Submitted:
27 April 2023
Posted:
28 April 2023
You are already at the latest version
Abstract
Keywords:
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
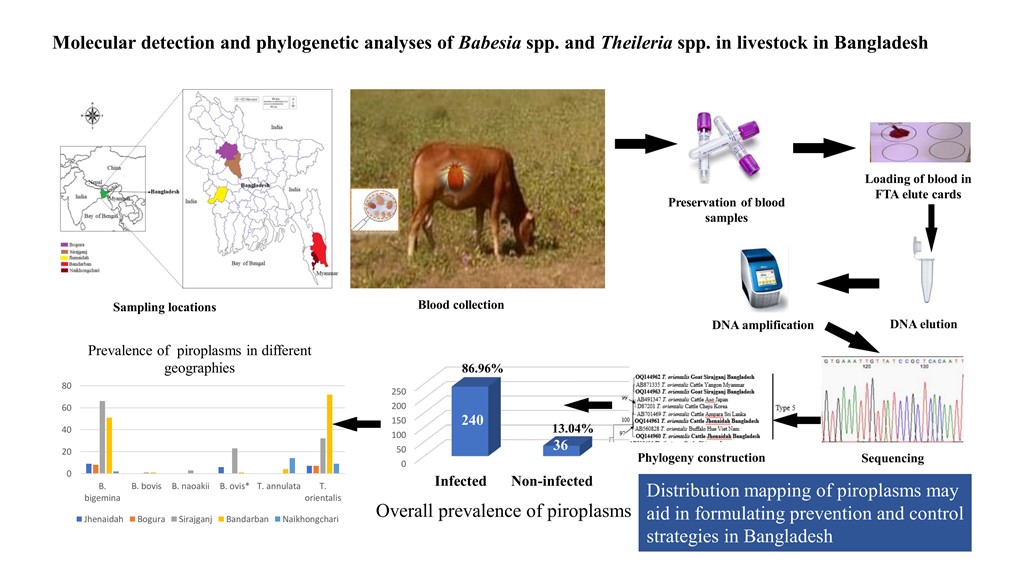
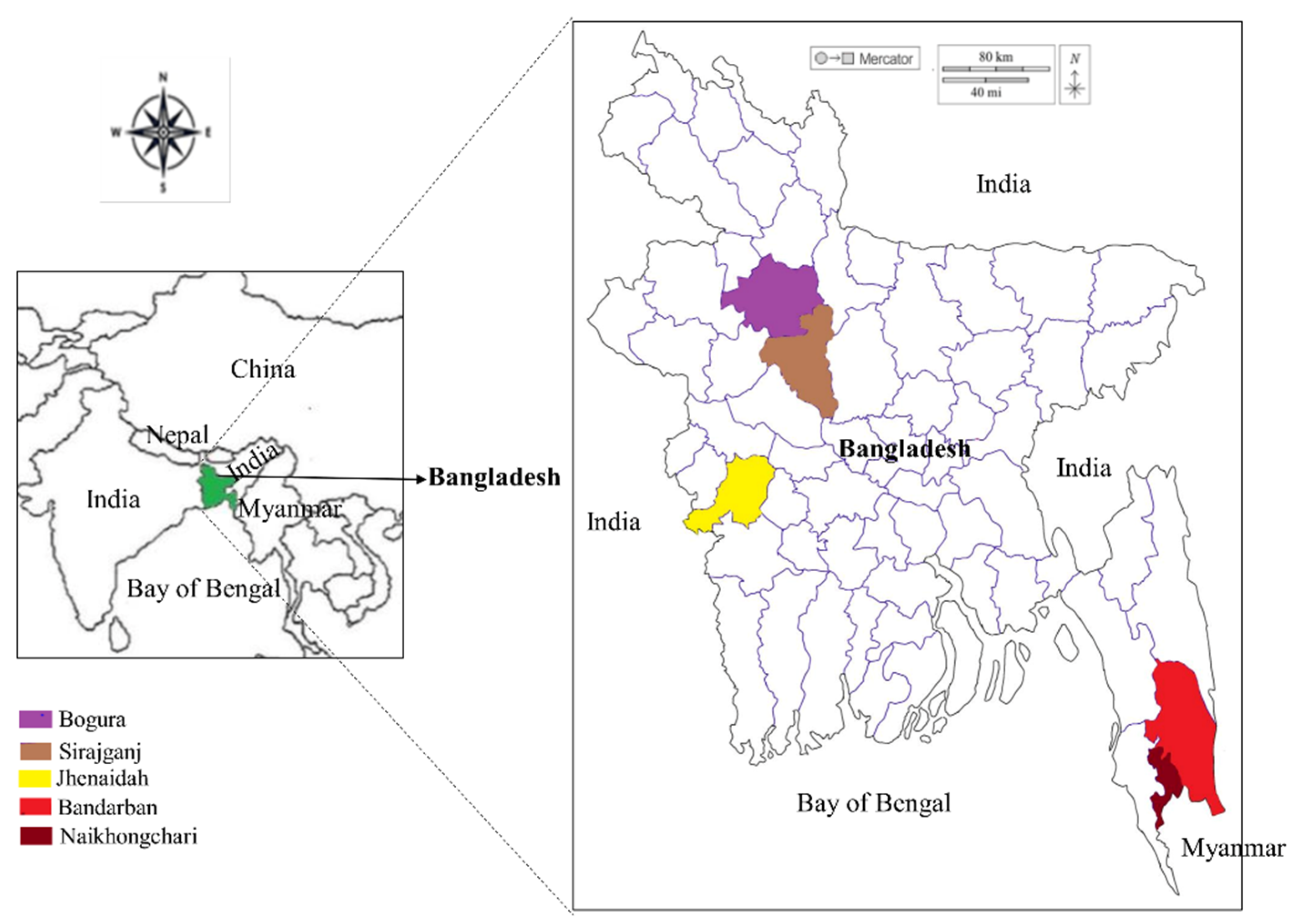
2.2. Study Sites and Sample Collection
2.3. Dried Blood Spot Preparation on FTATM Elute Micro Card and DNA Elution
2.4. Molecular Detection of Piroplasms
2.5. Sequencing of the PCR-Positive Samples
2.6. Phylogenetic Analyses
2.7. GenBank Accession Numbers
2.8. Statistical Analyses
3. Results
3.1. Overall Prevalence
3.2. Co‒Infections with Different Piroplasms
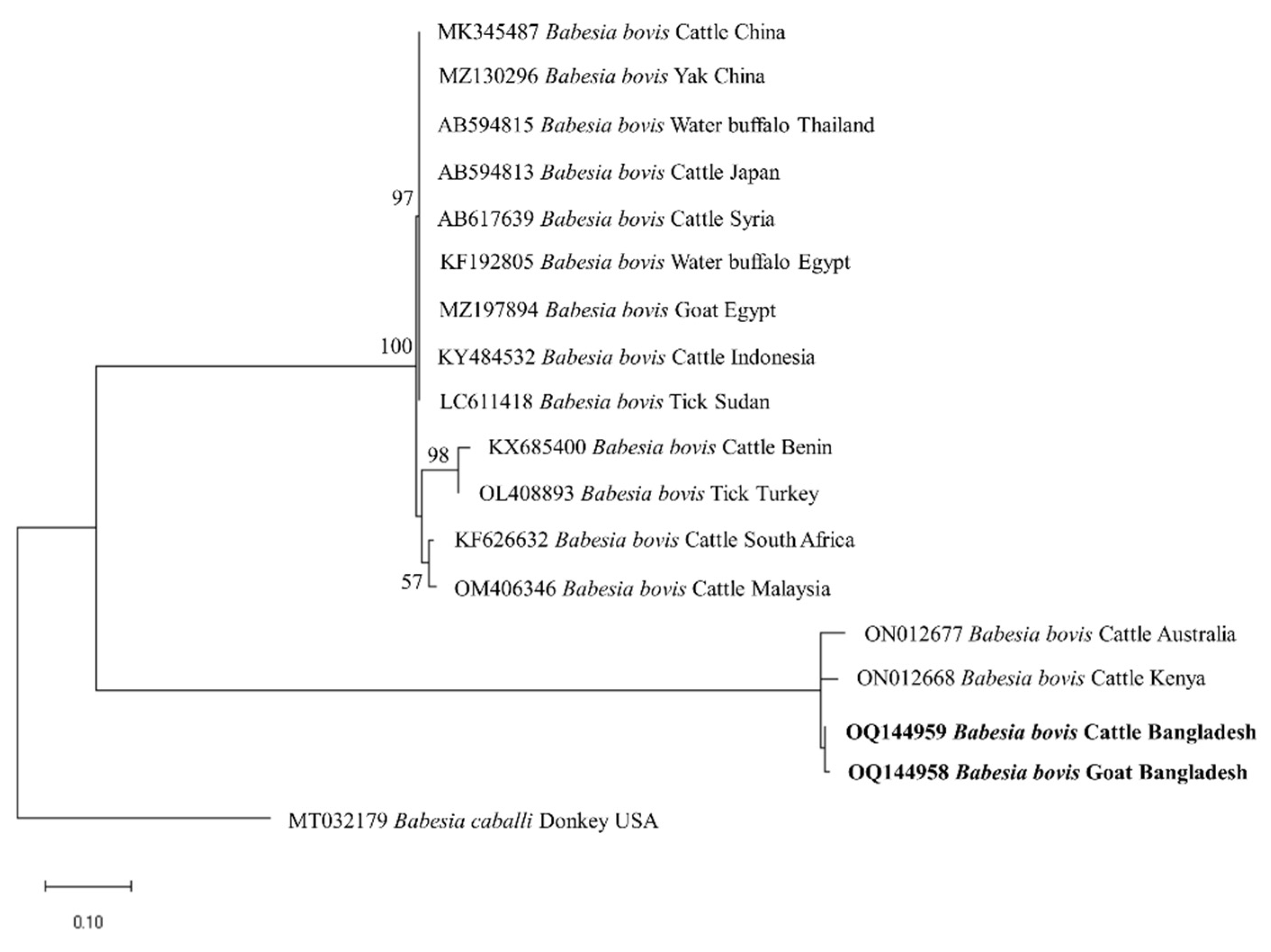
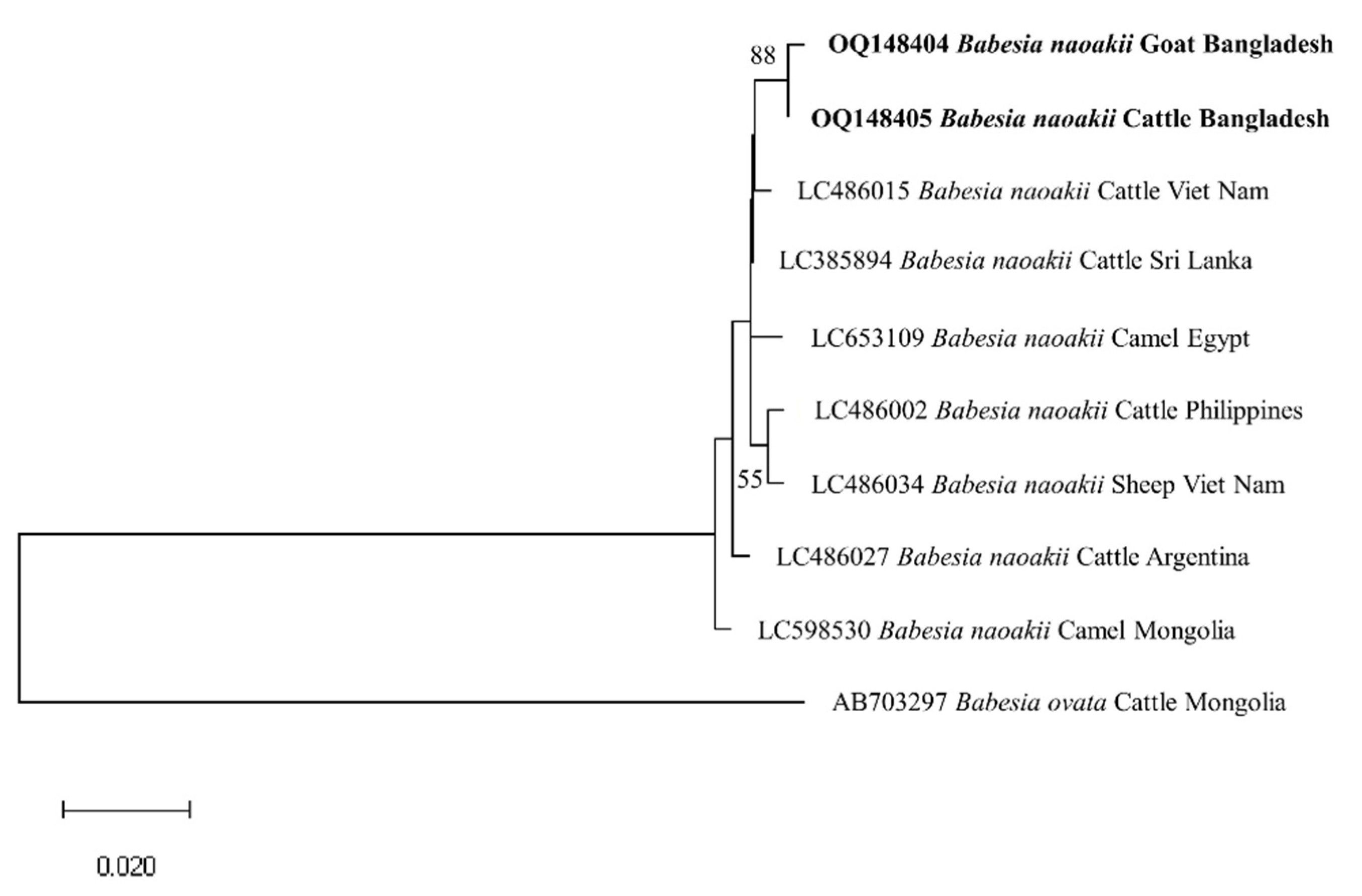
