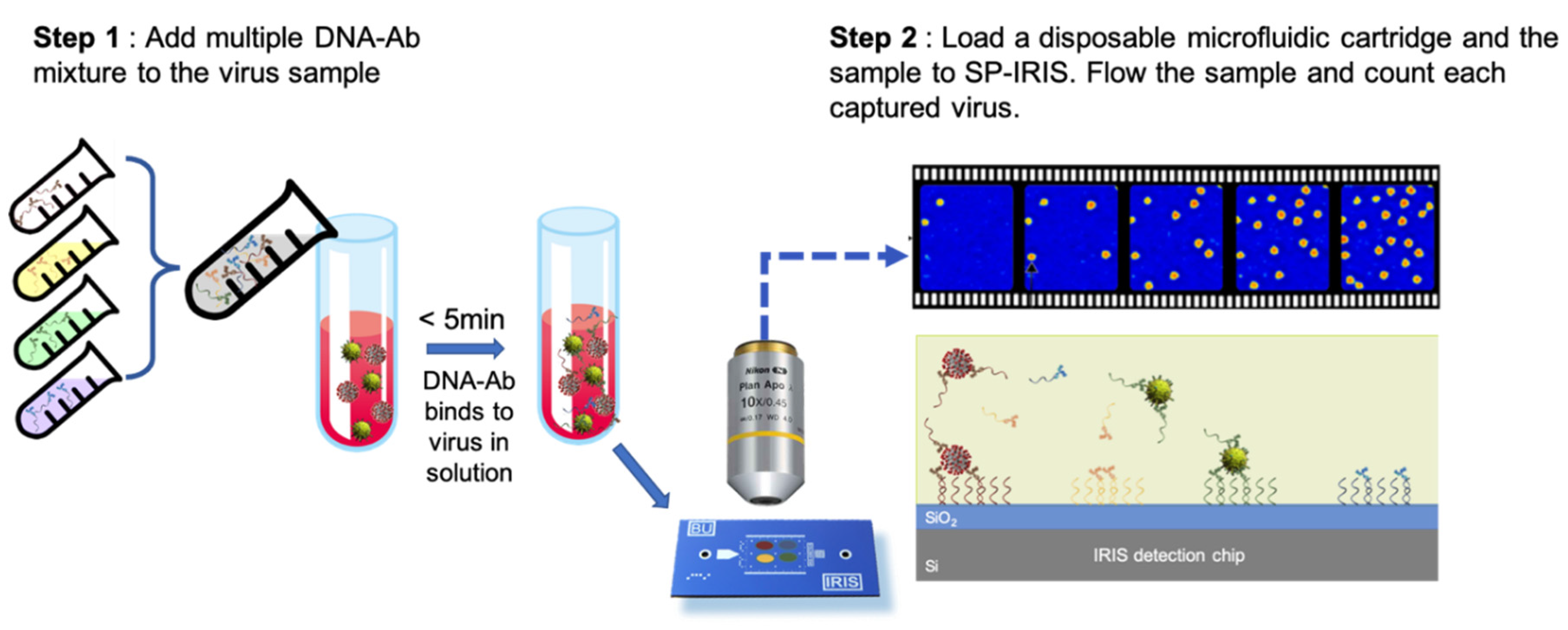
103]. This new surface preparation technique was shown to provide a 16-fold increase in sensitivity for rVSV-EBOV detection for a 15-min incubation period. This improvement is most likely due to the increased accessibility of the antibodies for virus binding as well as increased functionality due to less surface attachment points in the antibody structure. This work was recently extended to a novel approach of mixing DNA-antibody conjugates and the virus sample in the solution phase before incubating the chip (
Figure 19). This homogeneous method achieved a slightly better sensitivity than conventional DDI while decreasing the assay time [
100]. Other advantages offered by this approach includes configurable sensor surface, decreased antibody amount needed for the assay, and long shelf-life of dried DNA-Ab conjugates.
SP-IRIS offers significant advantages compared to the other optical biosensing platforms mentioned in this review. First of all, SP-IRIS has a comparable sensitivity to SPR, the most commonly used label-free biosensor, while having higher multiplexing capability, substantially less expensive substrates, and shorter analysis time [
86]. Moreover, the detection principle of SP-IRIS is immune to bulk-effect, a major problem of SPR-based systems that is caused by the changes in the refractive index of the solution. SP-IRIS overcomes any background-related affect by imaging only the nanoparticles that are bound to the surface. Moreover, unlike optical resonator-based sensors, SP-IRIS signal is not affected by environmental factors such as temperature changes or binding position of the particles on the sensor. Thus, SP-IRIS combines robust and reliable signal transduction mechanism with high-sensitivity, high-throughput detection in a cost-effective and easy-to-use platform. All of these features make SP-IRIS a perfect candidate for virus diagnostics applications especially for POC applications.