3.3. Gene Sequence Analyses
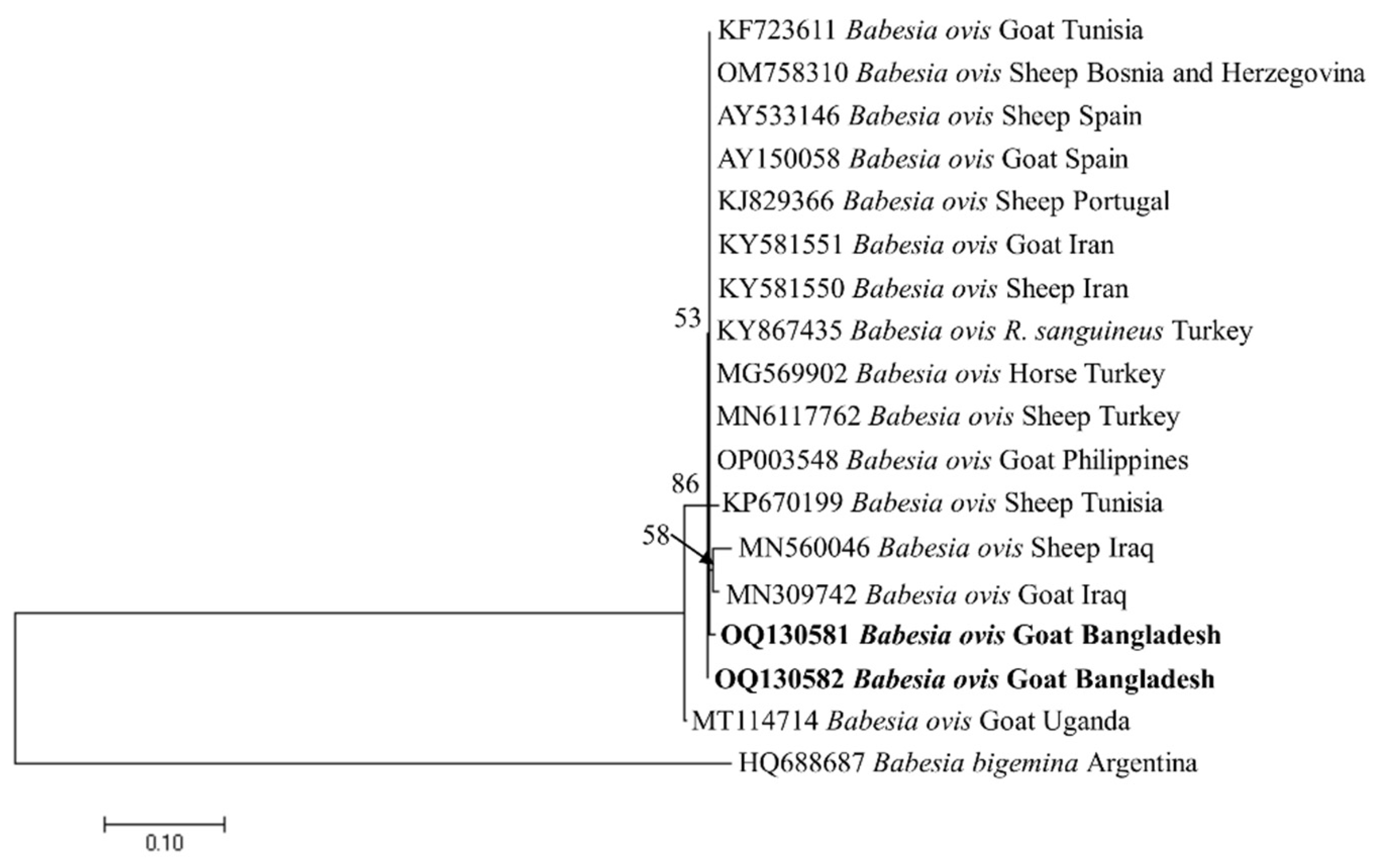
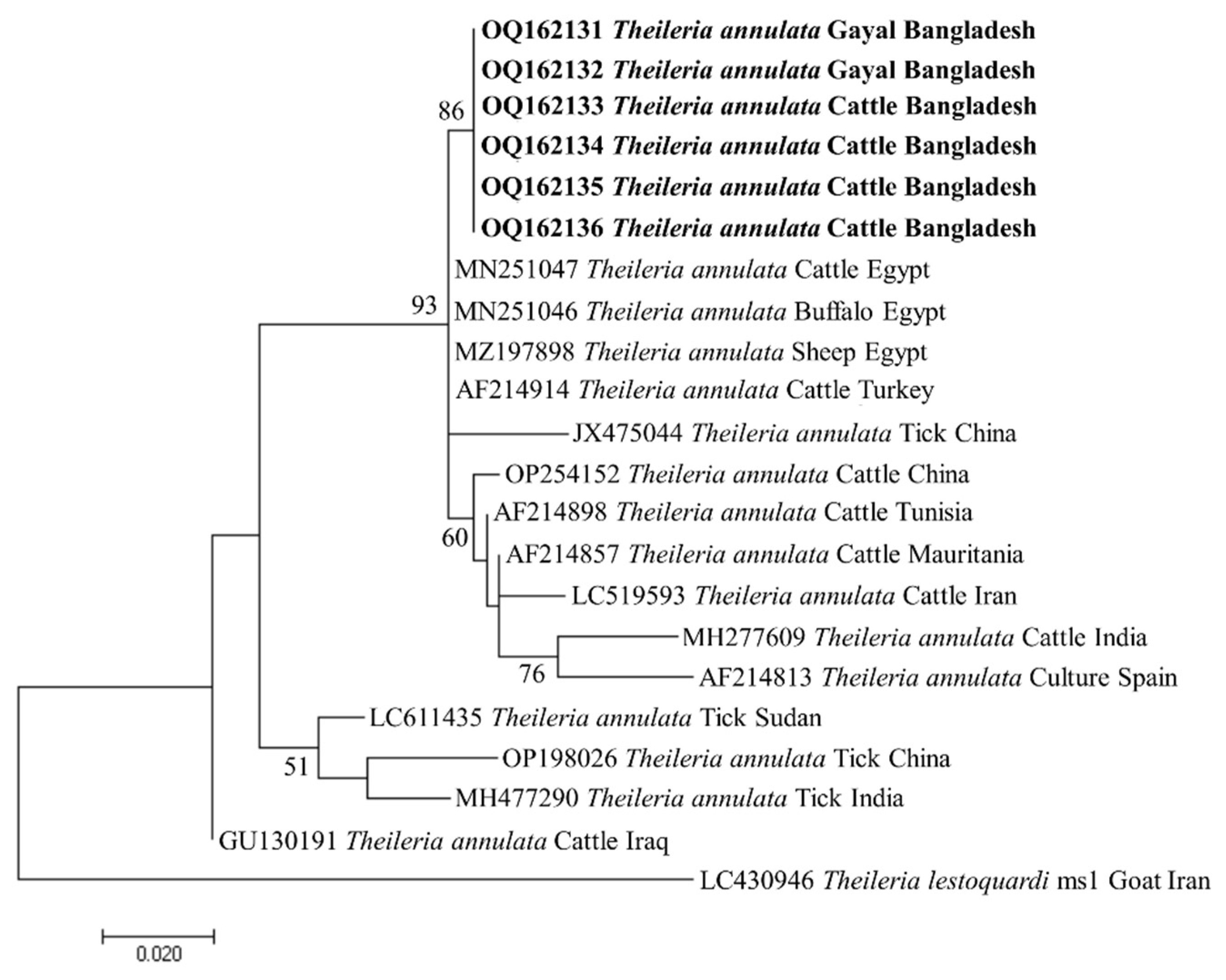
3.4. Phylogenetic Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Uilenberg, G. International collaborative research: Significance of Tick-Borne Hemoparasitic Diseases to World Animal Health. Vet. Parasitol. 1995, 57, 19–41. [CrossRef]
- Uilenberg, G. Babesia-A Historical Overview. Vet. Parasitol. 2006, 138, 3–10. [CrossRef]
- Minjauw, B.; McLeod, A. Tick-Borne Diseases and Poverty: The Impact of Ticks and Tick-Borne Diseases on the Livelihoods of Small-Scale and Marginal Livestock Owners in India and Eastern and Southern Africa. DFID Animal Health Programme, Centre for Tropical Veterinary Medicine: Edinburgh, 2003.
- McLeod, R.; Kristjanson, P. Economic Impact of Ticks and Tick-Borne Diseases to Livestock in Africa, Asia and Australia; Report to the International Livestock Research Institute, Nairobi, Kenya, 1999.
- Hunfeld, K.; Hildebrandt, A.; Gray, J. Babesiosis: Recent Insights into an Ancient Disease. Int. J. Parasitol. 2008, 38, 1219–1237. [CrossRef]
- Bock, R.; Jackson, L.; De Vos, A.; Jorgensen, W. Babesiosis of Cattle. Parasitology. 2004, 129. [CrossRef]
- Aktaş, M.; Altay, K.; Dumanli, N. Development of a Polymerase Chain Reaction Method for Diagnosis of Babesia ovis Infection in Sheep and Goats. Vet. Parasitol. 2005, 133, 277–281. [CrossRef]
- Zhao, S.; Liu, J.; Zhao, H.; Li, Y.; Xie, J.; Liu, A.; Hassan, M.A.; Yin, H.; Guan, G.; Luo, J. Evaluating an Indirect RMPSP Enzyme-Linked Immunosorbent Assay for the Detection of Bovine Theileria Infection in China. Parasitol. Res. 2017, 116, 667–676. [CrossRef]
- Lorusso, V.; Wijnveld, M.; Majekodunmi, A.O.; Dongkum, C.; Fajinmi, A.; Dogo, A.G.; Thrusfield, M.; Mugenyi, A.; Vaumourin, E.; Igweh, A.C.; et al. Tick-Borne Pathogens of Zoonotic and Veterinary Importance in Nigerian Cattle. Parasit. Vectors. 2016, 9. [CrossRef]
- Ringo, A.E.; Nonga, H.E.; Galon, E.M.; Ji, S.; Rizk, M.A.; El-Sayed, S.A.E.S.; Mohanta, U.K.; Ma, Z.; Chikufenji, B.; Do, T.T.; et al. Molecular Investigation of Tick-Borne Haemoparasites Isolated From Indigenous Zebu Cattle in the Tanga Region, Tanzania. Animals. 2022, 12. [CrossRef]
- D’Oliveira, C.; van der Weide, M.; Habela, M.A.; Jacquiet, P.; Jongejan, F. Detection of Theileria annulata in Blood Samples of Carrier Cattle by PCR. J. Clin. Microbiol. 1995, 33, 2665–2669. [CrossRef]
- Ziam, H.; Kernif, T.; Saidani, K.; Kelanemer, R.; Hammaz, Z.; Geysen, D. Bovine Piroplasmosis-anaplasmosis and Clinical Signs of Tropical Theileriosis in the Plains of Djurdjura (North Algeria). Vet. Med. Sci. 2020, 6, 720–729. [CrossRef]
- Neitz, W. 0 Theileriosis, Gonderioses and Cytauxzoonoses: A Review. Onderstepoort J. Vet. Res. 1957, 27.
- Minami, T.; Fujinaga, T.; Furuya, K.; Ishihara, T. Clinico-Hematologic and Serological Comparison of Japanese and Russian Strains of Theileria sergenti. Natl. Inst. Anim. Health. Q. (Tokyo). 1980, 20, 44–52.
- Sugimoto, C.; Fujisaki, K. Non-transforming Theileria Parasites of Ruminants. In Theileria. World Class Parasites; Dobbelaere, D.A.E., McKeever, D.J., Eds.; Springer: Boston, MA, 2002; Vol. 3, pp. 93–106.
- Perera, P.K.; Gasser, R.B.; Firestone, S.M.; Anderson, G.A.; Malmo, J.; Davis, G.; Beggs, D.S.; Jabbar, A. Oriental Theileriosis in Dairy Cows Causes a Significant Milk Production Loss. Parasit. Vectors. 2014, 7, 73. [CrossRef]
- Aparna, M.; Ravindran, R.; Vimalkumar, M.B.; Lakshmanan, B.; Rameshkumar, P.; Kumar, K.G.A.; Promod, K.; Ajithkumar, S.; Ravishankar, C.; Devada, K.; et al. Molecular Characterization of Theileria orientalis Causing Fatal Infection in Crossbred Adult Bovines of South India. Parasitol. Int. 2011, 60, 524–529. [CrossRef]
- Patial, V.; Gupta, T.; Angaria, S.; Bali, D.; Katoch, A.; Gautam, M.; Singh, N.K.; Sharma, M.; Chahota, R. Theileria orientalis Outbreak in an Organized Cattle Breeding Farm. Vet. Parasitol. Reg. Stud. Reports. 2021, 24. [CrossRef]
- McFadden, A.; Rawdon, T.; Meyer, J.; Makin, J.; Morley, C.; Clough, R.; Tham, K.; Müllner, P.; Geysen, D. An Outbreak of Haemolytic Anaemia Associated with Infection of Theileria Orientalis in Naïve Cattle. N. Z. Vet. J. 2011, 59, 79–85. [CrossRef]
- Eamens, G.J.; Gonsalves, J.R.; Jenkins, C.; Collins, D.; Bailey, G. Theileria Orientalis MPSP Types in Australian Cattle Herds Associated with Outbreaks of Clinical Disease and Their Association with Clinical Pathology Findings. Vet. Parasitol. 2013, 191, 209–217. [CrossRef]
- BBS (Bangladesh Bureau of Statistics) Statistical Yearbook of Bangladesh 2020; 40th ed.; Statistics & Informatics Division (SID), Ministry of Planning, Government of The People`s Republic of Bangladesh, Dhaka, Bangladesh, 2021.
- Ghosh, S.; Bansal, G.C.; Gupta, S.C.; Ray, D.; Khan, M.Q.; Irshad, H.; Shahiduzzaman, M.; Seitzer, U.; Ahmed, J.S. Status of Tick Distribution in Bangladesh, India and Pakistan. Parasitol. Res. 2007, 101 Suppl 2, S207-16. [CrossRef]
- Islam, M.K.; Alim, M.A.; Tsuji, N.; Mondal, M.M.H. An Investigation into the Distribution, Host-Preference and Population Density of Ixodid Ticks Affecting Domestic Animals in Bangladesh. Trop. Anim. Health. Prod. 2006, 38, 485–490. [CrossRef]
- Kabir, M.H.B.; Mondal, M.M.H.; Eliyas, M.; Mannan, M.A.; Hashem, M.A.; Debnath, N.C.; Miazi, O.F.; Mohiuddin, C.; Kashem, M.A.; Islam, M.R.; Elahi, M.F. An epidemiological survey on investigation of tick infestation in cattle at Chittagong District, Bangladesh. Afr. J. Microbiol. Res. 2011, 5, 346-352. [CrossRef]
- Al-Mahmud, M.A.; Belal, S.S.H.; Hossain, M.A. Prevalence of Theileriosis and Babesiosis in Cattle in Sirajganj District of Bangladesh. Res. Agric. Livest. Fish. 2015, 2, 79-86. [CrossRef]
- Alim, M.A.; Das, S.; Roy, K.; Masuduzzaman, M.; Sikder, S.; Hassan, M.M.; Siddiki, A.Z.; Hossain, M.A. Prevalence of Hemoprotozoan Diseases in Cattle Population of Chittagong Division, Bangladesh. Pak. Vet. J. 2012, 32, 221–224.
- Chowdhury, S.; Hossain, M.; Barua, S.; Islam, S. Occurrence of Common Blood Parasites of Cattle in Sirajgonj Sadar Area of Bangladesh. Bang. J. Vet. Med. 2006, 4, 143–145. [CrossRef]
- Belal, S.S.H.; Mahmud, Md.A. al; Ferdous, M.J. Prevalence of Anaplasmosis in Cattle in Sirajganj District of Bangladesh. Res. Agric. Livest. Fish. 2015, 1, 97–103. [CrossRef]
- Mohanta, U.K.; Anisuzzaman; Mondal, M.M.H. Tick and Tick Borne Protozoan Diseases of Livestock in the Selected Hilly Areas of Bangladesh. Int. J. Agril. Res. Innov. & Tech. 2011, 1, 60–63. [CrossRef]
- Roy, B.C.; Krücken, J.; Ahmed, J.S.; Majumder, S.; Baumann, M.P.; Clausen, P.H.; Nijhof, A.M. Molecular Identification of Tick-Borne Pathogens Infecting Cattle in Mymensingh District of Bangladesh Reveals Emerging Species of Anaplasma and Babesia. Transbound. Emerg. Dis. 2018, 65, e231–e242. [CrossRef]
- Hossain, M.J.; Raut, S.; Singh, R.P.; Mishra, P.; Hossain, M.S.; Dey, A.R.; Kabir, A.; Anisuzzaman; Talukder, M.H.; Shahiduzzaman, M. Molecular Detection of Babesia and Theileria from Crossbred Cattle in Sirajganj and Rangpur Districts of Bangladesh. Vet. Med. Sci. 2022,1-8. [CrossRef]
- Sivakumar, T.; Tuvshintulga, B.; Zhyldyz, A.; Kothalawala, H.; Yapa, P.R.; Kanagaratnam, R.; Vimalakumar, S.C.; Abeysekera, T.S.; Weerasingha, A.S.; Yamagishi, J.; et al. Genetic Analysis of Babesia Isolates from Cattle with Clinical Babesiosis in Sri Lanka. J. Clin. Microbiol. 2018, 56. [CrossRef]
- Ota, N.; Mizuno, D.; Kuboki, N.; Igarashi, I.; Nakamura, Y.; Yamashina, H.; Hanzaike, T.; Fujii, K.; Onoe, S.; Hata, H.; et al. Epidemiological Survey of Theileria Orientalis Infection in Grazing Cattle in the Eastern Part of Hokkaido, Japan. J. Vet. Med. Sci. 2009, 71, 937-944. [CrossRef]
- Terkawi, M.A.; Huyen, N.X.; Shinuo, C.; Inpankaew, T.; Maklon, K.; Aboulaila, M.; Ueno, A.; Goo, Y.K.; Yokoyama, N.; Jittapalapong, S.; et al. Molecular and Serological Prevalence of Babesia Bovis and Babesia Bigemina in Water Buffaloes in the Northeast Region of Thailand. Vet. Parasitol. 2011, 178, 201–207. [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [CrossRef]
- Quddus, M.A. Crop Production Growth in Different Agro-Ecological Zones of Bangladesh. J. Bang. Agril. Univ. 2009, 7, 351–360.
- Faruque, M.; Rahaman, M.; Hoque, M.; Ikeya, K.; Amano, T.; Han, J.; Dorji, T.; Omar, A. Present Status of Gayal (Bos Frontalis) in the Home Tract of Bangladesh. Bang. J. Anim. Sci. 2015, 44, 75–84. [CrossRef]


| Target gene | Assays |
Primer sequences |
Annealing temp. (℃) |
References | |
| Forward 5' → 3' Reverse | |||||
| B. bigemina (BbigRAP-1a) | PCR | GAGTCTGCCAAATCCTTAC | TCCTCTACAGCTGCTTCG | 55 | [34] |
| nPCR | AGCTTGCTTTCACAACTCGCC | TTGGTGCTTTGACCGACGACAT | 50 | ||
| B. bovis (BboSBP-4) | PCR | AGTTGTTGGAGGAGGCTAAT | TCCTTCTCGGCGTCCTTTTC | 55 | [34] |
| nPCR | GAAATCCCTGTTCCAGAG | TCGTTGATAACACTGCAA | 50 | ||
|
B. naoakii (AMA-1) |
PCR | TGGCGCCGACTTCCTGGAGCCCATCTCCAA | AGCTGGGGCCCTCCTTCGATGAACCGTCGG | 64 | [32] |
|
B. ovis (ssu rRNA) |
PCR | TGGGCAGGACCTTGGTTCTTCT | CCGCGTAGCGCCGGCTAAATA | 62 | [7] |
|
T. annulata (Tams-1) |
PCR | GTAACCTTTAAAAACGT | GTTACGAACATGGGTTT | 54 | [11] |
| nPCR | CACCTCAACATACCCC | TGACCCACTTATCGTCC | 54 | ||
|
T. orientalis (MPSP) |
PCR | CTTTGCCTAGGATACTTCCT | ACGGCAAGTGGTGAGAACT | 58 | [33] |
| Pathogens | Locations | Total n=276 |
P-value | ||||
| Jhenaidah n=29 |
Bogura n=14 |
Sirajganj n=107 |
Bandarban n=105 |
Naikhongchari n=21 |
|||
| B. bigemina | 9 (31.03%) | 8 (57.14%) | 66 (61.68%) | 51 (48.57% | 2 (9.52%) | 136 (49.28%) | <0.001 |
| B. bovis | n.d. | n.d. | 1 (0.93%) | 1 (0.95%) | n.d. | 2 (0.72%) | NA |
| B. naoakii | n.d. | n.d. | 3 (2.80%) | n.d. | n.d. | 3 (1.09%) | NA |
| B. ovis* | 6 (27.27%) | n.d. | 23 (37.09%) | 1 (14.29%) | n.d. | 30 (32.26%) | NA |
| T. annulata | n.d. | n.d. | n.d. | 4 (3.81%) | 14 (66.67%) | 18 (6.52%) | NA |
| T. orientalis | 7 (24.13%) | 7 (50%) | 32 (29.90%) | 72 (68.57% | 9 (42.86%) | 127 (46.01%) | <0.001 |
| Pathogens | Animal species | Total (n=276) | p-value | ||
| Cattle (n=174) | Gayals (n=9) | Goat (n=93) | |||
| B. bigemina | 95 (54.60%) | 1 (11.11%) | 40 (43.01%) | 136 (49.28%) | ˂0.05 |
| B. bovis | 1 (0.57%) | n.d. | 1 (1.08%) | 2 (0.72%) | NA |
| B. naoakii | 1 (0.57%) | n.d. | 2 (2.15%) | 3 (1.09%) | NA |
| B. ovis* | n.s. | n.s. | 30 (32.26%) | 30 (32.26%) | NA |
| T. annulata | 9 (5.17%) | 7 (77.78%) | 2 (2.15%) | 18 (6.52%) | NA |
| T. orientalis | 117 (67.24%) | 4 (44.44%) | 7 (7.53%) | 128 (46.38%) | <0.001 |
| Pathogens | Animal species | Total (n=276) | |||||
| Cattle (n=174) | Gayals (n=9) | Goats (n=93) | |||||
| <2 yrs (n=64) | ≥2 yrs (n=110) | <2 yrs (n=5) | ≥2 yrs (n=4) | <2 yrs (n=63) | ≥2 yrs (n=30) | ||
| B. bigemina | 40 (22.99%) | 55 (50.00%) | 1 (20.00%) | n.d. | 25 (39.68%) | 15 (50.00%) | 136 (49.28%) |
| B. bovis | 1 (0.57%) | n.d. | n.d. | n.d. | 1 (1.59%) | n.d. | 2 (0.72%) |
| B. naoakii | 1 (0.57%) | n.d. | n.d. | n.d. | n.d. | 2 (6.67%) | 3 (1.09%) |
| B. ovis† | n.s. | n.s. | n.d. | n.s. | 24 (38.10%) | 6 (20.00%) | 30 (32.26%) |
| T. annulata | 4 (2.30) | 5 (4.55%) | 5 (100.00%) | 2 (50.00%) | 1 (1.59%) | 1 (3.33%) | 18 (6.52%) |
| T. orientalis | 46 (26.44%) | 71 (64.55%) | 3 (60.00%) | 1 (25.00%) | 2 (3.17%) * | 5 (16.67%) * | 128 (46.01%) |
| Pathogens | Animal species | Total | |||||
| Cattle | Goats | Gayals | |||||
| Male (n=46) | Female (n=128) | Male (n=24) | Female (n=69) | Male (n=3) | Female (n=6) | ||
| B. bigemina | 30 (65.22%) | 65 (50.78%) | 6 (25.00%) | 34 (49.28%) | n.d. | 1 (16.67%) | 136 (49.28%) |
| B. bovis | n.d. | 1 (0.78%) | n.d. | 1 (1.45%) | n.d. | n.d. | 2 (0.72%) |
| B. naoakii | 1 (2.17%) | n.d. | n.d. | 2 (2.90%) | n.d. | n.d. | 3 (1.09%) |
| B. ovis* | n.s. | n.s. | 8 (33.33%) | 22 (31.88%) | n.s. | n.s. | 30 (32.26%) |
| T. annulata | 1 (2.17%) | 8 (6.25%) | n.d. | 2 (2.90%) | 2 (66.66%) | 5 (83.33%) | 18 (6.52%) |
| T. orientalis | 34 (73.91%) | 83 (64.84%) | 1 (4.17%) | 6 (8.70) | 1 (33.33%) | 3 (50.00%) | 128 (46.01%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).





